Genome-Wide Methylation Patterns in Androgen-Independent Prostate Cancer Cells: A Comprehensive Analysis Combining MeDIP-Bisulfite, RNA, and microRNA Sequencing Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. MeDIP-bisulfite Sequencing
2.3. Raw Data Preprocessing and Reads Mapping
2.4. Differentially Methylated Regions Screening
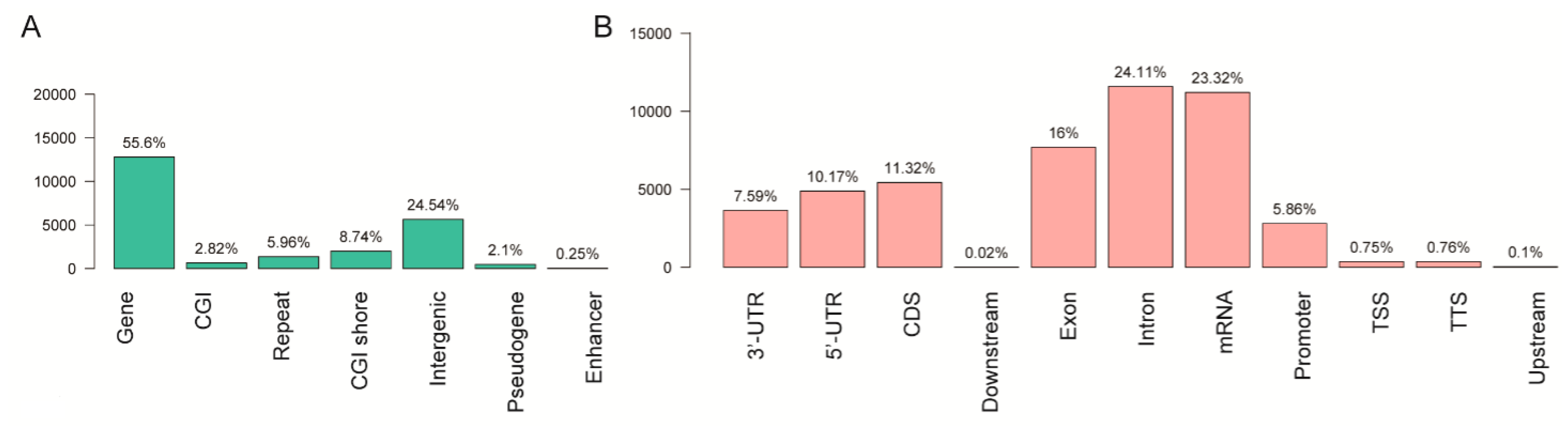
2.5. Annotation and Distribution of Differentially Methylated Regions
2.6. Analysis of Transcription Factor-Binding Motifs in Differentially Methylated Regions -Promoter-Overlapping Regions
2.7. Comprehensive Analysis of Differentially Methylated Regions and Transcriptome
2.8. Pathway and Functional Enrichment Analyses of Differentially Expressed Genes with DMRs (MDEGs)
2.9. Construction of Transcription Factor-Target Network
2.10. Construction of micro RNA–Target Network
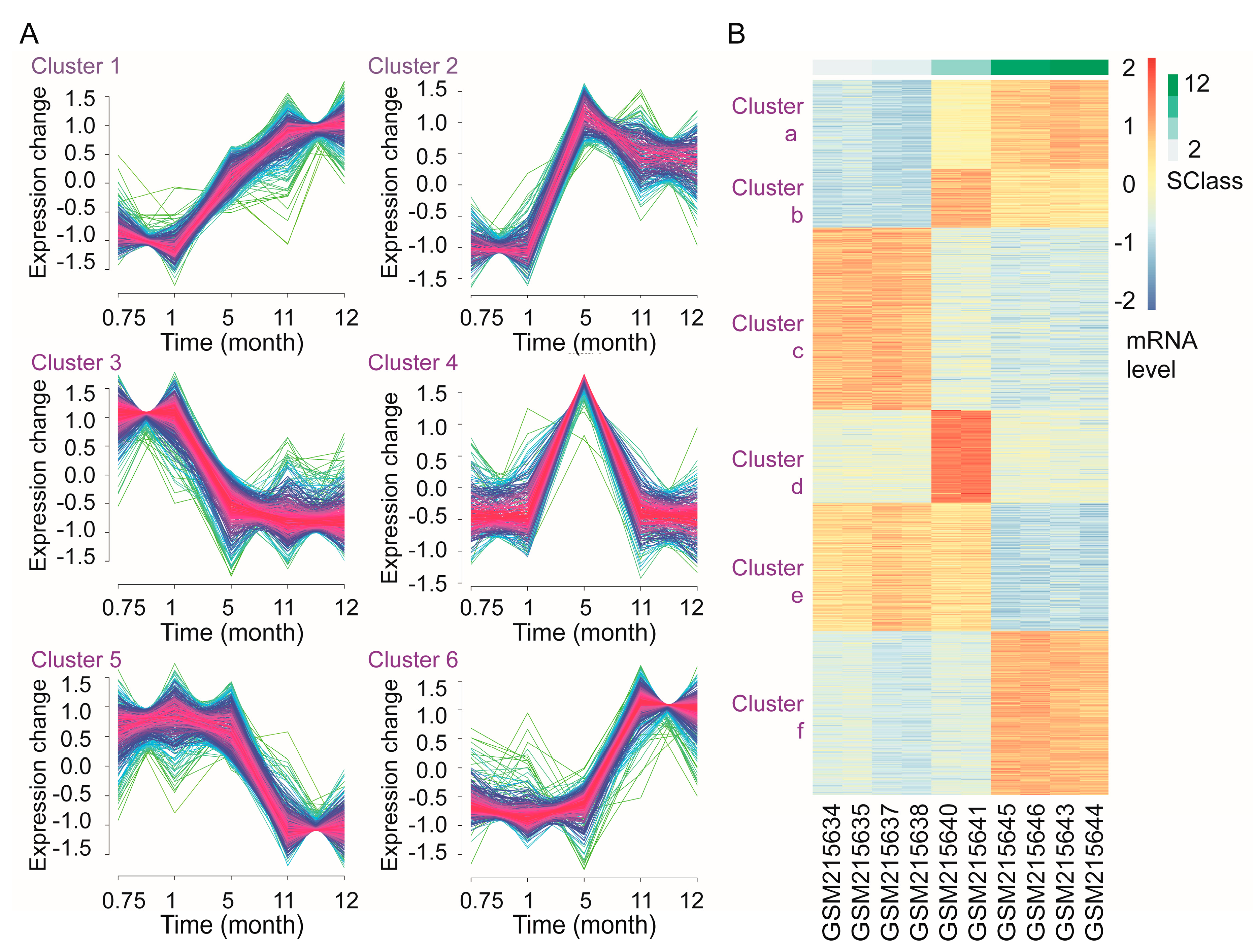
2.11. Time-Course Analysis of Gene Transcription during Androgen Deprivation
2.12. Comprehensive Differentially Expressed Genes Analysis of Sequencing Data and Microarray Data
2.13. Data Validation of the Identified Differentially Methylated Regions and Differentially Expressed micro RNAs
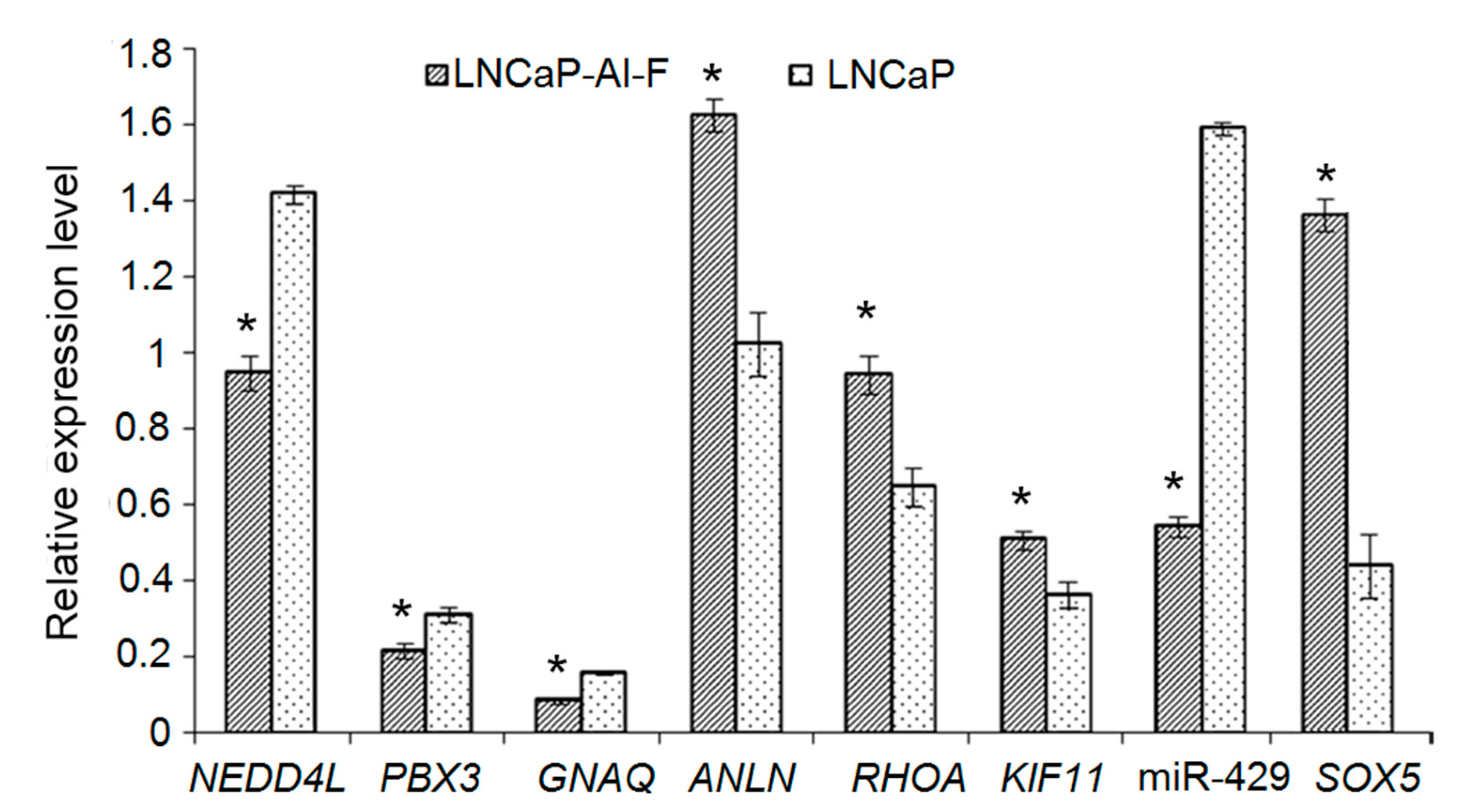
2.14. Quantitative Reverse Transcription Polymerase Chain Reaction Assay
2.15. Statistical Method
3. Results
3.1. Overview of the MeDIP-bisulfite Sequencing Data
3.2. Annotation and Distribution of Differentially Methylated Regions
