Physiological and Comparative Genomic Analysis of Arthrobacter sp. SRS-W-1-2016 Provides Insights on Niche Adaptation for Survival in Uraniferous Soils
Abstract
:1. Introduction
2. Results and Discussion
2.1. Uranium and Nickel Depletion Potential of Arthrobacter sp. SRS-1-W-2016
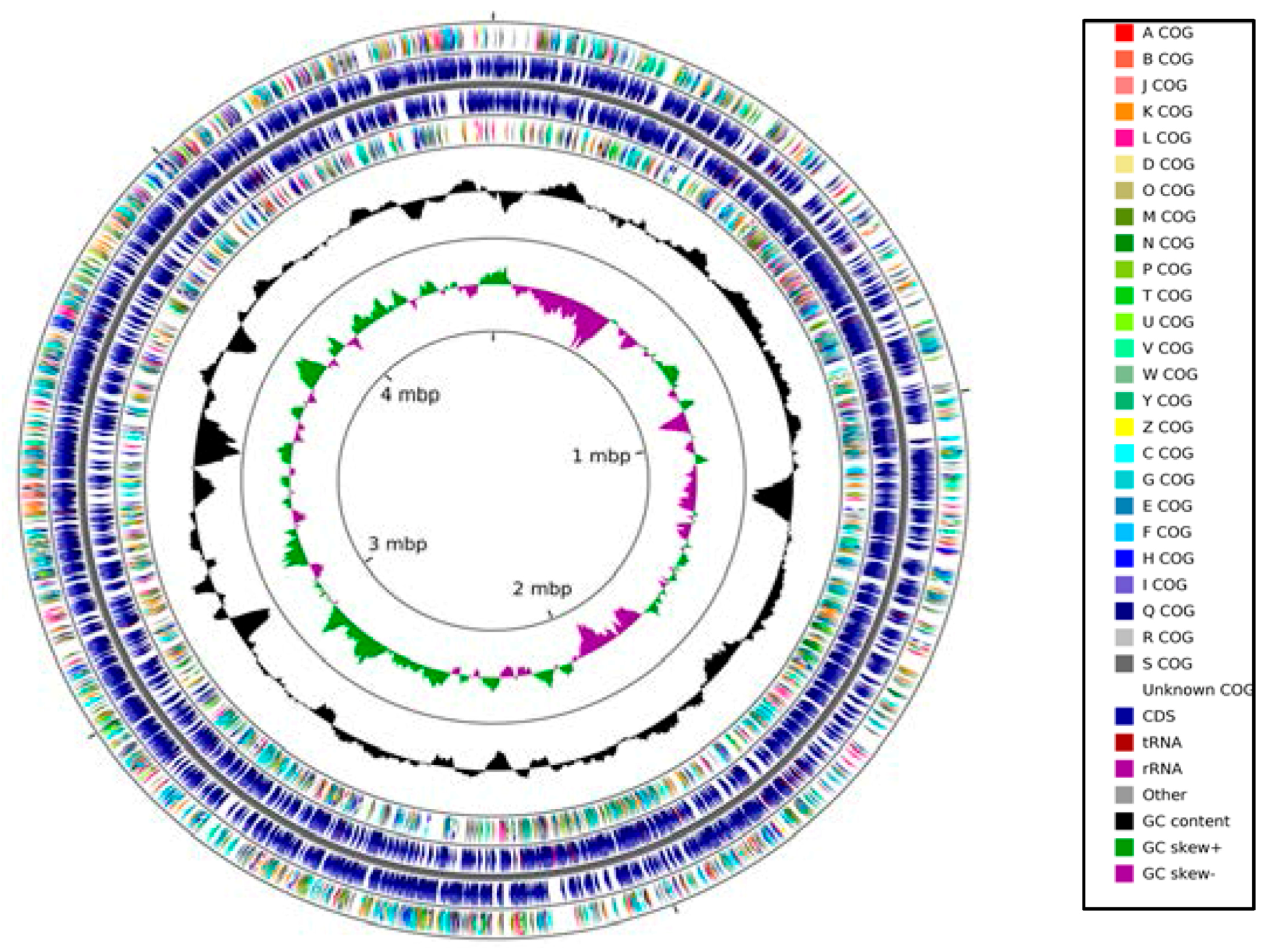
2.2. Genome-Centric Evaluation of Strain SRS-W-1-2016
2.3. Genes Potentially Conferring Metal Resistance in Strain SRS-W-1-2016
2.4. Resistome of Arthrobacter sp. Strain SRS-W-1-2016
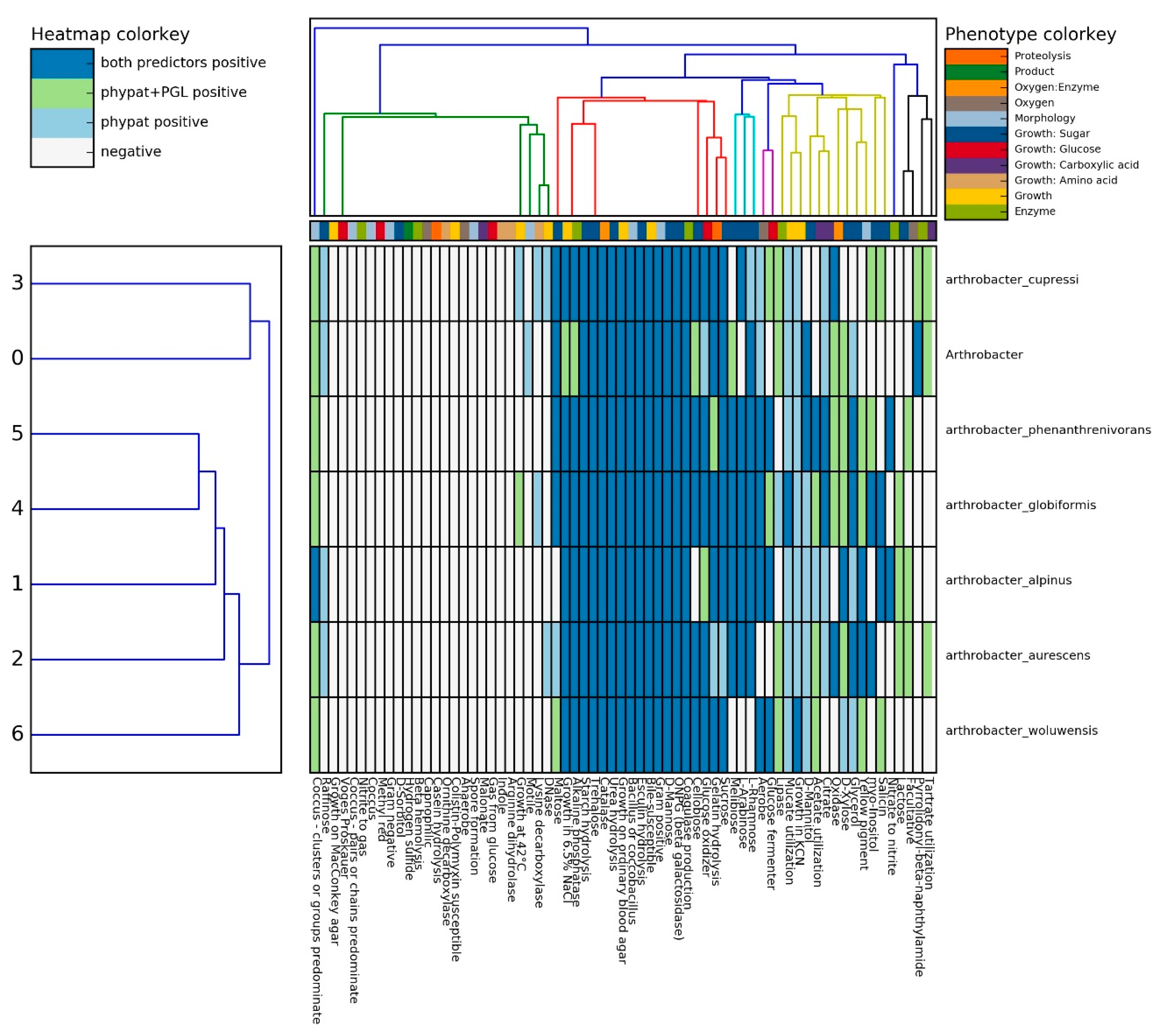
2.5. Prediction of Phenotypic Traits in Arthrobacter sp. Strain SRS-W-1-2016
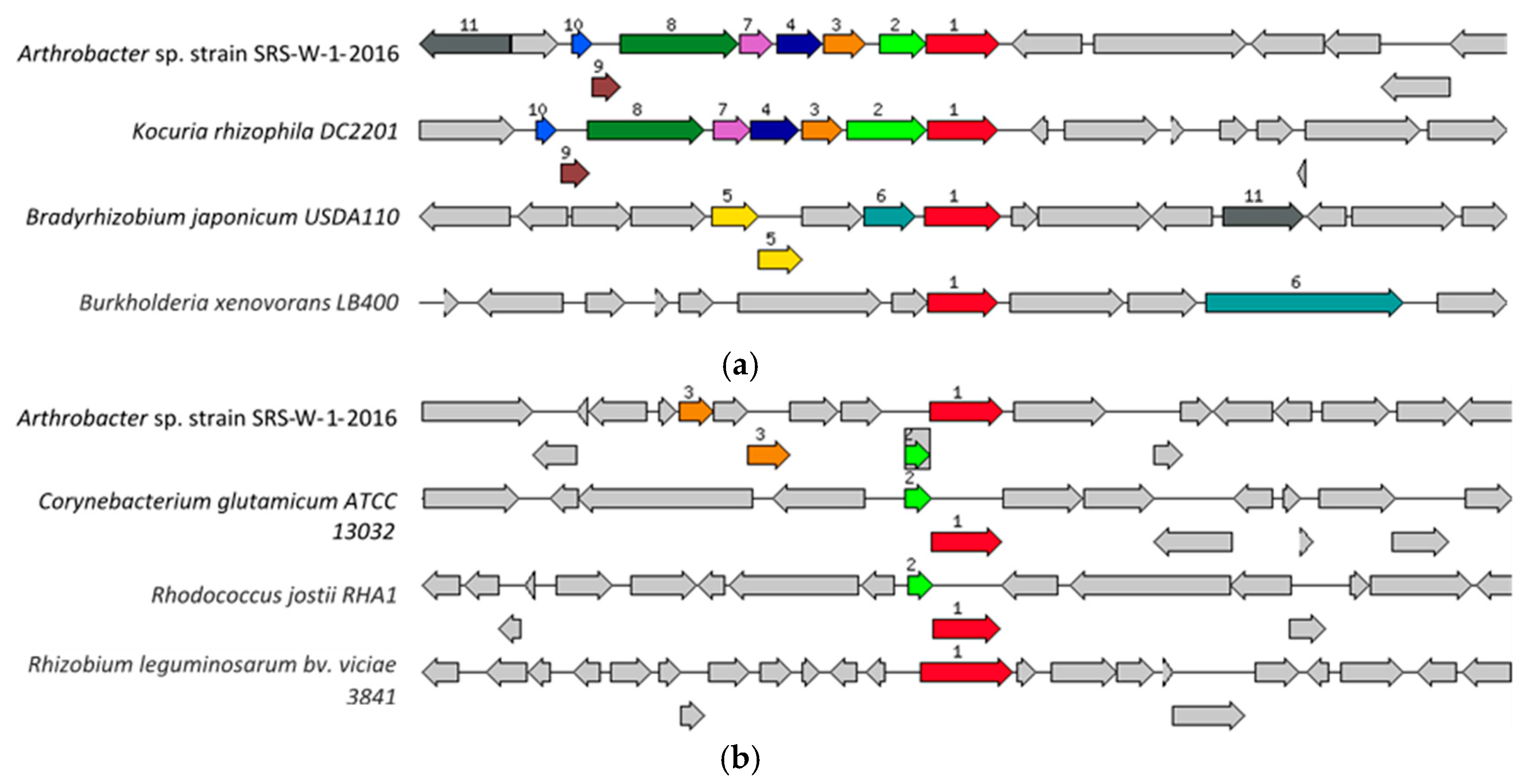
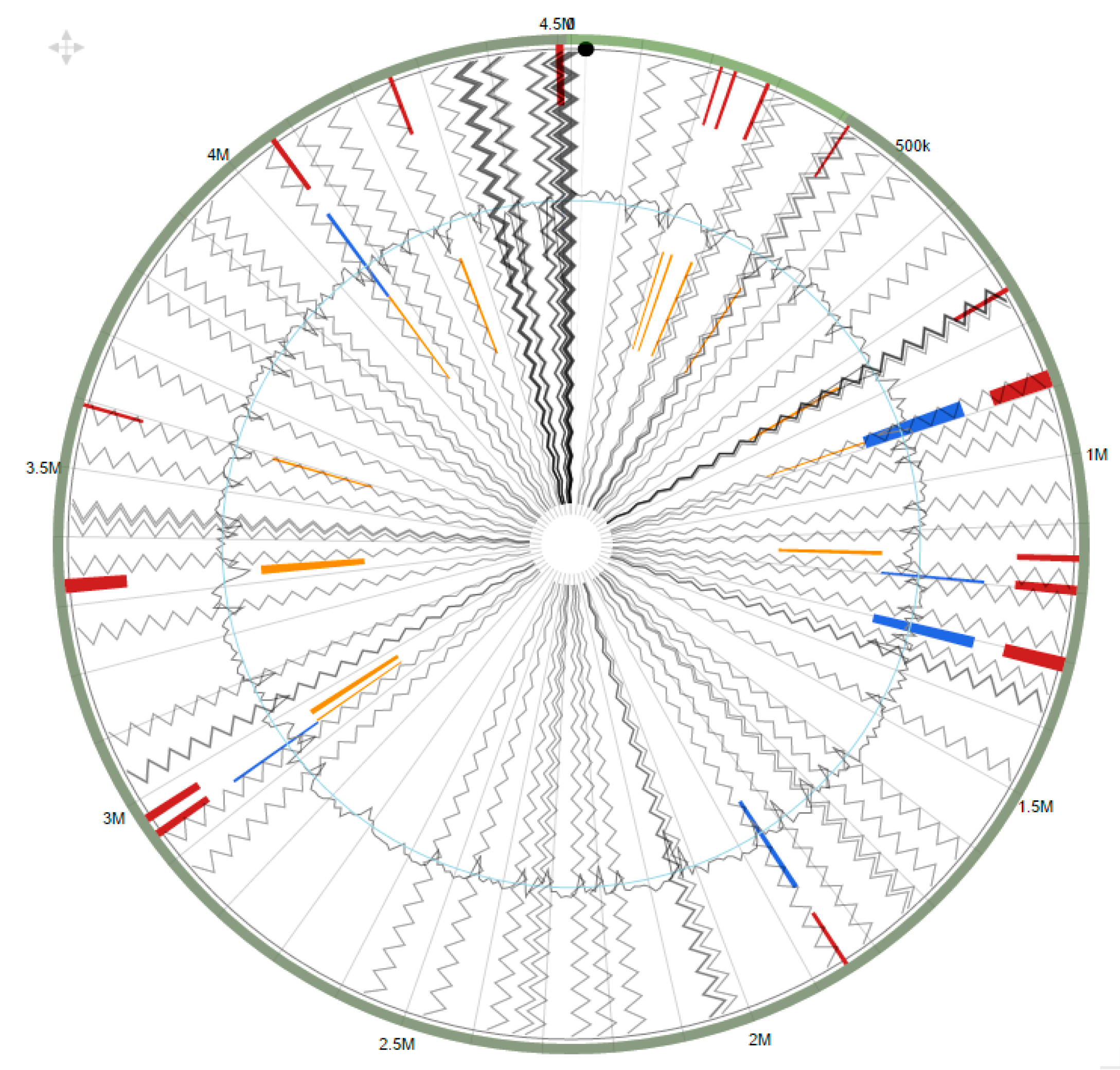
2.6. Genomic Islands in Arthrobacter sp. Strain SRS-W-1-2016
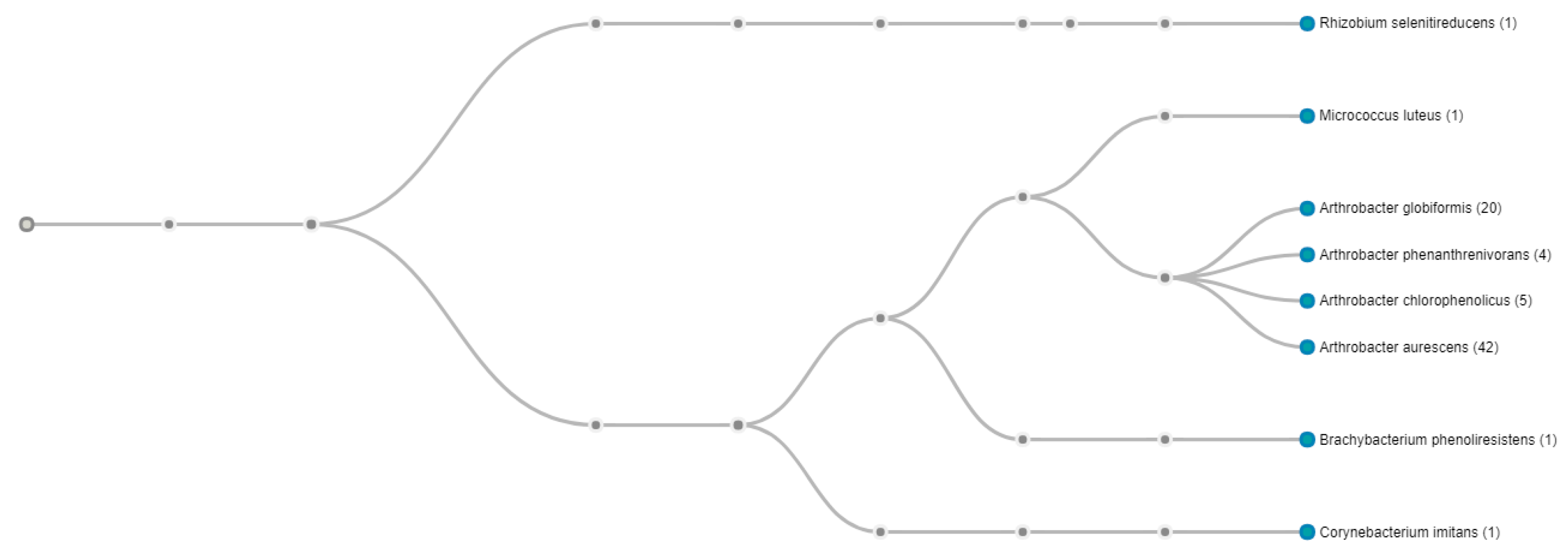
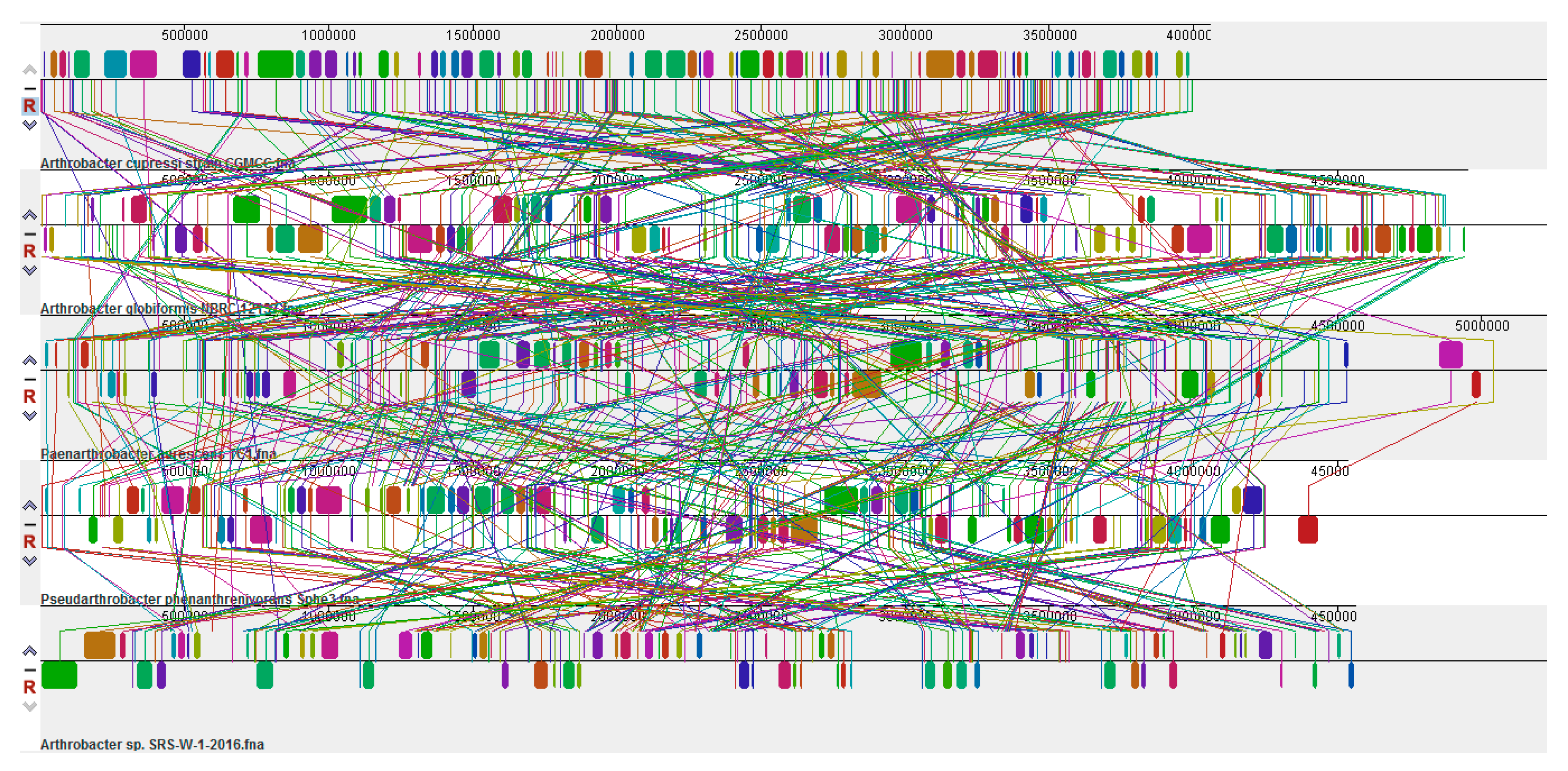
2.7. Genomic Comparison of Arthrobacter sp. Strain SRS-W-1-2016
3. Experimental Section
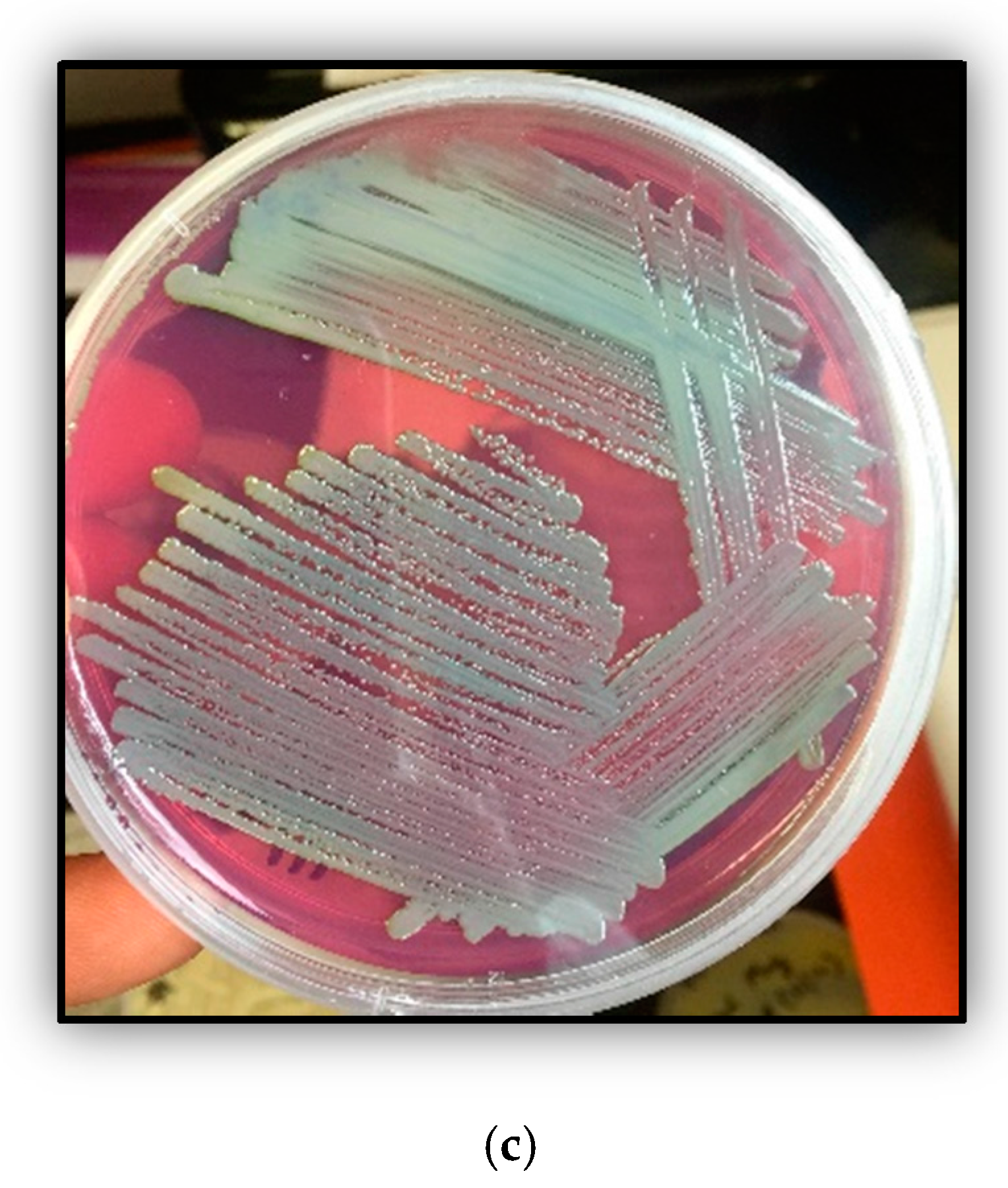
3.1. Isolation and Physiological Growth Studies of Strain SRS-W-1-2016
3.2. Uranium Depletion by Strain SRS-W-1-2016
3.3. Histochemical Screening of Strain SRS-W-1-2016 for Phosphatase Enzyme
3.4. Nucleotide Sequence Accession Number
3.5. Genomic Characterization of Strain SRS-W-1-2016
3.6. Characterization of the Resistome of Strain SRS-W-1-2016
3.7. Comparative Genomics of Strain SRS-W-1-2016
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Riley, R.G.; Zachara, J. Chemical Contaminants on Doe Lands and Selection of Contaminant Mixtures for Subsurface Science Research; Pacific Northwest Lab.: Richland, WA, USA, 1992. [Google Scholar]
- Evans, A.; Bauer, L.; Haselow, J.; Hayes, D.; Martin, H.; McDowell, W.; Pickett, J. Uranium in the Savannah River Site Environment; Westinghouse Savannah River Co.: Aiken, SC, USA, 1992. [Google Scholar]
- Martinez, R.J.; Wu, C.H.; Beazley, M.J.; Andersen, G.L.; Conrad, M.E.; Hazen, T.C.; Taillefert, M.; Sobecky, P.A. Microbial community responses to organophosphate substrate additions in contaminated subsurface sediments. PLoS ONE 2014, 9, e100383. [Google Scholar] [CrossRef] [PubMed]
