Time to Spread Your Wings: A Review of the Avian Ancient DNA Field
Abstract
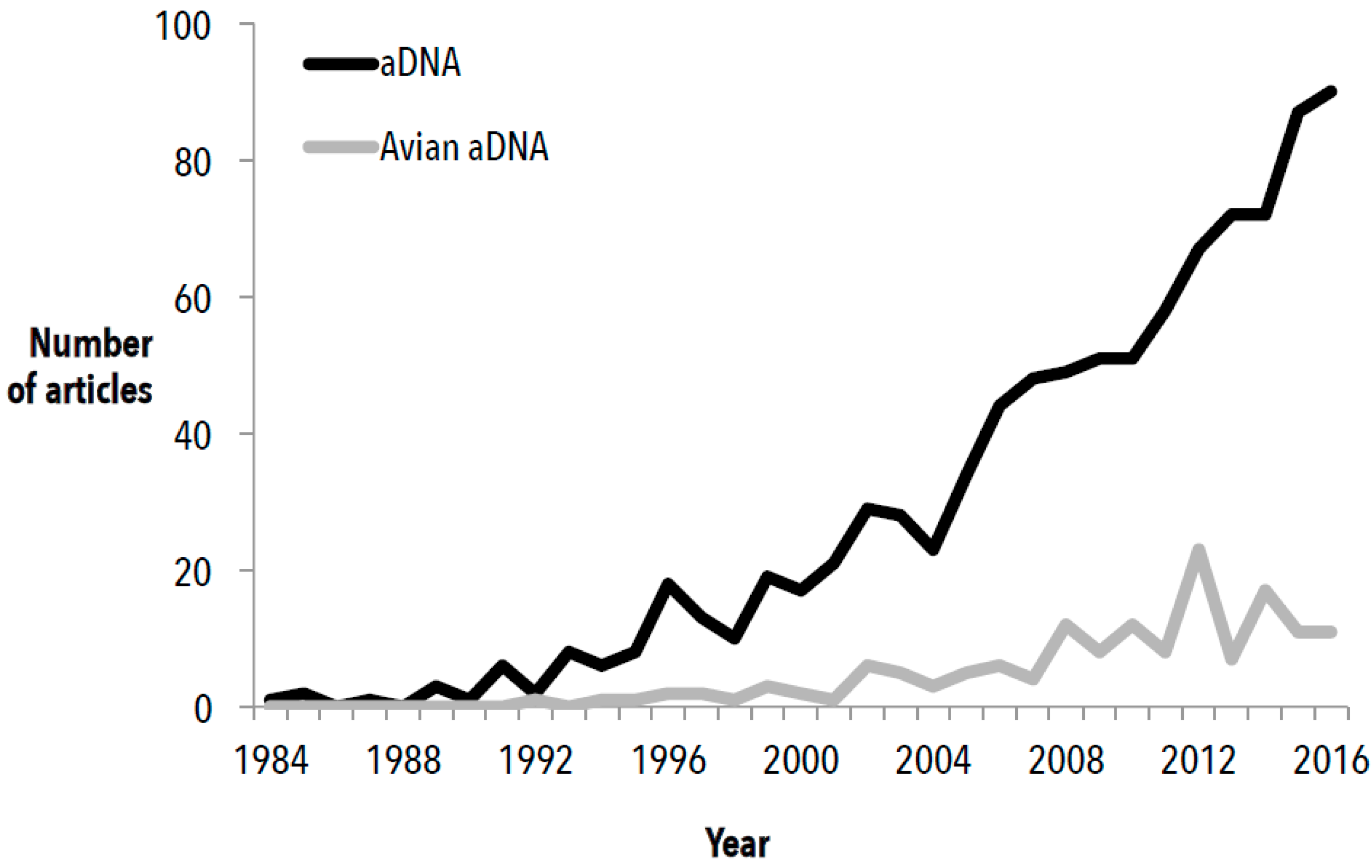
:1. Introduction
2. Avian Ancient DNA Applications
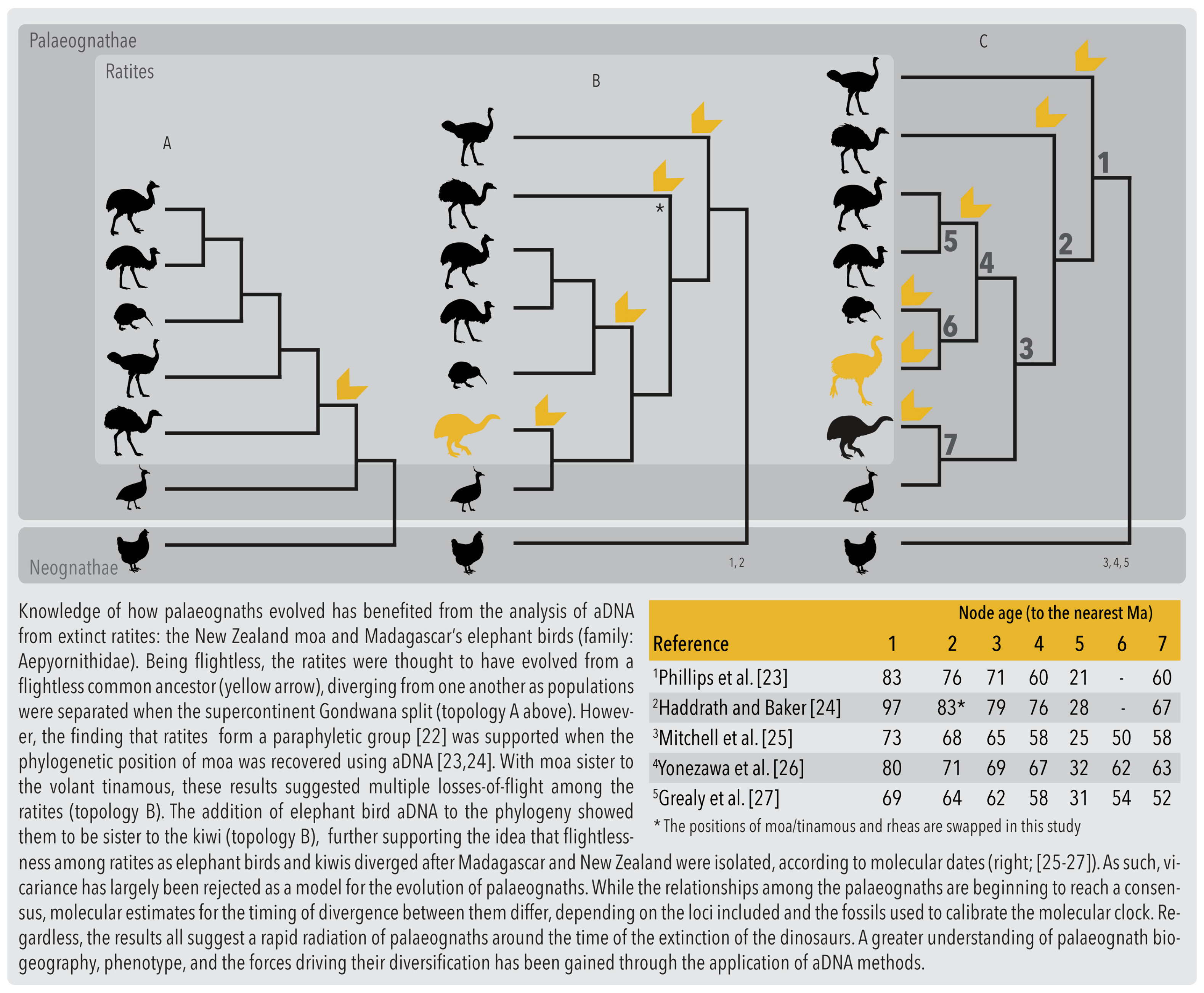
2.1. Phylogenetics and Biogeography
2.2. Taxonomy
2.3. Domestication
2.4. Palaeoenvironmental Reconstruction
2.5. Zooarchaeology
2.6. Conservation
3. The Scope of the Field
3.1. Substrates and Species
3.2. Molecular Techniques
3.3. Target Loci
4. Future Research Trajectories
4.1. Genomes and Metagenomes
4.2. Palaeo-Functional Genomics
4.3. De-Extinction—A Viable Pursuit?
4.4. From Ancient DNA to Action
5. Conclusions
Supplementary Materials
Conflicts of Interest
References
- Anderson, A. Prodigious Birds; University of Otago Press: Dunedin, New Zealand, 1989. [Google Scholar]
- Unwin, M. The Atlas of Birds; Princeton University Press: Princeton, NJ, USA, 2011. [Google Scholar]
- Boyer, A.G. Extinction patterns in the avifauna of the Hawai’ian Islands. Biodivers. Res. 2008, 14, 509–517. [Google Scholar]
- Hume, J.P.; Walters, M. Extinct Birds; Bloomsbury Publishing: London, UK, 2012. [Google Scholar]
- Lovette, I.J.; Rubenstein, D.R. A comprehensive molecular phylogeny of the starlings (Aves: Sturnidae) and mockingbirds (Aves: Mimidae): Congruent mtDNA and nuclear trees for a cosmopolitan avian radiation. Mol. Phylogenet. Evol. 2007, 44, 1031–1056. [Google Scholar] [CrossRef] [PubMed]