3.3. Transcription Factor-Binding Motifs in Differentially Methylated Regions -Promoter-Overlapping Regions
3.4. Comprehensive Analysis of Differentially Methylated Regions and Transcriptome Data
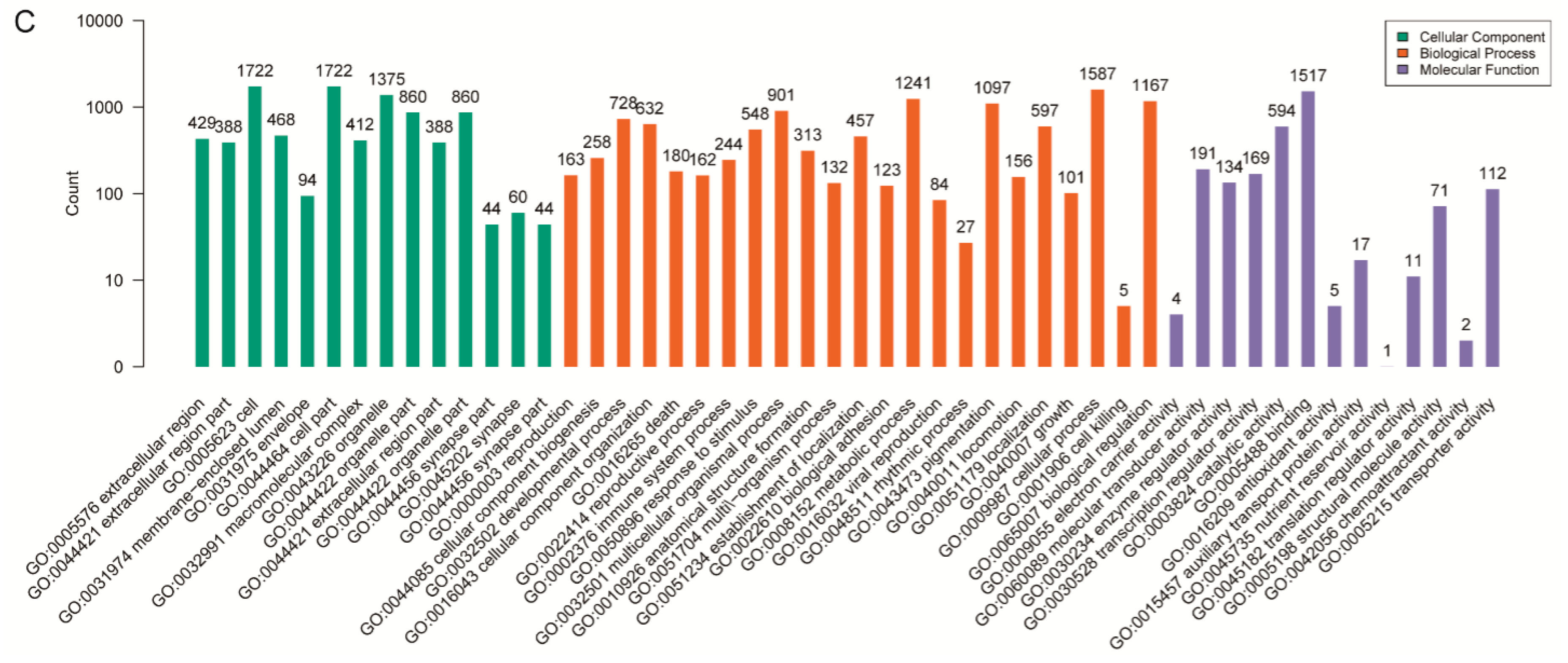
3.5. Pathway and Functional Enrichment Analyses of Differentially Expressed Genes with DMRs
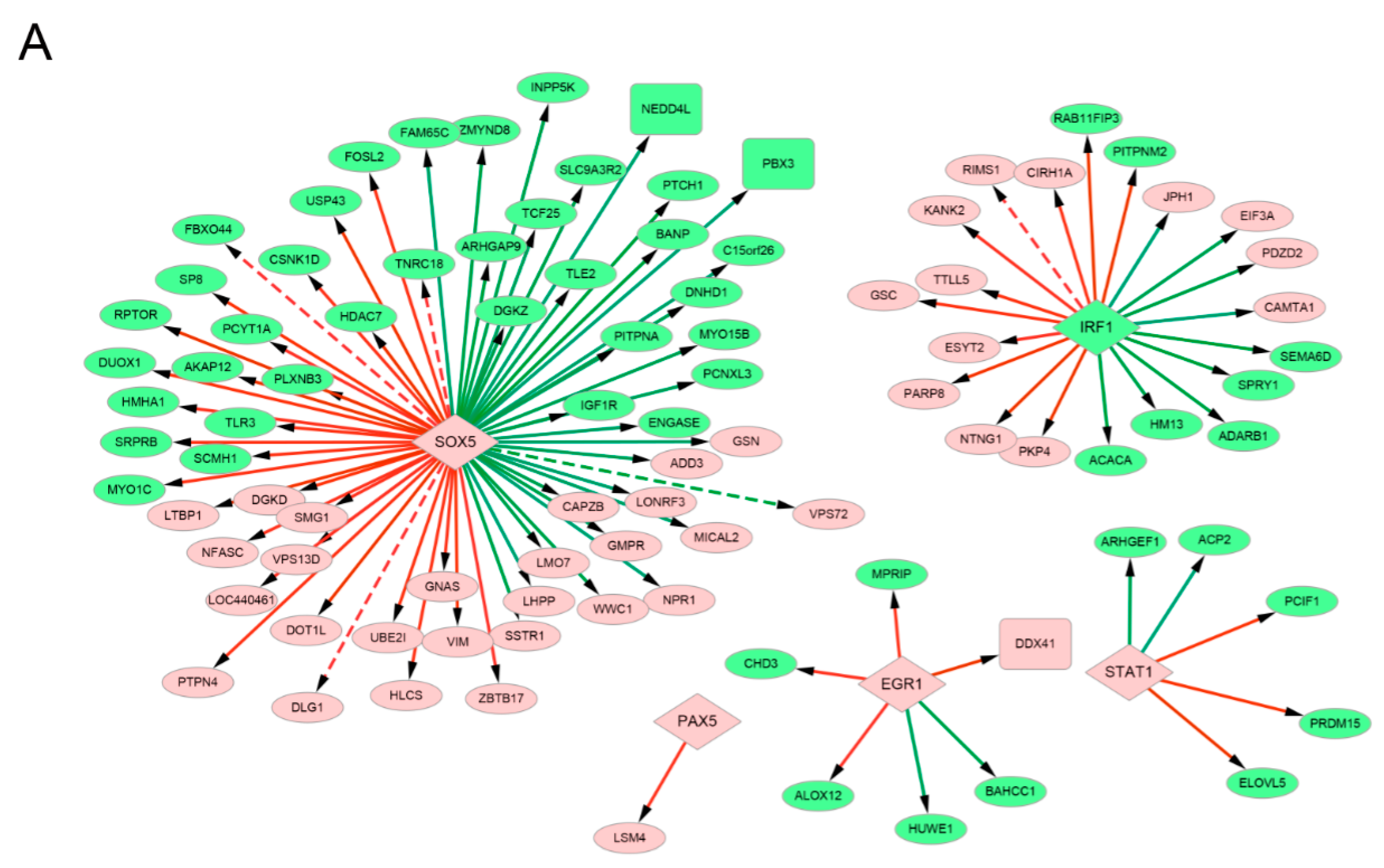
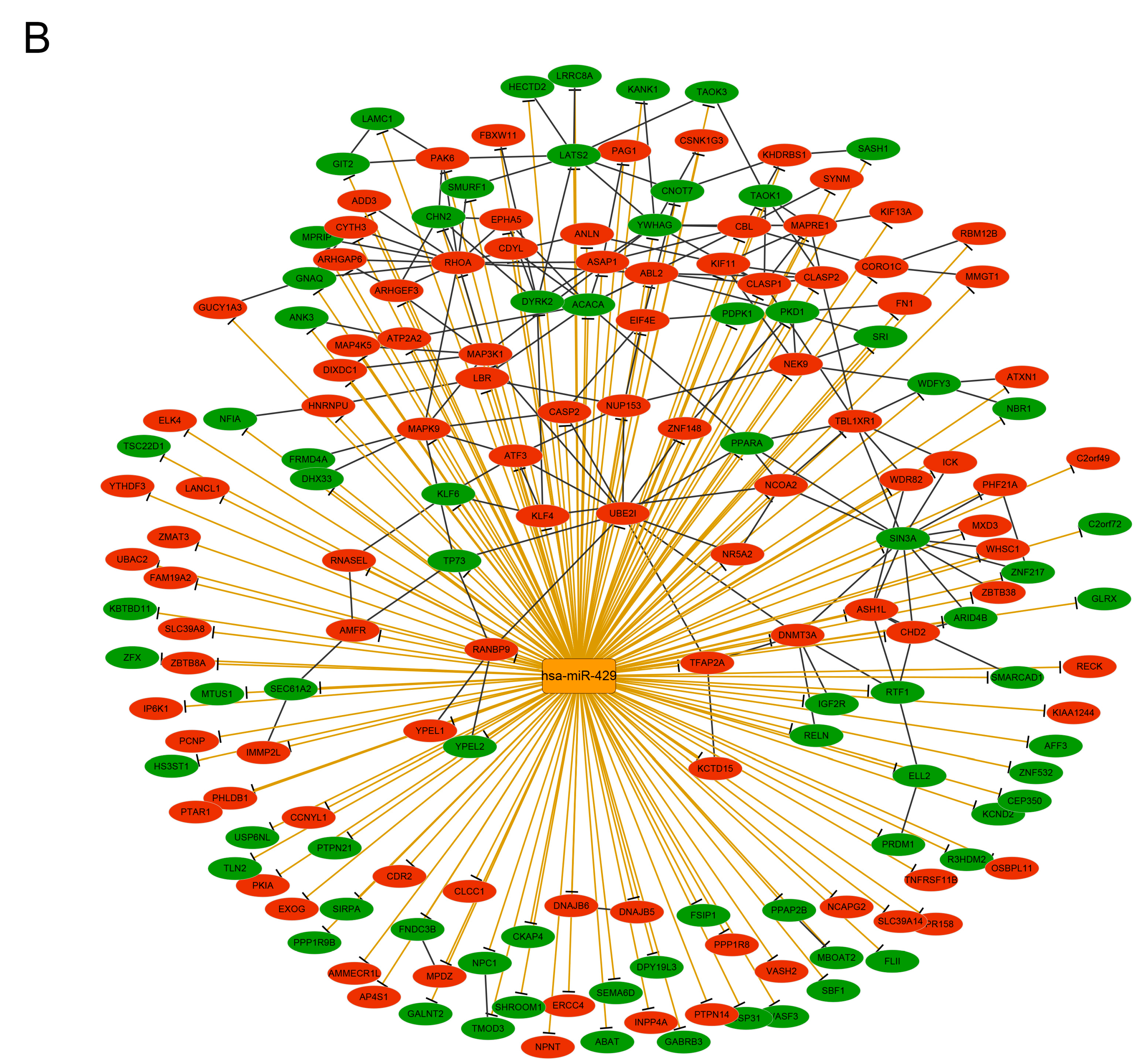
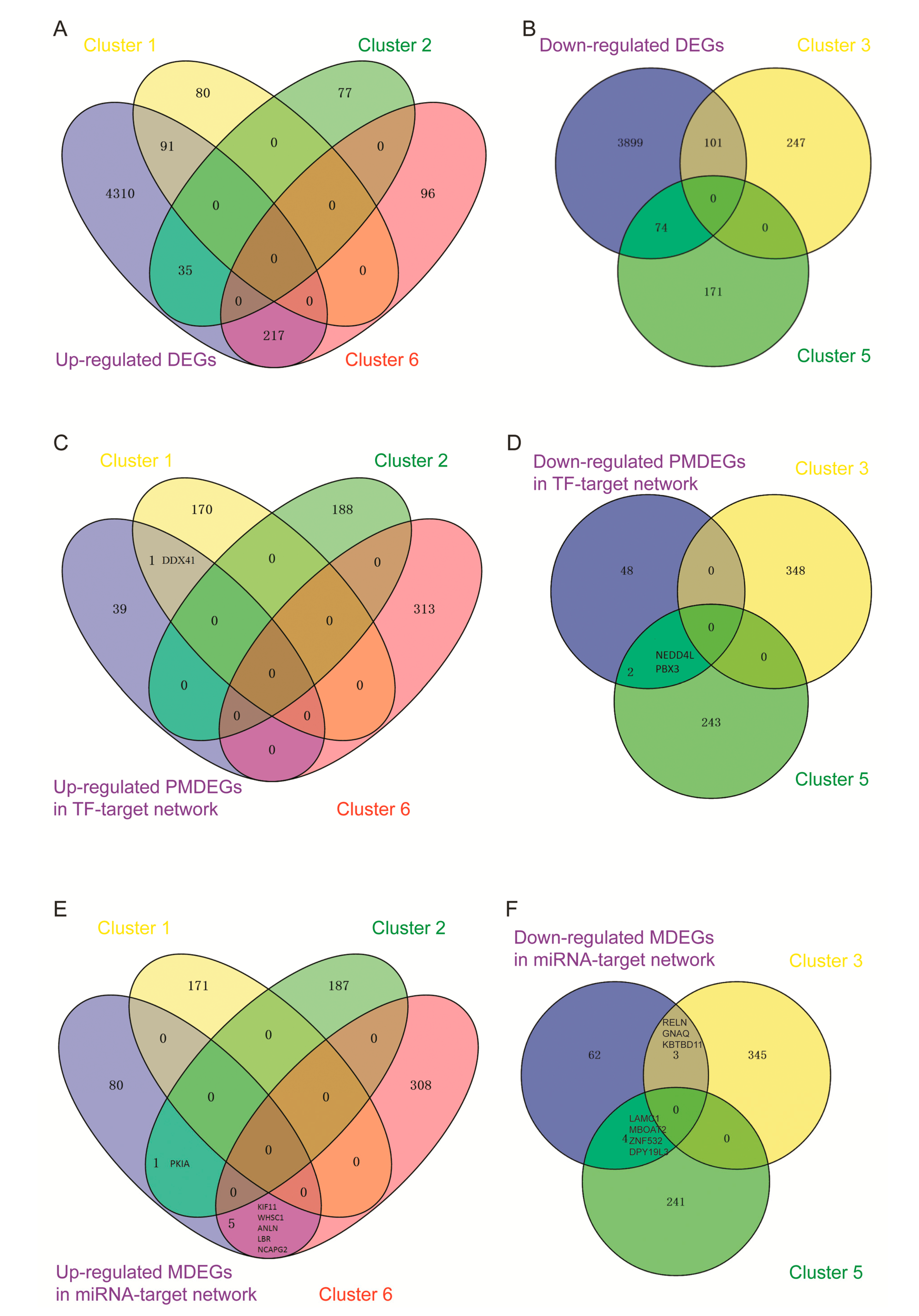
3.6. Analysis of the Transcription Factor-Target and micro RNA–Target Networks
3.7. Time-Course Analysis of Gene Transcription during Androgen Deprivation
3.8. Comprehensive Differentially Expressed Genes Analysis of Sequencing Data and Microarray Data
3.9. Validated DMRs and Differentially Expressed micro RNAs in Public Datasets
3.10. Validation of Differentially Expressed Genes with DMRs and Differentially Expressed micro RNAs by Quantitative Real-Time Polymerase Chain Reaction
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- De La Taille, A.; Vacherot, F.; Salomon, L.; Druel, C.; De Medina, S.G.D.; Abbou, C.; Buttyan, R.; Chopin, D. Hormone-refractory prostate cancer: A multi-step and multi-event process. Prostate Cancer Prostatic Dis. 2001, 4, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Kahn, B.; Collazo, J.; Kyprianou, N. Androgen receptor as a driver of therapeutic resistance in advanced prostate cancer. Int. J. Biol. Sci. 2014, 10, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Karantanos, T.; Evans, C.P.; Tombal, B.; Thompson, T.C.; Montironi, R.; Isaacs, W.B. Understanding the mechanisms of androgen deprivation resistance in prostate cancer at the molecular level. Eur. Urol. 2015, 67, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.G.; Yegnasubramanian, S. Resistance emerges to second-generation antiandrogens in prostate cancer. Cancer Discov. 2013, 3, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Amaral, T.M.S.; Macedo, D.; Fernandes, I.; Costa, L. Castration-resistant prostate cancer: Mechanisms, targets, and treatment. Prostate Cancer 2012, 2012, 327253. [Google Scholar] [CrossRef] [PubMed]
- Sonpavde, G.; Matveev, V.; Burke, J.; Caton, J.R.; Fleming, M.T.; Hutson, T.E.; Galsky, M.D.; Berry, W.R.; Karlov, P.; Holmlund, J.T.; et al. Randomized phase II trial of docetaxel plus prednisone in combination with placebo or AT-101, an oral small molecule Bcl-2 family antagonist, as first-line therapy for metastatic castration-resistant prostate cancer. Ann. Oncol. 2012, 23, 1803–1808. [Google Scholar] [CrossRef] [PubMed]
- Arora, V.K.; Schenkein, E.; Murali, R.; Subudhi, S.K.; Wongvipat, J.; Balbas, M.D.; Shah, N.; Cai, L.; Efstathiou, E.; Logothetis, C.; et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013, 155, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Q.; Xu, G.; Mao, F.; Qin, T.; Teng, H.; Cai, W.; Yu, P.; Cai, T.; Zhao, M.; et al. Comparative RNA-Seq analysis reveals potential