- Newsome, L.; Morris, K.; Lloyd, J.R. The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem. Geol. 2014, 363, 164–184. [Google Scholar] [CrossRef]
- Ye, Q. Microbial Diversity Associated with Metal- and Radionuclide-Contamination at the Doe Savannah River Site (SRS), South Carolina, USA; University of Georgia: Athens, GA, USA, 2007. [Google Scholar]
- Mirete, S.; De Figueras, C.G.; González-Pastor, J.E. Novel nickel resistance genes from the rhizosphere metagenome of plants adapted to acid mine drainage. Appl. Environ. Microbiol. 2007, 73, 6001–6011. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, S.; Streten-Joyce, C.; Parry, D.L.; McGuinness, K.A.; Lu, P.; Gibb, K.S. Fungi outcompete bacteria under increased uranium concentration in culture media. J. Environ. Radioact. 2013, 120, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Luo, S.; Chen, L.; Xiao, X.; Xi, Q.; Wei, W.; Zeng, G.; Liu, C.; Wan, Y.; Chen, J. Bioremediation of heavy metals by growing hyperaccumulaor endophytic bacterium Bacillus sp. L14. Bioresour. Technol. 2010, 101, 8599–8605. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dash, H.R.; Chakraborty, J. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbiol. Biotechnol. 2016, 100, 2967–2984. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.S. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegrad. 2003, 51, 271–276. [Google Scholar] [CrossRef]
- Perron, K.; Caille, O.; Rossier, C.; van Delden, C.; Dumas, J.-L.; Köhler, T. Czcr-czcs, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 8761–8768. [Google Scholar] [CrossRef] [PubMed]