- Lerner, H.R.L.; Meyer, M.; James, H.F.; Hofreiter, M.; Fleischer, R.C. Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawai’ian honeycreepers. Curr. Biol. 2011, 21, 1838–1844. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Watson, R.T.; Minde, D.P. Prioritizing species conservation: Does the cape verde kite exist? Proc. R. Soc. B-Biol. Sci. 2005, 272, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Irestedt, M.; Ohlson, J.I.; Zuccon, D.; Kallersjo, M.; Ericson, P.G.P. Nuclear DNA from old collections of avian study skins reveals the evolutionary history of the old world suboscines (Aves, Passeriformes). Zool. Scr. 2006, 35, 567–580. [Google Scholar] [CrossRef]
- McKay, B.D.; Mays, H.L.; Yao, C.T.; Wan, D.M.; Higuchi, H.; Nishiumi, I. Incorporating color into integrative taxonomy: Analysis of the varied tit (Sittiparus varius) complex in east Asia. Syst. Biol. 2014, 63, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Rawlence, N.J.; Scofield, R.P.; Spencer, H.G.; Lalas, C.; Easton, L.J.; Tennyson, A.J.D.; Adams, M.; Pasquet, E.; Fraser, C.; Waters, J.M.; et al. Genetic and morphological evidence for two species of leucocarbo shag (Aves, Pelecaniformes, Phalacrocoracidae) from southern South Island of New Zealand. Zool. J. Linn. Soc. 2016, 177, 676–694. [Google Scholar] [CrossRef]
- Leonard, J.A. Ancient DNA applications for wildlife conservation. Mol. Ecol. 2008, 19, 4186–4196. [Google Scholar] [CrossRef] [PubMed]
- “Anthropocene” and “Data Mining”. 2017. Merriam-Webster’s Dictionary. Available online: https://merriam-webster.com/dictionary (accessed on 15 May 2017).