mechanisms mediating the conversion to androgen independence in an LNCaP progression cell model. Cancer Lett. 2014, 342, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Durojaiye, V.; Ilboudo, A.; Levine, F.; Osborne, J.; Park, J.Y.; Ogunwobi, O.O. miR-1205/FRYL as a novel regulatory mechanism in androgen-independent prostate cancer. Cancer Res. 2015, 75 (Suppl. 15). [Google Scholar] [CrossRef]
- Yang, K.; Handorean, A.M.; Iczkowski, K.A. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int. J. Clin. Exp. Pathol. 2009, 2, 361–369. [Google Scholar] [PubMed]
- Ribas, J.; Ni, X.; Haffner, M.; Wentzel, E.A.; Salmasi, A.H.; Chowdhury, W.H.; Kudrolli, T.A.; Yegnasubramanian, S.; Luo, J.; Rodriguez, R.; et al. miR-21: An androgen receptor–regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009, 69, 7165–7169. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wu, J.; Zhou, L.; Chen, B.; Sun, Z.; Zhao, F.; Tao, Z. Characterization of the small RNA transcriptomes of androgen dependent and independent prostate cancer cell line by deep sequencing. PLoS ONE 2010, 5, e15519. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, T.W.; Roy, R.; Tomlins, S.A.; Ngo, V.T.; Kobayashi, Y.; Azameera, A.; Rubin, M.A.; Pienta, K.J.; Chinnaiyan, A.; Ittmann, M.M.; et al. Common structural and epigenetic changes in the genome of castration-resistant prostate cancer. Cancer Res. 2012, 72, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.K.; Chen, Z.; Wu, Y.; Sarkissyan, M.; Koeffler, H.P.; Vadgama, J.V. Global