- Pathak, A.; Chauhan, A.; Stothard, P.; Green, S.; Maienschein-Cline, M.; Jaswal, R.; Seaman, J. Genome-centric evaluation of Burkholderia sp. Strain srs-w-2-2016 resistant to high concentrations of uranium and nickel isolated from the savannah river site (srs), USA. Genom. Data 2017, 12, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Banfield, J.F. Resistance to, and accumulation of, uranium by bacteria from a uranium-contaminated site. Geomicrobiol. J. 2004, 21, 113–121. [Google Scholar] [CrossRef]
- Katsenovich, Y.; Carvajal, D.; Guduru, R.; Lagos, L.; Li, C.-Z. Assessment of the resistance to uranium (vi) exposure by Arthrobacter sp. Isolated from hanford site soil. Geomicrobiol. J. 2013, 30, 120–130. [Google Scholar]
- Fredrickson, J.K.; Zachara, J.M.; Balkwill, D.L.; Kennedy, D.; Li, S.-m.W.; Kostandarithes, H.M.; Daly, M.J.; Romine, M.F.; Brockman, F.J. Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the hanford site, washington state. Appl. Environ. Microbiol. 2004, 70, 4230–4241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Duan, C.; Shao, Q.; Ren, W.; Sha, T.; Chen, L.; Zhao, Z.; Hu, B. Genetic and physiological diversity of phylogenetically and geographically distinct groups of Arthrobacter isolated from lead–zinc mine tailings. FEMS Microbiol. Ecol. 2004, 49, 333–341. [Google Scholar]
- Macur, R.E.; Jackson, C.R.; Botero, L.M.; Mcdermott, T.R.; Inskeep, W.P. Bacterial populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environ. Sci. Technol. 2004, 38, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Swer, P.B.; Joshi, S.R.; Acharya, C. Cesium and strontium tolerant Arthrobacter sp. Strain kmszp6 isolated from a pristine uranium ore deposit. AMB Express 2016, 6, 69. [Google Scholar] [PubMed]
- Van Waasbergen, L.G.; Balkwill, D.L.; Crocker, F.H.; Bjornstad, B.N.; Miller, R.V. Genetic diversity among Arthrobacter species collected across a heterogeneous series of terrestrial deep-subsurface sediments as determined on the basis of 16s rrna and reca gene sequences. Appl. Environ. Microbiol. 2000, 66, 3454–3463. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Nongkhlaw, M.; Acharya, C.; Joshi, S.R. Uranium (u)-tolerant bacterial diversity from u ore deposit of domiasiat in north-east india and its prospective utilisation in bioremediation. Microbes Environ. 