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.; Mourerchauvire, C.; Chambers, G.K.; Vonhaeseler, A.; Wilson, A.C.; Paabo, S. Independent origins of New-Zealand moas and kiwis. Proc. Natl. Acad. Sci. USA 1992, 89, 8741–8744. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.J.; Wood, J.R.; Llamas, B.; McLenachan, P.A.; Kardailsky, O.; Scofield, R.P.; Worthy, T.H.; Cooper, A. Ancient mitochondrial genomes clarify the evolutionary history of New Zealand’s enigmatic acanthisittid wrens. Mol. Phylogenet. Evol. 2016, 102, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.E.; Novak, B.J.; Haile, J.; Heupink, T.H.; Fjeldsa, J.; Gilbert, M.T.; Poinar, H.; Church, G.M.; Shapiro, B. Complete mitochondrial genomes of living and extinct pigeons revise the timing of the columbiform radiation. BMC Evol. Biol. 2016, 16, 230. [Google Scholar] [CrossRef] [PubMed]
- Scofield, R.P.; Mitchell, K.J.; Wood, J.R.; De Pietri, V.L.; Jarvie, S.; Llamas, B.; Cooper, A. The origin and phylogetic relationships of the New Zealand ravens. Mol. Phylogenet. Evol. 2017, 106, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.C.; Trewick, S.A. Dispersal and speciation in purple swamphens (Rallidae: Porphyrio). Auk 2015, 132, 140–155. [Google Scholar] [CrossRef]
- Boessenkool, S.; Austin, J.J.; Worthy, T.H.; Scofield, P.; Cooper, A.; Seddon, P.J.; Waters, J.M. Relict or colonizer? Extinction and range expansion of penguins in southern New Zealand. Proc. R. Soc. B-Biol. Sci. 2009, 276, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Rawlence, N.J.; Perry, G.L.W.; Smith, I.W.G.; Scofield, R.P.; Tennyson, A.J.D.; Matisoo-Smith, E.A.; Boessenkool, S.; Austin, J.J.; Waters, J.M. Radiocarbon-dating and ancient DNA reveal rapid replacement of extinct prehistoric penguins. Quat. Sci. Rev. 2015, 112, 59–65. [Google Scholar] [CrossRef]
- Ritchie, P.A.; Millar, C.D.; Gibb, G.C.; Baroni, C.; Lambert, D.M. Ancient DNA enables timing of the pleistocene origin and holocene expansion of two adelie penguin lineages in Antarctica. Mol. Biol. Evol. 2004, 21, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Harshman, J.; Braun, E.L.; Braun, M.J.; Huddleston, C.J.; Bowie, R.C.; Chojnowski, J.L.; Hackett, S.J.; Han, K.L.; Kimball, R.T.; Marks, B.D.; et al. Phylogenomic evidence for multiple losses of flight in ratite birds. Proc. Natl. Acad. Sci. USA 2008, 105, 13462–13467. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Gibb, G.C.; Crimp, E.A.; Penny, D. Tinamous and moa flock together: Mitochondrial genome sequence analysis reveals independent losses of flight among ratites. Syst. Biol. 2010, 59, 90–107. [Google Scholar] [CrossRef] [PubMed]
- Haddrath, O.; Baker, A.J. Complete mitochondrial DNA genome sequences of extinct birds: Ratite phylogenetics and the vicariance biogeography hypothesis. Proc. R. Soc. B-Biol. Sci. 2001, 268, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.J.; Llamas, B.; Soubrier, J.; Rawlence, N.J.; Worthy, T.H.; Wood, J.; Lee, M.S.Y.; Cooper, A. Ancient DNA reveals elephant birds and kiwi are sister taxa and clarifies ratite bird evolution. Science 2014, 344, 898–900. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, T.; Segawa, T.; Mori, H.; Campos, P.F.; Hongoh, Y.; Endo, H.; Akiyoshi, A.; Kohno, N.; Nishida, S.; Wu, J.; et al. Phylogenomics and morphology of extinct palaeognaths reveal the origin and evolution of the ratites. Curr. Biol. 2016, 27, 1–10. [Google Scholar]
- Grealy, A.; Phillips, M.; Miller, G.; Gilbert, M.T.P.; Rouillard, J.M.; Lambert, D.; Bunce, M.; Haile, J. Eggshell palaeogenomics: Palaeognath evolutionary history revealed through ancient nuclear and mitochondrial DNA from Madagascan elephant bird (Aepyornis sp.) eggshell. Mol. Phylogenet. Evol. 2017, 109, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Seabrook-Davison, M.; Huynen, L.; Lambert, D.M.; Brunton, D.H. Ancient DNA resolves identity and phylogeny of New Zealand’s extinct and living quail (Coturnix sp.). PLoS ONE 2009, 4, e6400. [Google Scholar] [CrossRef] [PubMed]
- Steeves, T.E.; Holdaway, R.N.; Hale, M.L.; McLay, E.; McAllan, I.A.W.; Christian, M.; Hauber, M.E.; Bunce, M. Merging ancient and modern DNA: Extinct seabird taxon rediscovered in the North Tasman Sea. Biol. Lett. 2010, 6, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.R.; Mitchell, K.J.; Scofield, R.P.; De Pietri, V.L.; Rawlence, N.J.; Cooper, A. Phylogenetic relationships and terrestrial adaptations of the extinct laughing owl, Sceloglaux albifacies (Aves: Strigidae). Zool. J. Linn. Soc. 2016. [Google Scholar] [CrossRef]