methylation pattern of genes in androgen-sensitive and androgen-independent prostate cancer cells. Mol. Cancer Ther. 2010, 9, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Taiwo, O.; Wilson, G.A.; Morris, T.; Seisenberger, S.; Reik, W.; Pearce, D.; Beck, S.; Butcher, L.M. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat. Protoc. 2012, 7, 617–636. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.A.; Wang, T.; Coarfa, C.; Nagarajan, R.P.; Hong, C.; Downey, S.L.; Johnson, B.E.; Fouse, S.D.; Delaney, A.; Zhao, Y.; et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010, 28, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Mao, F.; Teng, H.; Cai, T.; Zhao, F.; Wu, J.; Sun, Z.S. MBRidge: An accurate and cost-effective method for profiling DNA methylome at single-base resolution. J. Mol. Cell Biol. 2015, 7, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Advani, A.; Melefors, O.; Glavas, S.; Nordström, H.; Ye, W.; Engstrand, L.; Andersson, A.F. Titration-free 454 sequencing using Y adapters. Nat. Protoc. 2011, 6, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Ruike, Y.; Imanaka, Y.; Sato, F.; Shimizu, K.; Tsujimoto, G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genom. 2010, 11, 137. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Davies, J.J.; Wittig, D.; Oakeley, E.J.; Haase, M.; Lam, W.L.; Schuebeler, D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005, 37, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 2000, 25, 60–83. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Mathelier, A.; Zhao, X.; Zhang, A.W.; Parcy, F.; Worsley-Hunt, R.; Arenillas, D.J.; Buchman, S.; Chen, C.; Chou, A.; Ienasescu, H.; et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014, 42, D142–D147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tansley, D.S. Linux and Unix Shell Programming; Addison-Wesley Professional: Boston, MA, USA, 2000. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Jupe, S.; Akkerman, J.W.; Soranzo, N.; Ouwehand, W.H. Reactome-a curated knowledgebase of biological pathways: Megakaryocytes and platelets. J. Thromb. Haemost. 2012, 10, 2399–2402. [Google Scholar] [CrossRef] [PubMed]
- Kelder, T.; van Iersel, M.P.; Hanspers, K.; Kutmon, M.; Conklin, B.R.; Evelo, C.T.; Pico, A.R. WikiPathways: Building research communities on biological pathways. Nucleic Acids Res. 2012, 40, D1301–D1307. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The pathway interaction database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Gene ontology annotations and resources. Nucleic Acids Res. 2013, 41, D530–D535. [Google Scholar]
- Shannon, P.; Richards, M. MotifDb: An Annotated Collection of Protein-DNA Binding Sequence Motifs. R package version 1.2. 2014. Available online: http://bioconductor.org/packages/release/bioc/html/MotifDb.html (accessed on 30 November 2017).