2013, 28, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Misra, C.S.; Gupta, A.; Ballal, A.; Apte, S.K. Interaction of uranium with bacterial cell surfaces: Inferences from phosphatase-mediated uranium precipitation. Appl. Environ. Microbiol. 2016, 82, 4965–4974. [Google Scholar] [CrossRef] [PubMed]
- Koribanics, N.M.; Tuorto, S.J.; Lopez-Chiaffarelli, N.; McGuinness, L.R.; Häggblom, M.M.; Williams, K.H.; Long, P.E.; Kerkhof, L.J. Spatial distribution of an uranium-respiring betaproteobacterium at the rifle, co field research site. PLoS ONE 2015, 10, e0123378. [Google Scholar] [CrossRef] [PubMed]
- Alboghobeish, H.; Tahmourespour, A.; Doudi, M. The study of nickel resistant bacteria (nirb) isolated from wastewaters polluted with different industrial sources. J. Environ. Health Sci. Eng. 2014, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-M.; Wu, C.-H.; James, E.K.; Chang, J.-S. Metal biosorption capability of Cupriavidus taiwanensis and its effects on heavy metal removal by nodulated mimosa pudica. J. Hazard. Mater. 2008, 151, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Osman, S.; Peeters, Z.; La Duc, M.T.; Mancinelli, R.; Ehrenfreund, P.; Venkateswaran, K. Effect of shadowing on survival of bacteria under conditions simulating the martian atmosphere and uv radiation. Appl. Environ. Microbiol. 2008, 74, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Mongodin, E.F.; Shapir, N.; Daugherty, S.C.; DeBoy, R.T.; Emerson, J.B.; Shvartzbeyn, A.; Radune, D.; Vamathevan, J.; Riggs, F.; Grinberg, V. Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens tc1. PLoS Genet. 2006, 2, e214. [Google Scholar] [CrossRef] [PubMed]
- Camargo, F.; Bento, F.; Okeke, B.; Frankenberger, W. Hexavalent chromium reduction by an actinomycete, Arthrobacter crystallopoietes es 32. Biol. Trace Elem. Res. 2004, 97, 183–194. [Google Scholar] [CrossRef]