- Mitchell, K.J.; Wood, J.R.; Scofield, R.P.; Llamas, B.; Cooper, A. Ancient mitochondrial genome reveals unsuspected taxonomic affinity of the extinct Chatham duck (Pachyanas chathamica) and resolves divergence times for New Zealand and sub-Antarctic brown teals. Mol. Phylogenet. Evol. 2014, 70, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Bunce, M.; Worthy, T.H.; Ford, T.; Hoppitt, W.; Willerslev, E.; Drummond, A.; Cooper, A. Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature 2003, 425, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Huynen, L.; Millar, C.D.; Scofield, R.P.; Lambert, D.M. Nuclear DNA sequences detect species limits in ancient moa. Nature 2003, 425, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Worthy, T.H.; Bunce, M.; Cooper, A.; Scofield, P. Dinornis—An Insular Oddity, A Taxonomic Conundrum Revealed. In Proceedings of the International Symposium “Insular Vertebrate Evolution: The Palaeontological Approach Monografies de la Societat d’Història Natural de les Balears”; Alcover, J.A., Bover, P., Eds.; 2005; pp. 377–390. [Google Scholar]
- White, N.E.; Bunce, M.; Mawson, P.R.; Dawson, R.; Saunders, D.A.; Allentoft, M.E.; Austin, J. Identifying conservation units after large-scale land clearing: A spatio-temporal molecular survey of endangered white-tailed black cockatoos (Calyptorhynchus spp.). Divers. Distrib. 2014, 20, 1208–1220. [Google Scholar] [CrossRef]
- Thomson, V.A.; Lebrasseur, O.; Austin, J.J.; Hunt, T.L.; Burney, D.A.; Denham, T.; Rawlence, N.J.; Wood, J.R.; Gongora, J.; Flink, L.G.; et al. Using ancient DNA to study the origins and dispersal of ancestral Polynesian chickens across the Pacific. Proc. Natl. Acad. Sci. USA 2014, 111, 4826–4831. [Google Scholar] [CrossRef] [PubMed]
- Storey, A.A.; Spriggs, M.; Bedford, S.; Hawkins, S.C.; Robins, J.H.; Huynen, L.; Matisoo-Smith, E. Mitochondrial DNA from 3000-year old chickens at the Teouma site, Vanuatu. J. Archaeol. Sci. 2010, 37, 2459–2468. [Google Scholar] [CrossRef]
- Storey, A.A.; Athens, J.S.; Bryant, D.; Carson, M.; Emery, K.; deFrance, S.; Higham, C.; Huynen, L.; Intoh, M.; Jones, S.; et al. Investigating the global dispersal of chickens in prehistory using ancient mitochondrial DNA signatures. PLoS ONE 2012, 7, e39171. [Google Scholar] [CrossRef] [PubMed]
- Storey, A.A.; Ramírez, J.M.; Quiroz, D.; Burley, D.V.; Addison, D.J.; Walter, R.; Anderson, A.J.; Hunt, T.L.; Athens, J.S.; Huynen, L.; et al. Radiocarbon and DNA evidence for a pre-Columbian introduction of Polynesian chickens to Chile. Proc. Natl. Acad. Sci. USA 2007, 104, 10335–10339. [Google Scholar] [CrossRef] [PubMed]
- Storey, A.A.; Matisoo-Smith, E.A. No evidence against Polynesian dispersal of chickens to pre-Columbian South America. Proc. Natl. Acad. Sci. USA 2014, 111, e3583. [Google Scholar] [CrossRef] [PubMed]
- Loog, L.; Thomas, M.G.; Barnett, R.; Allen, R.; Skykes, N.; Paxinos, P.D.; Lebrasseur, O.; Dobney, K.; Peters, J.; Manica, A.; et al. Inferring allele frequency trajectories from ancient DNA indicates that selection on a chicken gene coincided with changes in medieval husbandry practices. Mol. Biol. Evol. 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- Haile, J.; Holdaway, R.; Oliver, K.; Bunce, M.; Gilbert, M.T.P.; Nielsen, R.; Munch, K.; Ho, S.Y.; Shapiro, B.; Willerslev, E. Ancient DNA chronology within sediment deposits: Are palaeobiological reconstructions possible and is DNA a leaching factor? Mol. Biol. Evol. 2007, 24, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.R.; Rawlence, N.J.; Rogers, G.M.; Austin, J.J.; Worthy, T.H.; Cooper, A. Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes). Quat. Sci. Rev. 2008, 27, 2593–2602. [Google Scholar] [CrossRef]
- Wood, J.R.; Wilmshurst, J.M.; Richardson, S.J.; Rawlence, N.J.; Wagstaff, S.J.; Worthy, T.H.; Cooper, A. Resolving lost herbivore community structure using coprolites of four sympatric moa species (Aves: Dinornithiformes). Proc. Natl. Acad. Sci. USA 2013, 110, 16910–16915. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.R.; Wilmshurst, J.M.; Wagstaff, S.J.; Worthy, T.H.; Rawlence, N.J.; Cooper, A. High-resolution coproecology: Using coprolites to reconstruct habits and habitats of New Zealand’s extinct upland moa (Megalapteryx didinus). PLoS ONE 2012, 7, e40025. [Google Scholar] [CrossRef] [PubMed]
- Murray, D.C.; Haile, J.; Dortch, J.; White, N.E.; Haouchar, D.; Bellgard, M.I.; Allcock, R.J.; Prideaux, G.J.; Bunce, M. Scrapheap challenge: A novel bulk-bone metabarcoding method to investigate ancient DNA in faunal assemblages. Sci. Rep. 2013, 3, 3371. [Google Scholar] [CrossRef] [PubMed]
- Haouchar, D.; Haile, J.; McDowell, M.C.; Murray, D.C.; White, N.E.; Allcock, R.J.N.; Phillips, M.J.; Prideaux, G.J.; Bunce, M. Thorough assessment of DNA preservation from fossil bone and sediments exavated from a late Pleistocene-Holocene cave deposit on Kangaroo Island, South Australia. Quat. Sci. Rev. 2014, 84, 56–64. [Google Scholar] [CrossRef]
- Grealy, A.; Macken, A.; Allentoft, M.E.; Rawlence, N.J.; Reed, E.; Bunce, M. An assessment of ancient DNA preservation in Holocene-Pleistocene fossil bone excavated from the world hertiage Naracoorte Caves, South Australia. J. Quat. Sci. 2016, 31, 33–45. [Google Scholar] [CrossRef]