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Xiao, W.; Leek, J.T.; Tompkins, R.G.; Davis, R.W. Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. USA 2005, 102, 12837–12842. [Google Scholar] [CrossRef] [PubMed]
- D’Antonio, J.M.; Ma, C.; Monzon, F.A.; Pflug, B.R. Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate 2008, 68, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Futschik, M.E.; Carlisle, B. Noise-robust soft clustering of gene expression time-course data. J. Bioinform. Comput. Biol. 2005, 3, 965–988. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap: Pretty Heatmaps. R package version 0.7.7. Available online: https://cran.r-project.org/src/contrib/Archive/pheatmap/ (accessed on 30 November 2017).
- Oliveros, J.C. VENNY—An Interactive Tool for Comparing Lists with Venn Diagrams. 2007. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 30 November 2017).
- Lin, P.C.; Giannopoulou, E.G.; Park, K.; Mosquera, J.M.; Sboner, A.; Tewari, A.K.; Garraway, L.A.; Beltran, H.; Rubin, M.A.; Elemento, O.; et al. Epigenomic alterations in localized and advanced prostate cancer. Neoplasia 2013, 15, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Grenier, J.; Fournier, A.; Labrie, C. Androgens differentially regulate the expression of NEDD4L transcripts in LNCaP human prostate cancer cells. Mol. Cell. Endocrinol. 2003, 210, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Pang, L.; Ren, C.; Ma, T. Decreased expression of Nedd4L correlates with poor prognosis in gastric cancer patient. Med. Oncol. 2012, 29, 1733–1738. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, J.P.; Chen, X.; Coffey, R.J. NEDD4L is downregulated in colorectal cancer and inhibits canonical WNT signaling. PLoS ONE 2013, 8, e81514. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.-H.; Han, C.; Srivastava, A.K.; Cui, T.; Zou, N.; Gao, Z.-Q.; Wang, Q.E. MiR-93 promotes TGF-β-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumor Biol. 2016, 37, 5645–5651. [Google Scholar] [CrossRef] [PubMed]
- Ramberg, H.; Alshbib, A.; Berge, V.; Svindland, A.; Tasken, K.A. Regulation of PBX3 expression by androgen and Let-7d in prostate cancer. Mol. Cancer 2011, 10, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Sun, Z.; Zhu, Z.; Zhang, J.; Sun, X.; Xu, H. PBX3 is overexpressed in gastric cancer and regulates cell proliferation. Tumor Biol. 2014, 35, 4363–4368. [Google Scholar] [CrossRef] [PubMed]
- Han, H.-B.; Gu, J.; Ji, D.-B.; Li, Z.-W.; Zhang, Y.; Zhao, W.; Wang, L.M.; Zhang, Z.Q. PBX3 promotes migration and invasion of colorectal cancer cells via activation of MAPK/ERK signaling pathway. World J. Gastroenterol. 2014, 20, 18260. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chan, Y.P.; Woolcock, B.; Hu, L.; Wong, K.Y.; Ling, M.T.; Bainbridge, T.; Webber, D.; Chan, T.H.M.; Guan, X.Y.; et al. DNA fingerprinting tags novel altered chromosomal regions and identifies the involvement of SOX5 in the progression of prostate cancer. Int. J. Cancer 2009, 124, 2323–2332. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Y.; Lin, Y.T.; Jan, P.S.; Hwang, Y.C.; Liang, S.T.; Peng, Y.; Huang, C.Y.; Wu, H.C.; Lin, C.T. Transcription factor SOX-5 enhances nasopharyngeal carcinoma progression by down-regulating SPARC gene expression. J. Pathol. 2008, 214, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Han, S.; Wang, X.; Peng, R.; Li, X. SOX5 promotes epithelial-mesenchymal transition and cell invasion via regulation of Twist1 in hepatocellular carcinoma. Med. Oncol. 2015, 32, 461. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; McKnight, N.C.; Zhang, T.; Lu, M.L.; Balk, S.P.; Yuan, X. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 2007, 67, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Leav, I.; Ibaragi, S.; Wegner, M.; Hu, G.F.; Lu, M.L.; Balk, S.P.; Yuan, X. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 2008, 68, 1625–1630. [Google Scholar] [CrossRef] [PubMed]