- Westerberg, K.; Elväng, A.M.; Stackebrandt, E.; Jansson, J.K. Arthrobacter chlorophenolicus sp. Nov., a new species capable of degrading high concentrations of 4-chlorophenol. Int. J. Syst. Evol. Microbiol. 2000, 50, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- O’loughlin, E.J.; Sims, G.K.; Traina, S.J. Biodegradation of 2-methyl, 2-ethyl, and 2-hydroxypyridine by an Arthrobacter sp. Isolated from subsurface sediment. Biodegradation 1999, 10, 93–104. [Google Scholar] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The rast server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Blom, J.; Albaum, S.P.; Doppmeier, D.; Pühler, A.; Vorhölter, F.-J.; Zakrzewski, M.; Goesmann, A. Edgar: A software framework for the comparative analysis of prokaryotic genomes. BMC Bioinform. 2009, 10, 154. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Weimann, A.; Mooren, K.; Frank, J.; Pope, P.B.; Bremges, A.; McHardy, A.C. From genomes to phenotypes: Traitar, the microbial trait analyzer. mSystems 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Yung, M.C.; Park, D.M.; Overton, K.W.; Blow, M.J.; Hoover, C.A.; Smit, J.; Murray, S.R.; Ricci, D.P.; Christen, B.; Bowman, G.R. Transposon mutagenesis paired with deep sequencing of Caulobacter crescentus under uranium stress reveals genes essential for detoxification and stress tolerance. J. Bacteriol. 2015, 197, 3160–3172. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Moreno, Z.R.; Caballero-Mellado, J.; Coutinho, B.G.; Mendonça-Previato, L.; James, E.K.; Venturi, V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012, 63, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Marx, R.B.; Aitken, M.D. Bacterial chemotaxis enhances naphthalene degradation in a heterogeneous aqueous system. Environ. Sci. Technol. 2000, 34, 3379–3383. [Google Scholar] [CrossRef]
