Detection of G1138A Mutation of the FGFR3 Gene in Tooth Material from a 180-Year-Old Museological Achondroplastic Skeleton
Abstract
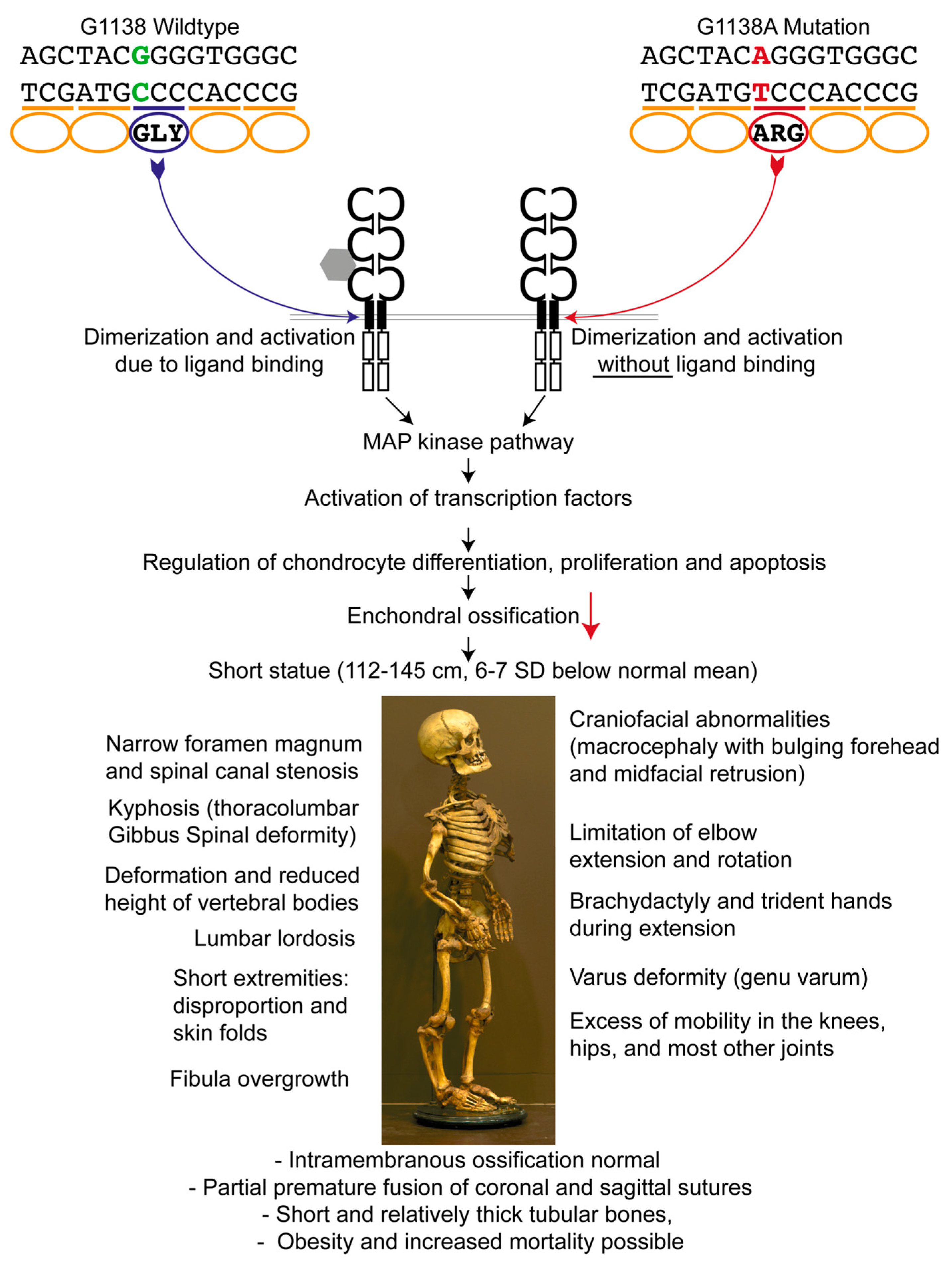
:1. Introduction
2. Materials and Methods
2.1. Achondroplastic Skeleton
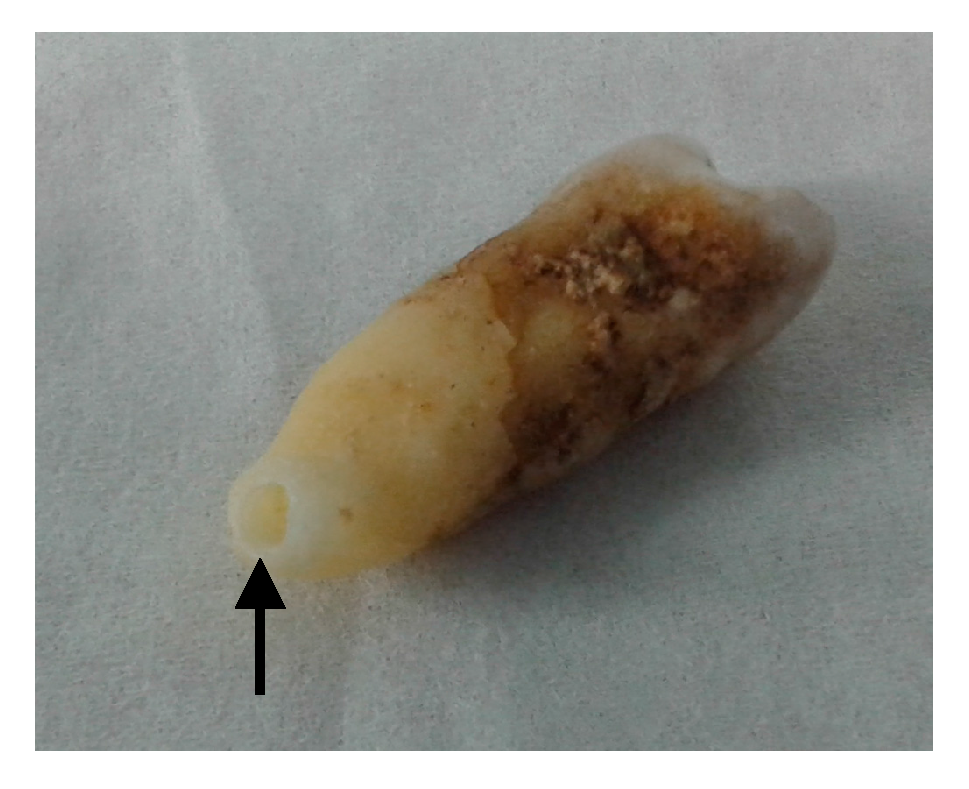
2.2. Sample Preparation
2.3. DNA Extraction
2.4. Initial PCR Testing for the Presence of DNA
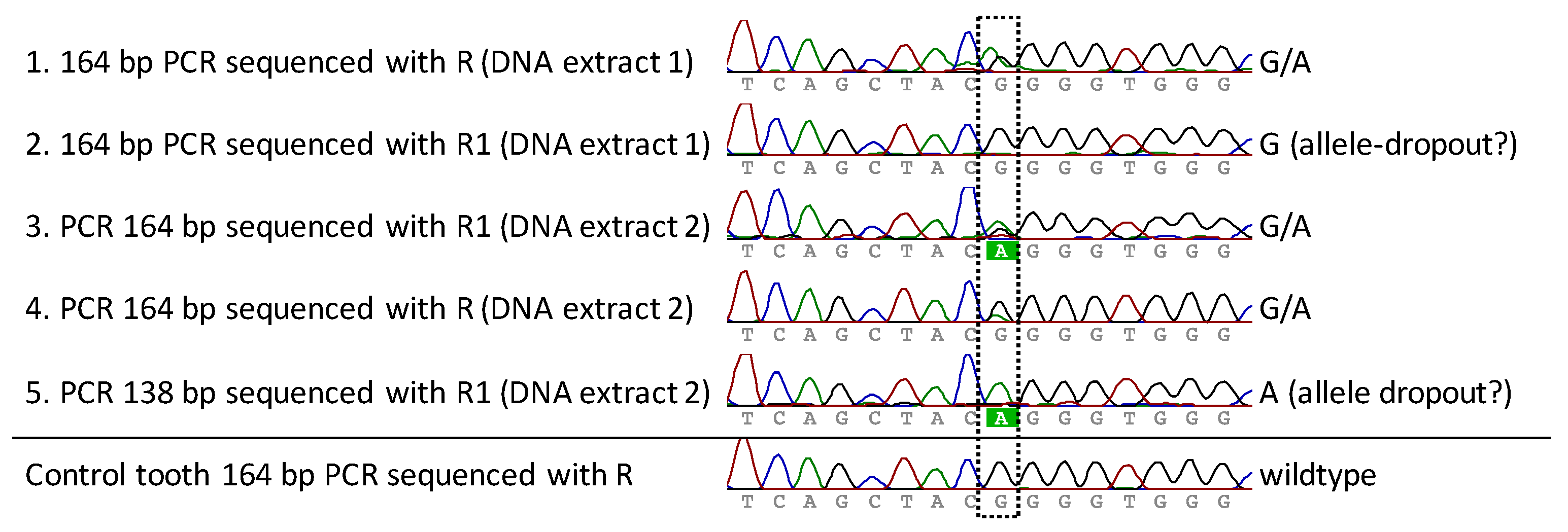
2.5. Mutation Detection Using PCR and Sanger Sequencing
3. Results
3.1. Obtained Material for Analysis and Pre-Testing
3.2. FGFR3 Analysis for Mutation Detection
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
Funding
References
- Beighton, P.; Sujansky, E.; Patzak, B.; Portele, K.A. Genetic skeletal dysplasias in the museum of pathological anatomy, Vienna. Am. J. Med. Genet. 1993, 47, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Beighton, P.; Sujansky, E.; Patzak, B.; Portele, K.A. Bone dysplasias of infancy in the Vienna collection. Pediatr. Radiol. 1994, 24, 384–386. [Google Scholar] [CrossRef] [PubMed]
- Oostra, R.J.; Baljet, B.; Dijkstra, P.F.; Hennekam, R.C. Congenital anomalies in the teratological collection of Museum Vrolik in Amsterdam, The Netherlands. II: Skeletal dysplasias. Am. J. Med. Genet. 1998, 77, 116–134. [Google Scholar] [CrossRef]
- Boer, L.L.; Schepens-Franke, A.N.; van Asten, J.J.A.; Bosboom, D.G.H.; Kamphuis-van Ulzen, K.; Kozicz, T.L.; Ruiter, D.J.; Oostra, R.J.; Klein, W.M. Radiological imaging of teratological fetuses: What can we learn? Insights Imaging 2017, 3, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Legeai-Mallet, L. Achondroplasia: Development, pathogenesis, and therapy. Dev. Dyn. 2017, 246, 291–309. [Google Scholar] [CrossRef] [PubMed]
- Horton, W.A.; Hall, J.G.; Hecht, J.T. Achondroplasia. Lancet 2007, 370, 162–172. [Google Scholar] [CrossRef]