- Wood, J.R. Moa (Aves: Dinornithiformes) nesting material from rockshelters in the semi-arid interior of South Island, New Zealand. J. R. Soc. N. Z. 2008, 38, 115–129. [Google Scholar] [CrossRef]
- Wood, J.R.; Wilmshurst, J.M.; Worthy, T.H.; Cooper, A. First coprolite evidence for the diet of Anomalopteryx didiformis, an extinct forest ratite from New Zealand. N. Z. J. Ecol. 2012, 36, 164–170. [Google Scholar]
- Wood, J.R.; Wilmshurst, J.M. Late quaternary terrestrial vertebrate coprolites from New Zealand. Quat. Sci. Rev. 2014, 98, 33–44. [Google Scholar] [CrossRef]
- Seddon, P.J.; Griffiths, C.J.; Soorae, P.S.; Armstrong, D.P. Reversing defaunation: Restoring species in a changing world. Science 2014, 345, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Nogues-Bravo, D.; Simberloff, D.; Rahbek, C.; Sanders, N.J. Rewilding is the new pandora’s box in conservation. Curr. Biol. 2016, 26, R87–R91. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.R.; Wilmshurst, J.M.; Worthy, T.H.; Holzapfel, A.S.; Cooper, A. A lost link between a flightless parrot and a parasitic plant and the potential role of coprolites in conservation paleobiology. Conserv. Biol. 2012, 26, 1091–1099. [Google Scholar] [CrossRef] [PubMed]
- Hofreiter, M.; Mead, J.I.; Martin, P.; Poinar, H.N. Molecular caving. Curr. Biol. 2003, 13, R693–R695. [Google Scholar] [CrossRef] [PubMed]
- Rawlence, N.J.; Metcalf, J.L.; Wood, J.R.; Worthy, T.H.; Austin, J.J.; Cooper, A. The effect of climate and environmental change on the megafaunal moa of New Zealand in the absence of humans. Quat. Sci. Rev. 2012, 50, 141–153. [Google Scholar] [CrossRef]
- Rawlence, N.J.; Wood, J.R.; Bocherens, H.; Rogers, K.M. Dietary interpretations for extinct megafauna using coprolites, intestinal contents and stable isotopes: Complimentary or contradictory? Quat. Sci. Rev. 2016, 142, 173–178. [Google Scholar] [CrossRef]
- Grealy, A.; McDowell, M.; Scofield, P.; Murray, D.; Fusco, D.; Haile, J.; Prideaux, G.; Bunce, M. A critical evaluation of how ancient DNA bulk bone metabarcoding complements traditional palaeontological methods. Quat. Sci. Rev. 2015, 128, 37–47. [Google Scholar] [CrossRef]
- Hartnup, K.; Huynen, L.; Te Kanawa, R.; Shepherd, L.D.; Millar, C.D.; Lambert, D.M. Ancient DNA recovers the origins of Māori feather cloaks. Mol. Biol. Evol. 2011, 28, 2741–2750. [Google Scholar] [CrossRef] [PubMed]
- Oskam, C.L.; Allentoft, M.E.; Walter, R.; Scofield, R.P.; Haile, J.; Holdaway, R.N.; Bunce, M.; Jacomb, C. Ancient DNA analyses of early archaeological sites in New Zealand reveal extreme exploitation of moa (Aves: Dinornithiformes) at all life stages. Quat. Sci. Rev. 2012, 52, 41–48. [Google Scholar] [CrossRef]
- Oskam, C.L.; Jacomb, C.; Allentoft, M.E.; Walter, R.; Scofield, R.P.; Haile, J.; Holdaway, R.N.; Bunce, M. Molecular and morphological analyses of avian eggshell excavated from a late thirteenth century earth oven. J. Archaeol. Sci. 2011, 38, 2589–2595. [Google Scholar] [CrossRef]
- Eda, M.; Koike, H.; Higuchi, H. Understanding prehistoric maritime adaptations in northern Japan: Indirect evidence from ancient DNA and historical observations of albatross (Aves: Diomedeidae) bones. Quat. Int. 2016, 419, 159–164. [Google Scholar] [CrossRef]
- Cooper, A.; Rhymer, J.; James, H.F.; Olson, S.L.; McIntosh, C.E.; Sorenson, M.D.; Fleischer, R.C. Ancient DNA and island endemics. Nature 1996, 381, 484. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.M.; Fraser, C.I.; Maxwell, J.J.; Rawlence, N.J. Did interaction between human pressure and Little Ice Age drive biological turnover in New Zealand? J. Biogeogr. 2017, in press. [Google Scholar] [CrossRef]
- Shepherd, L.D.; Lambert, D.M. Ancient DNA and conservation: Lessons from the endangered kiwi of New Zealand. Mol. Ecol. 2008, 17, 2174–2184. [Google Scholar] [CrossRef] [PubMed]
- Behrensmeyer, A.K.; Stayton, C.T.; Chapman, R.E. Taphonomy and ecology of modern avifaunal remains from Amboseli Park, Kenya. Paleobiology 2003, 29, 52–70. [Google Scholar] [CrossRef]
- Clark, G.R.; Petchey, P.; McGlone, M.S.; Bristow, P. Faunal and floral remains from Earnscleugh cave, central Otago, New Zealand. J. R. Soc. N. Z. 1996, 263, 363–380. [Google Scholar] [CrossRef]
- Worthy, T.H.; Holdaway, R.N. Taphonomy of two Holocene microvertebrate deposits, Takaka Hill, Nelson, New Zealand and identification of the avian predator responsible. Hist. Biol. 1996, 12, 1–24. [Google Scholar] [CrossRef]
- Gaston, K.J.; Blackburn, T.M. Birds, body size, and the threat of extinction. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1995, 347, 205–212. [Google Scholar] [CrossRef]
- Duncan, R.P.; Blackburn, T.M. Causes of extinction in island birds. Animal Conserv. 2007, 10, 149–150. [Google Scholar] [CrossRef]
- Gill, B.J.; Bell, B.D.; Chambers, G.K.; Medway, D.G.; Palma, R.L.; Scofield, R.P.; Tennyson, A.J.D.; Worthy, T.H. Checklist of the Birds in New Zealand, Norfolk and Macquarie Islands, and the Ross Dependency Antarctica, 4th ed.; Te Papa Press: Wellington, New Zealand, 2010. [Google Scholar]