- Perez-Pantoja, D.; Nikel, P.I.; Chavarría, M.; de Lorenzo, V. Endogenous stress caused by faulty oxidation reactions fosters evolution of 2, 4-dinitrotoluene-degrading bacteria. PLoS Genet. 2013, 9, e1003764. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, J.D.; Khijniak, T.J.; Neely, B.; Sattar, M.A.; Sowder, A.G.; Mills, G.; Bertsch, P.M.; Morris, P.J. Reduction of nickel and uranium toxicity and enhanced trichloroethylene degradation to Burkholderia vietnamiensis pr1301 with hydroxyapatite amendment. Environ. Sci. Technol. 2007, 41, 1877–1882. [Google Scholar] [CrossRef] [PubMed]
- Riccio, M.; Rossolini, G.; Lombardi, G.; Chiesurin, A.; Satta, G. Expression cloning of different bacterial phosphatase encoding genes by histochemical screening of genomic libraries onto an indicator medium containing phenolphthalein diphosphate and methyl green. J. Appl. Microbiol. 1997, 82, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Moat, A.G.; Foster, J.W.; Spector, M.P. Nitrogen metabolism. Microb. Physiol. Fourth Ed. 1995, 475–502. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Arantes, A.S.; Stothard, P. Comparing thousands of circular genomes using the cgview comparison tool. BMC Genom. 2012, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.-M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P. Img: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2011, 40, D115–D122. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. Ncbi prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-M.A.; Markowitz, V.M.; Palaniappan, K.; Szeto, E.; Chu, K.; Huang, J.; Ratner, A.; Pillay, M.; Hadjithomas, M.; Huntemann, M. Supporting community annotation and user collaboration in the integrated microbial genomes (img) system. BMC Genom. 2016, 17, 307. [Google Scholar] [CrossRef] [PubMed]