- Vajo, Z.; Francomano, C.A.; Wilkin, D.J. The molecular and genetic basis of fibroblast growth factor receptor 3 disorders: The achondroplasia family of skeletal dysplasias, Muenke craniosynostosis, and Crouzon syndrome with acanthosis nigricans. Endocr. Rev. 2000, 21, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Stanescu, R.; Stanescu, V.; Maroteaux, P. Homozygous achondroplasia: Morphologic and biochemical study of cartilage. Am. J. Med. Genet. 1990, 37, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Parrot, J.M. L’achondroplasie et les lésions osseusses de la syphilis hereditaire et du rachitisme. Arch. Physiol. 1876, 2, 133–139. [Google Scholar]
- Le Merrer, M.; Rousseau, F.; Legeai-Mallet, L.; Landais, J.C.; Pelet, A.; Bonaventure, J.; Sanak, M.; Weissenbach, J.; Stoll, C.; Munnich, A. A gene for achondroplasia-hypochondroplasia maps to chromosome 4p. Nat. Genet. 1994, 6, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Velinov, M.; Slaugenhaupt, S.A.; Stoilov, I.; Scott, C.I., Jr.; Gusella, J.F.; Tsipouras, P. The gene for achondroplasia maps to the telomeric region of chromosome 4p. Nat. Genet. 1994, 6, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Etlik, O.; Koksal, V.; Tugba Arican-Baris, S.; Baris, I. An improved tetra-primer PCR approach for the detection of the FGFR3 G380R mutation responsible for achondroplasia. Mol. Cell. Probes 2008, 22, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Richette, P.; Bardin, T.; Stheneur, C. Achondroplasia: From genotype to phenotype. Joint Bone Spine 2008, 75, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.K.; Donoghue, D.J. Constitutive activation of fibroblast growth factor receptor 3 by the transmembrane domain point mutation found in achondroplasia. EMBO J. 1996, 15, 520–527. [Google Scholar] [PubMed]
- Deng, C.; Wynshaw-Boris, A.; Zhou, F.; Kuo, A.; Leder, P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 1996, 84, 911–921. [Google Scholar] [CrossRef]