- Cristidis, L.; Boles, W.E. Systematics and Taxonomy of Australian Birds; CSIRO Publishing: Melbourne, Australia, 3 July 2008. [Google Scholar]
- Allentoft, M.E.; Rawlence, N.J. Moa’s ark or volant ghosts of Gondwana? Insights from nineteen years of ancient DNA research on the extinct moa (Aves: Dinornithiformes) of New Zealand. Ann. Anat.-Anat. Anz. 2012, 194, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.; Lalueza-Fox, C.; Anderson, S.; Rambaut, A.; Austin, J.; Ward, R. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature 2001, 409, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Allentoft, M.E.; Schuster, S.C.; Holdaway, R.N.; Hale, M.L.; McLay, E.; Oskam, C.; Gilbert, M.T.P.; Spencer, P.; Willerslev, E.; Bunce, M. Identification of microsatellites from an extinct moa species using high-throughput (454) sequence data. Biotechniques 2009, 46, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Allentoft, M.E.; Oskam, C.; Houston, J.; Hale, M.L.; Gilbert, M.T.P.; Rasmussen, M.; Spencer, P.; Jacomb, C.; Willerslev, E.; Holdaway, R.N.; et al. Profiling the dead: Generating microsatellite data from fossil bones of extinct megafauna-protocols, problems, and prospects. PLoS ONE 2011, 6, e16670. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.R.; De Pietri, V.L. Next-generation paleornithology: Technological and methodological advances allow new insights into the evolutionary and ecological histories of living birds. Auk 2015, 132, 486–506. [Google Scholar] [CrossRef]
- Millar, C.D.; Dodd, A.; Anderson, J.; Gibb, G.C.; Ritchie, P.A.; Baroni, C.; Woodhams, M.D.; Hendy, M.D.; Lambert, D.M. Mutation and evolutionary rates in Adelie penguins from the Antarctic. PLoS Genet. 2008, 4, e1000209. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Denver, D.R.; Millar, C.D.; Heupink, T.; Aschrafi, A.; Emslie, S.D.; Baroni, C.; Lambert, D.M. High mitotic evolutionary rates and time dependency. Trends Genet. 2009, 25, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Huynen, L.; Gill, B.J.; Millar, C.D.; Lambert, D.M. Ancient DNA reveals extreme egg morphology and nesting behavior in New Zealand’s extinct moa. Proc. Natl. Acad. Sci. USA 2010, 107, 16201–16206. [Google Scholar] [CrossRef] [PubMed]
- Oskam, C.L.; Haile, J.; McLay, E.; Rigby, P.; Allentoft, M.E.; Olsen, M.E.; Bengtsson, C.; Miller, G.H.; Schwenninger, J.L.; Jacomb, C.; et al. Fossil avian eggshell preserves ancient DNA. Proc. R. Soc. B-Biol. Sci. 2010, 277, 1991–2000. [Google Scholar] [CrossRef] [PubMed]
- Chilton, G.; Sorenson, M.D. Genetic identification of eggs purportedly from the extinct labrador duck (Camptorhynchus labradorius). Auk 2007, 124, 962–968. [Google Scholar] [CrossRef]
- Rawlence, N.J.; Wood, J.R.; Armstrong, K.N.; Cooper, A. DNA content and distribution in ancient feathers and potential to reconstruct the plumage of extinct avian taxa. Proc. R. Soc. B-Biol. Sci. 2009, 276, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Willerslev, E.; Hansen, A.J.; Binladen, J.; Brand, T.B.; Gilbert, M.T.P.; Shapiro, B.; Bunce, M.; Wiuf, C.; Gilichinsky, D.A.; Cooper, A. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 2003, 300, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Anmarkrud, J.A.; Lifjeld, J.T. Complete mitochondrial genomes of eleven extinct or possibly extinct bird species. Mol. Ecol. Res. 2016, 17, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.V. Is a new and general theory of molecular systematics emerging? Evolution 2009, 63, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Carstens, B.C.; Pelletier, T.A.; Reid, N.M.; Satler, J.D. How to fail at species delimitation. Mol. Ecol. 2013, 22, 4369–4383. [Google Scholar] [CrossRef] [PubMed]
- Rand, D.M. Thermal habit, metabolic rate and the evolution of mitochondrial DNA. Trends Ecol. Evol. 1994, 9, 125–131. [Google Scholar] [CrossRef]
- Ksepka, D.T.; Phillips, M.J. Avian diversification patterns across the K-Pg boundary: Influence of calibrations, datasets, and model misspecification. Ann. Mo. Bot. Garden 2015, 100, 300–328. [Google Scholar] [CrossRef]
- Ksepka, D.T.; Ware, J.L.; Lamm, K.S. Flying rocks and flying clocks: Disparity in fossil and molecular dates for birds. Proc. R. Soc. B-Biol. Sci. 2014, 281, 20140677. [Google Scholar] [CrossRef] [PubMed]
- Cibois, A.; Dekker, R.W.R.J.; Pasquet, E.; Thibault, J.C. New insights into the systematics of the enigmatic Polynesian sandpipers Aechmorhynchus parvirostris and Prosobonia leucoptera. IBIS 2012, 154, 756–767. [Google Scholar] [CrossRef]