- Minot, S.S.; Krumm, N.; Greenfield, N.B. One codex: A sensitive and accurate data platform for genomic microbial identification. BioRxiv 2015. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N. Card 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Hadjithomas, M.; Chen, I.-M.A.; Chu, K.; Ratner, A.; Palaniappan, K.; Szeto, E.; Huang, J.; Reddy, T.; Cimermančič, P.; Fischbach, M.A. Img-abc: A knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. MBio 2015, 6, e00932-15. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Duran, S.; Hernando, G.; de los Santos, E.L.; Kim, H.U. Antismash 4.0—Improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef]
- Juhas, M.; van der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servín-Garcidueñas, L.E.; Rogel, M.A.; Ormeño-Orrillo, E.; Zayas-del Moral, A.; Sánchez, F.; Martínez-Romero, E. Complete genome sequence of Bradyrhizobium sp. Strain ccge-la001, isolated from field nodules of the enigmatic wild bean phaseolus microcarpus. Genome Announc. 2016, 4, e00126-16. [Google Scholar]
- Auch, A.F.; Klenk, H.-P.; Göker, M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genom. Sci. 2010, 2, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-W.; Wang, J.-T.; Li, J.; Shi, X.-Z.; Ma, Y.-B.; Chen, D.; He, J.-Z. Long-term nickel contamination increases the occurrence of antibiotic resistance genes in agricultural soils. Environ. Sci. Technol. 2016, 51, 790–800. [Google Scholar] [CrossRef] [PubMed]

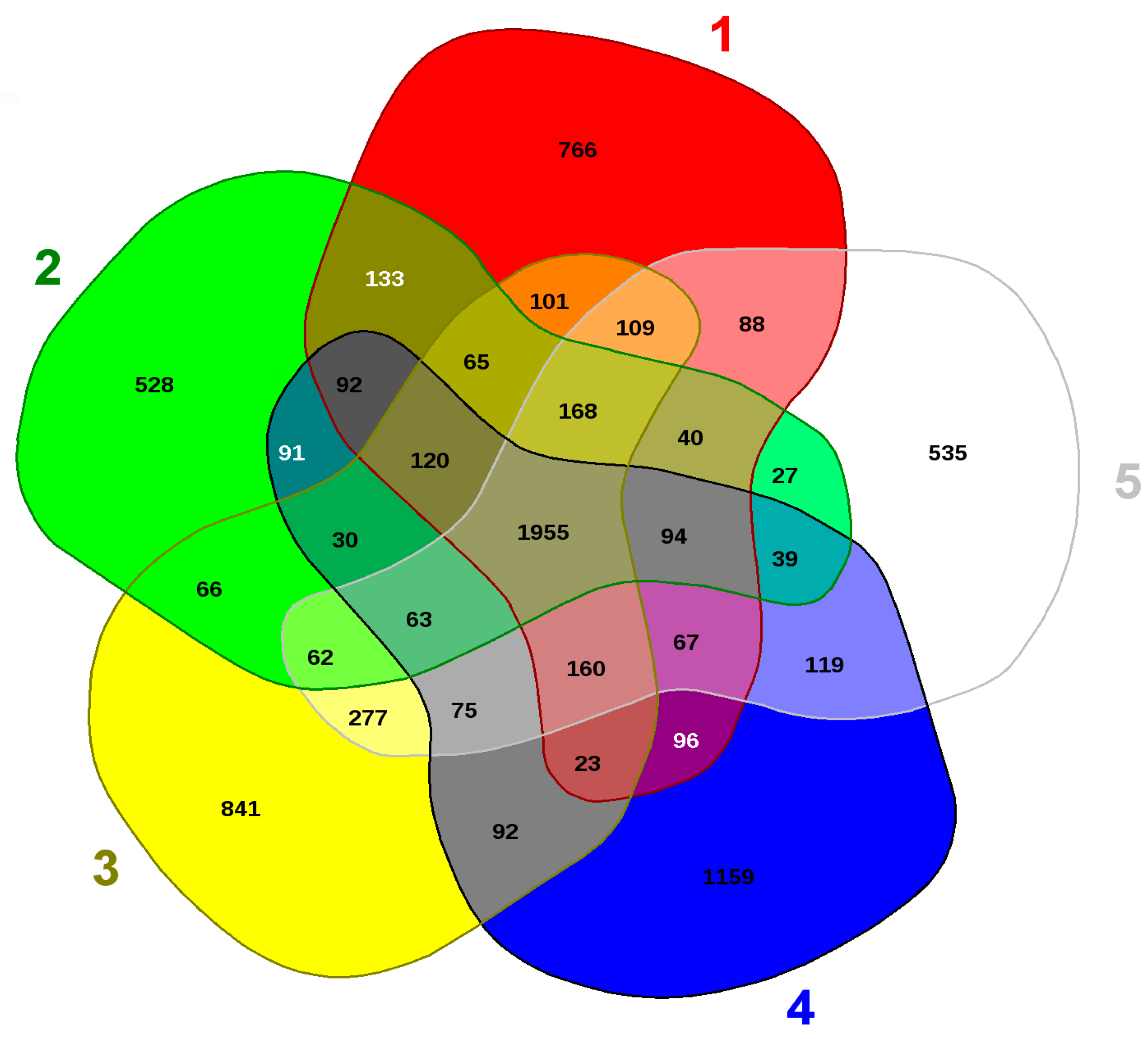
| KEGG Category | Percentage (%) |
|---|---|
| Global and Overview Maps | 63.08 |
| Carbohydrate Metabolism | 25.27 |
| Amino Acid Metabolism | 22.78 |
| Energy Metabolism | 12.01 |
| Metabolism of Cofactors and Vitamins | 11.39 |
| Membrane Transport | 9.34 |
| Nucleotide Metabolism | 8.72 |
| Lipid Metabolism | 7.56 |
| Translation | 7.21 |
| Xenobiotics Biodegradation and Metabolism | 6.32 |
| Signal Transduction | 5.87 |
| Metabolism of other Amino Acids | 4.8 |
| Replication and Repair | 4.54 |
| Folding, Sorting and Degradation | 4.18 |
| Metabolism of Terpenoids and Polyketides | 3.91 |
| Biosynthesis of other Secondary Metabolites | 3.2 |
| Glycan Biosynthesis and Metabolism | 2.85 |
| Cell Motility | 2.67 |