- Horton, W.A.; Rotter, J.I.; Rimoin, D.L.; Scott, C.I.; Hall, J.G. Standard growth curves for achondroplasia. J. Pediatr. 1978, 93, 435–438. [Google Scholar] [CrossRef]
- Baujat, G.; Legeai-Mallet, L.; Finidori, G.; Cormier-Daire, V.; Le Merrer, M. Achondroplasia. Best Pract. Res. Clin. Rheumatol. 2008, 22, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Pauli, R.M. Achondroplasia; Pagon, R.A., Adam, M.P., Ardinger, H.H., Wallace, S.E., Amemiya, A., Bean, L.J.H., Bird, T.D., Ledbetter, N., Mefford, H.C., Smith, R.J.H., et al., Eds.; GeneReviews®: Seattle, WA, USA, 1993–2017. [Google Scholar]
- Di Rocco, F.; Biosse Duplan, M.; Heuze, Y.; Kaci, N.; Komla-Ebri, D.; Munnich, A.; Mugniery, E.; Benoist-Lasselin, C.; Legeai-Mallet, L. FGFR3 mutation causes abnormal membranous ossification in achondroplasia. Hum. Mol. Genet. 2014, 23, 2914–2925. [Google Scholar] [CrossRef] [PubMed]
- Wynn, J.; King, T.M.; Gambello, M.J.; Waller, D.K.; Hecht, J.K. Mortality in achondroplasia study: A 42-year follow-up. Am. J. Med. Genet. A 2007, 143A, 2502–2511. [Google Scholar] [CrossRef] [PubMed]
- Cheema, J.I.; Grissom, L.E.; Harcke, H.T. Radiographic characteristics of lower-extremity bowing in children. Radiographics 2003, 23, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.; Song, H.R.; Mahajan, R.; Makwana, V.; Suh, S.W.; Lee, S.H. Development of genu varum in achondroplasia: Relation to fibular overgrowth. J. Bone Joint Surg. Br. 2007, 89, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Pusch, C.M.; Broghammer, M.; Nicholson, G.J.; Nerlich, A.G.; Zink, A.; Kennerknecht, I.; Bachmann, L.; Blin, N. PCR-induced sequence alterations hamper the typing of prehistoric bone samples for diagnostic achondroplasia mutations. Mol. Biol. Evol. 2004, 21, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Dusseau, J.L. Museé Vrolik. Catalogue de la Collection d’Anatomie Humaine, Compareé et Pathologique. De M.M. Ger. et W. Vrolik, professeurs à l’Athénée Illustre d’Amsterdam; J. Roever-Kröber: Amsterdam, The Netherlands, 1865; p. 303. [Google Scholar]