- Nylander, J.A.A.; Olsson, U.; Alstrom, P.; Sanmartin, I. Accounting for phylogenetic uncertainty in biogeography: A Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus). Syst. Biol. 2008, 57, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.P.; Clayton, D.H.; Dumbacher, J.P.; Fleischer, R.C. The flight of the passenger pigeon: Phylogenetics and biogeographic history of an extinct species. Mol. Phylogenet. Evol. 2010, 57, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.J.; Haddrath, O.; McPherson, J.D.; Cloutier, A. Genomic support for a moa-tinamou clade and adaptive morphological convergence in flightless ratites. Mol. Biol. Evol. 2014, 31, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, L.D.; Millar, C.D.; Ballard, G.; Ainley, D.G.; Wilson, P.R.; Haynes, G.D.; Baroni, C.; Lambert, D.M. Microevolution and mega-icebergs in the Antarctic. Proc. Natl. Acad. Sci. USA 2005, 102, 16717–16722. [Google Scholar] [CrossRef] [PubMed]
- Aidala, Z.; Huynen, L.; Brennan, P.L.R.; Musser, J.; Fidler, A.; Chong, N.; Capuska, G.E.M.; Anderson, M.G.; Talaba, A.; Lambert, D.; et al. Ultraviolet visual sensitivity in three avian lineages: Palaeognaths, parrots, and passerines. J. Comput. Physiol. A 2012, 198, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hofreiter, M.; Straube, N.; Corrigan, S.; Naylor, G.J.P. Capturing protein-coding genes across highly divergent species. Biotechniques 2013, 54, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.M.; Shaner, P.J.L.; Zink, R.M.; Liu, W.C.; Chu, T.C.; Huang, W.S.; Li, S.H. Drastic population fluctuations explain the rapid extinction of the passenger pigeon. Proc. Natl. Acad. Sci. USA 2014, 111, 10636–10641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appelt, S.; Fancello, L.; Le Bailly, M.; Raoult, D.; Drancourt, M.; Desnues, C. Viruses in a 14th-century coprolite. Appl. Environ. Microbiol. 2014, 80, 2648–2655. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.R.; Wilmshurst, J.M.; Rawlence, N.J.; Bonner, K.I.; Worthy, T.H.; Kinsella, J.M.; Cooper, A. A megafauna’s microfauna: Gastrointestinal parasites of New Zealand’s extinct moa (Aves: Dinornithiformes). PLoS ONE 2013, 8, e57315. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proc. Gatew. Comput. Environ. Workshop (GCE) 2010, 1–8. [Google Scholar]
- CYVERSE. Available online: http://www.cyverse.org/ (accessed on 15 May 2017).
- Blankenberg, D.; Von Kuster, G.; Coraro, N.; Ananda, G.; Lazarus, R.; Mangan, M.; Nekrutenko, A.; Taylor, J. Galaxy: A web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010. [Google Scholar] [CrossRef]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Elnitski, L.; Shah, P.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; The Galaxy Team. Galaxy: A comprehensive approach for supporting accessible, reproducible and transparent computational research in the life sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, S.; Visel, A.; Akiyama, J.A.; Shoukry, M.; Lewis, K.D.; Holt, A.; Plajzer-Frick, I.; Morrison, H.; FitzPatrick, D.R.; Afzal, V.; et al. Human-specific gain of function in a developmental enhancer. Science 2008, 321, 1346–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Rajewsky, N. The evolution of gene regulation by transcription factors and micrornas. Nat. Rev. Genet. 2007, 8, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Llamas, B.; Holland, M.L.; Chen, K.; Cropley, J.E.; Cooper, A.; Suter, C.M. High-resolution analysis of cytosine methylation in ancient DNA. PLoS ONE 2012, 7, e30226. [Google Scholar] [CrossRef] [PubMed]
- Pask, A.J.; Behringer, R.R.; Renfree, M.B. Resurrection of DNA function in vivo from an extinct genome. PLoS ONE 2008, 3, e2240. [Google Scholar] [CrossRef] [PubMed]
- Huynen, L.; Suzuki, T.; Ogura, T.; Watanabe, Y.; Millar, C.D.; Hofreiter, M.; Smith, C.; Mirmoeini, S.; Lambert, D.M. Reconstruction and in vivo analysis of the extinct tbx5 gene from ancient wingless moa (Aves: Dinornithiformes). BMC Evol. Biol. 2014, 14, 75. [Google Scholar] [CrossRef] [PubMed]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Smith, O.; Clapham, A.J.; Rose, P.; Liu, Y.; Wang, J.; Allaby, R.G. Genomic methylation patterns in archaeological barley show de-methylation as a time-dependent diagenetic process. Sci. Rep. 2014, 4, 5559. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.W.; Monroe, C.; Bolnick, D.A. Detection of cytosine methylation in ancient DNA from five native American populations using bisulfite sequencing. PLoS ONE 2015, 10, e0125344. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.W.A.; Bolnick, D.A. Analysis of cytosine methylation in native American ancient DNA. Am. J. Phys. Anthropol. 2013, 150, 258. [Google Scholar]
- Gokhman, D.; Lavi, E.; Prufer, K.; Fraga, M.F.; Riancho, J.A.; Kelso, J.; Paabo, S.; Meshorer, E.; Carmel, L. Reconstructing the DNA methylation maps of the neandertal and the denisovan. Science 2014, 344, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Gokhman, D.; Meshorer, E.; Carmel, L. Epigenetics: It’s getting old. Past meets future in paleoepigenetics. Trends Ecol. Evol. 2016, 31, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Genetic Rescue Foundation. Available online: www.geneticrescue.science (accessed on 15 May 2017).
- Revive and Restore. Available online: reviverestore.org (accessed on 15 May 2017).