| Cell Growth and Death | 1.42 |
| Drug Resistance | 1.33 |
| Transport and Catabolism | 0.98 |
| Transcription | 0.44 |
| Environmental Adaptation | 0.18 |
| Excretory System | 0.18 |
| Category | Gene Homologue |
|---|---|
| Transporter proteins | Low-affinity inorganic phosphate transporter |
| H(+)/Cl(-) exchange transporter ClcA | |
| Putative inner membrane transporter YhbE | |
| Divalent metal cation transporter MntH | |
| Stress proteins | General stress protein 14 |
| Universal stress protein/MT2085 | |
| Cytochromes | Cytochrome P450 107B |
| Cytochrome bc complex cytochrome b subunit | |
| Metal resistance proteins | Cadmim, cobalt and zinc/H(+)-K(+) antiporter |
| Arsenical resistance operon repressor | |
| Copper resistance protein A precursor | |
| Copper chaperone CopZ | |
| Drug resistance | Multidrug resistance operon repressor |
| Multidrug resistance protein |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chauhan, A.; Pathak, A.; Jaswal, R.; Edwards III, B.; Chappell, D.; Ball, C.; Garcia-Sillas, R.; Stothard, P.; Seaman, J. Physiological and Comparative Genomic Analysis of Arthrobacter sp. SRS-W-1-2016 Provides Insights on Niche Adaptation for Survival in Uraniferous Soils. Genes 2018, 9, 31. https://doi.org/10.3390/genes9010031
Chauhan A, Pathak A, Jaswal R, Edwards III B, Chappell D, Ball C, Garcia-Sillas R, Stothard P, Seaman J. Physiological and Comparative Genomic Analysis of Arthrobacter sp. SRS-W-1-2016 Provides Insights on Niche Adaptation for Survival in Uraniferous Soils. Genes. 2018; 9(1):31. https://doi.org/10.3390/genes9010031
Chicago/Turabian StyleChauhan, Ashvini, Ashish Pathak, Rajneesh Jaswal, Bobby Edwards III, Demario Chappell, Christopher Ball, Reyna Garcia-Sillas, Paul Stothard, and John Seaman. 2018. "Physiological and Comparative Genomic Analysis of Arthrobacter sp. SRS-W-1-2016 Provides Insights on Niche Adaptation for Survival in Uraniferous Soils" Genes 9, no. 1: 31. https://doi.org/10.3390/genes9010031