- Walker, J.A.; Hedges, D.J.; Perodeau, B.P.; Landry, K.E.; Stoilova, N.; Laborde, M.E.; Shewale, J.; Sinha, S.K.; Batzer, M.A. Multiplex polymerase chain reaction for simultaneous quantitation of human nuclear, mitochondrial, and male Y-chromosome DNA: Application in human identification. Anal. Biochem. 2005, 337, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Shiang, R.; Thompson, L.M.; Zhu, Y.Z.; Church, D.M.; Fielder, T.J.; Bocian, M.; Winokur, S.T.; Wasmuth, J.J. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell 1994, 78, 335–342. [Google Scholar] [CrossRef]
- Boer, L.L.; Morava, E.; Klein, W.M.; Schepens-Franke, A.N.; Oostra, R.J. Sirenomelia: A Multi-systemic Polytopic Field Defect with Ongoing Controversies. Birth Defects Res. 2017, 109, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Oostra, R.J.; Baljet, B.; Schutgens, R.B.; Hennekam, R.C. Smith-Lemli-Opitz syndrome diagnosed in a 130-year-old anatomical specimen. Am. J. Med. Genet. 1997, 31, 257–259. [Google Scholar] [CrossRef]
- Hansen, H.B.; Damgaard, P.B.; Margaryan, A.; Stenderup, J.; Lynnerup, N.; Willerslev, E.; Allentoft, M.E. Comparing Ancient DNA Preservation in Petrous Bone and Tooth Cementum. PLoS ONE 2017, 12, e0170940. [Google Scholar] [CrossRef] [PubMed]
- De Rooy, L.; van den Bogaard, H. Forces of Form: The Vrolik Museum; University Press: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Hofreiter, M. Nondestructive DNA Extraction from Museum Specimens. In Ancient DNA; Humana Press: New York, NY, USA, 2012; pp. 93–100. [Google Scholar] [CrossRef]