- Novak, B. Why Birds Are a Challenge. Revive and Restore: Genetic Rescue for Endangered and Extinct Species. 2013. Available online: http://reviverestore.org/why-birds-are-a-challenge/ (accessed on 15 May 2017).
- Sandler, R. The ethics of reviving long extinct species. Conserv. Biol. 2013, 28, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.R.; Maloney, R.F.; Steeves, T.E.; Brazill-Boast, J.; Possingham, H.P.; Seddon, P.J. Spending limited resources on de-extinction could lead to net biodiversity loss. Nat. Ecol. Evol. 2017, 1, 53. [Google Scholar] [CrossRef]
- Seddon, P.J. The ecology of De-extinction. Funct. Ecol. 2017, 31, 987–1172. [Google Scholar] [CrossRef]
- Taylor, H.R.; Colbourne, R.M.; Robertson, H.A.; Nelson, N.J.; Allendorf, F.W.; Ramstad, K.M. Cryptic inbreeding depression in a growing population of a long-lived species. Mol. Ecol. 2017, 26, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; Leary, R.F.; Hitt, N.P.; Knudsen, K.L.; Lunquist, L.L.; Spruell, P. Intercrosses and the US Endangered species act: Should hybridised populations be included as westslope cutthroat trout? Conserv. Biol. 2004, 18, 1203–1213. [Google Scholar] [CrossRef]
- Wayne, R.K.; Shaffer, B. Hybridisation and endangered species protection in the molecular era. Mol. Ecol. 2016, 25, 2680–2689. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.R.; Dussex, N.; van Heezik, Y. De-extinction needs consultation. Nat. Ecol. Evol. 2017, 1, 198. [Google Scholar] [CrossRef]
- Dietl, G.P.; Flessa, K.W. Conservation paleobiology: Putting the dead to work. Trends Ecol. Evol. 2011, 26, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, L.; Worthy, T.H.; Tennyson, A.J.D.; Scofield, R.P.; Ramstad, K.M.; Lambert, D.M. Ancient DNA analyses reveal constrasting phylogeographic patterns amongst kiwi (Apteryx spp.) and a recently extinct lineage of spotted kiwi. PLoS ONE 2012, 8, e42384. [Google Scholar]
- Taylor, H.R.; Dussex, N.; van Heezik, Y. Bridging the conservation genetics gap by identifying barriers to implementation for conservation practitioners. Global Ecol. Conserv. 2017, 10, 231–242. [Google Scholar] [CrossRef]

| Term | Definition |
|---|---|
| Palaeognath | A clade of extant birds, sister to Neognaths; retain a “primitive” palate. |
| Neognath | A clade of extant birds, sister to Palaeognaths; differ from Palaeognathae in the structure of their palate. |
| Vicariance | The process by which new species are generated through the formation of a geographical barrier to gene flow between populations. |
| Sexual dimorphism | Disparity in the morphology (typically size) between the males and females of a species. |
| Coprolite | Fossil faeces. |
| Palynology | The study of pollen. |
| Midden | A refuse heap. |
| Anthropocene | “The period of time during which human activities have had an environmental impact on the Earth regarded as constituting a distinct geological age” [12]. |
| Volant | Possessing the ability to fly. |
| Predator naïvety | The indifference of island species to potential predators making them vulnerable to predation and extinction. |
| Next-generation sequencing | NGS; also known as “high-throughput” and “second-generation” sequencing. Short fragments of DNA (typically 50–500 bp) can be sequenced in parallel. |
| Metabarcoding | Involves the use of highly conserved primers that are able to bind to DNA from multiple different species in a mixed sample, yet amplify a region (a DNA “barcode”) that is variable enough to distinguish between species within the sample based on its sequence [13]. |
| PCR | Polymerase chain reaction; the method by which specific target regions of DNA are amplified. |
| Sanger sequencing | Also known as “first-generation” sequencing; employs a “chain-termination” chemistry to sequence typically long fragments (400 bp +) with high accuracy, one-at-a-time. |
| Shotgun sequencing | All DNA fragments within an extract are built into a sequencing library through the ligation of sequences adapters to either end; sequence reads are then overlapped to a continuous sequence. For aDNA, both endogenous and contaminating DNA is sequenced. |
| Transposable elements | “Jumping genes”; gene sequences that can copy, excise, and reinsert themselves throughout the genome. |
| Microsatellites | Sequences consisting of short tandem repeats; different alleles are characterised by the number of repeats at a locus. |
| Hybridisation capture | A method by which to enrich target DNA prior to sequencing through the use of probes from a modern species to “bait” DNA from it’s extinct relative, leaving contaminating DNA behind. |
| Data mining | “The practice of searching through large amounts of computerised data to find useful patterns or trends” [12]. |
| Front-end analysis | Analysis that occurs prior to the out-set of project in order to plan the most effective way to meet the project’s end-goals. |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grealy, A.; Rawlence, N.J.; Bunce, M. Time to Spread Your Wings: A Review of the Avian Ancient DNA Field. Genes 2017, 8, 184. https://doi.org/10.3390/genes8070184
Grealy A, Rawlence NJ, Bunce M. Time to Spread Your Wings: A Review of the Avian Ancient DNA Field. Genes. 2017; 8(7):184. https://doi.org/10.3390/genes8070184
Chicago/Turabian StyleGrealy, Alicia, Nicolas J. Rawlence, and Michael Bunce. 2017. "Time to Spread Your Wings: A Review of the Avian Ancient DNA Field" Genes 8, no. 7: 184. https://doi.org/10.3390/genes8070184
APA StyleGrealy, A., Rawlence, N. J., & Bunce, M. (2017). Time to Spread Your Wings: A Review of the Avian Ancient DNA Field. Genes, 8(7), 184. https://doi.org/10.3390/genes8070184