- Gomes, C.; Palomo-Díez, S.; Roig, J.; López-Parra, A.M.; Baeza-Richer, C.; Esparza-Arroyo, A.; Gibaja, J.; Arroyo-Pardo, E. Nondestructive extraction DNA method from bones or teeth, true or false? Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e279–e282. [Google Scholar] [CrossRef]
- Bolnick, D.A.; Bonine, H.M.; Mata-Míguez, J.; Kemp, B.M.; Snow, M.H.; LeBlanc, S.A. Nondestructive sampling of human skeletal remains yields ancient nuclear and mitochondrial DNA. Am. J. Phys. Anthropol. 2012, 147, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Higgins, D.; Austin, J.J. Teeth as a source of DNA for forensic identification of human remains: A Review. Sci. Justice 2013, 53, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Adler, C.J.; Haak, W.; Donlon, D.; Cooper, A. Survival and recovery of DNA from ancient teeth and bones. J. Archaeol. Sci. 2011, 38, 956–964. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boer, L.L.; Naue, J.; De Rooy, L.; Oostra, R.-J. Detection of G1138A Mutation of the FGFR3 Gene in Tooth Material from a 180-Year-Old Museological Achondroplastic Skeleton. Genes 2017, 8, 214. https://doi.org/10.3390/genes8090214
Boer LL, Naue J, De Rooy L, Oostra R-J. Detection of G1138A Mutation of the FGFR3 Gene in Tooth Material from a 180-Year-Old Museological Achondroplastic Skeleton. Genes. 2017; 8(9):214. https://doi.org/10.3390/genes8090214
Chicago/Turabian StyleBoer, Lucas L., Jana Naue, Laurens De Rooy, and Roelof-Jan Oostra. 2017. "Detection of G1138A Mutation of the FGFR3 Gene in Tooth Material from a 180-Year-Old Museological Achondroplastic Skeleton" Genes 8, no. 9: 214. https://doi.org/10.3390/genes8090214
APA StyleBoer, L. L., Naue, J., De Rooy, L., & Oostra, R.-J. (2017). Detection of G1138A Mutation of the FGFR3 Gene in Tooth Material from a 180-Year-Old Museological Achondroplastic Skeleton. Genes, 8(9), 214. https://doi.org/10.3390/genes8090214