- Murugan, A.K.; Dong, J.; Xie, J.; Xing, M. Uncommon GNAQ, MMP8, AKT3, EGFR, and PIK3R1 mutations in thyroid cancers. Endocr. Pathol. 2011, 22, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Degese, M.S.; Iglesias-Bartolome, R.; Vaque, J.P.; Molinolo, A.A.; Rodrigues, M.; Zaidi, M.R.; Ksander, B.R.; Merlino, G.; Sodhi, A.; et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 2014, 25, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, C.; Daigo, Y.; Ishikawa, N.; Kato, T.; Hayama, S.; Ito, T.; Tsuchiya, E.; Nakamura, Y. ANLN plays a critical role in human lung carcinogenesis through the activation of RHOA and by involvement in the phosphoinositide 3-kinase/AKT pathway. Cancer Res. 2005, 65, 11314–11325. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, Z.; Shen, N.; Pi, W.; Jiang, W.; Huang, J.; Hu, Y.; Li, X.; Sun, L. Knockdown of ANLN by lentivirus inhibits cell growth and migration in human breast cancer. Mol. Cell. Biochem. 2015, 398, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Furihata, M.; Tsunoda, T.; Ashida, S.; Takata, R.; Obara, W.; Yoshioka, H.; Daigo, Y.; Nasu, Y.; Kumon, H.; et al. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007, 67, 5117–5125. [Google Scholar] [CrossRef] [PubMed]
- Kamai, T.; Yamanishi, T.; Shirataki, H.; Takagi, K.; Asami, H.; Ito, Y.; Yoshida, K.I. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin. Cancer Res. 2004, 10, 4799–4805. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Ling, M.T.; Kin-Wah Lee, T.; Man, K.; Wang, X.; Wong, Y.C. FTY720, a fungus metabolite, inhibits invasion ability of androgen-independent prostate cancer cells through inactivation of RhoA-GTPase. Cancer Lett. 2006, 233, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Mongroo, P.S.; Rustgi, A.K. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 2010, 10, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, L.; Matyunina, L.V.; Hill, C.G.; McDonald, J.F. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecol. Oncol. 2011, 121, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Du, L.; Yang, Y.; Wang, C.; Liu, H.; Wang, L.; Zhang, X.; Li, W.; Zheng, G.; Dong, Z. MiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer Lett. 2013, 329, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Daigo, K.; Takano, A.; Thang, P.M.; Yoshitake, Y.; Shinohara, M.; Tohnai, I.; Murakami, Y.; Maegawa, J.; Daigo, Y. Characterization of KIF11 as a novel prognostic biomarker and therapeutic target for oral cancer. Int. J. Oncol. 2018, 52, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Oue, N.; Nishioka, M.; Mukai, S.; Oshima, T.; Sakamoto, N.; Sentani, K.; Matsusaki, K.; Yoshida, K.; Yasui, W. Overexpression of KIF11 in gastric cancer with intestinal mucin phenotype. Pathobiology 2017, 84, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Koller, E.; Fazli, L.; Gleave, M.E. The role of the miR-200 family in epithelial-mesenchymal transition. Prostate 2008, 68, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
| Motif | Site | Width | E-Value | Known or Similar Motifs |
|---|---|---|---|---|
| 1 | 67 | 30 | 3.00 × 10−69 | IRF1 (MA0050.2) |
| FOXP1 (MA0481.1) | ||||
| 2 | 1709 | 5 | 2.30 × 10−18 | STAT1 (MA0137.3) |
| STAT4 (MA0518.1) | ||||
| STAT2::STAT1 (MA0517.1) | ||||
| 3 | 97 | 29 | 1.40 × 10−15 | ZNF263 (MA0528.1) |
| EGR1 (MA0162.2) | ||||
| 4 | 1207 | 5 | 1.60 × 10−9 | ZEB1 (MA0103.2) |
| T (MA0009.1) | ||||
| SMAD2::SMAD3::SMAD4 (MA0513.1) | ||||
| 5 | 1615 | 5 | 1.30 × 10−5 | GATA4 (MA0482.1) |
| 6 | 1000 | 6 | 2.90 × 10−5 | NHLH1 (MA0048.1) |
| MYOG (MA0500.1) | ||||
| PAX5 (MA0014.2) | ||||
| 7 | 104 | 8 | 1.20 × 10−3 | EGR1 (MA0162.2) |
| 8 | 13 | 29 | 1.70 × 10−3 | SOX5 (MA0087.1) |
| 9 | 47 | 8 | 4.20 × 10−3 | RUNX2 (MA0511.1) |
| 10 | 510 | 5 | 7.50 × 10−3 | ARID3A (MA0151.1) |
| MDEGs | Category | Term ID | Term Description | Adjusted p-Value | Gene Count |
|---|---|---|---|---|---|
| Up | Pathway | R-HSA-69278 | Cell Cycle, Mitotic [R-HSA-69278] | 4.29 × 10−6 | 85 |
| Pathway | R-HSA-1640170 | Cell Cycle [R-HSA-1640170] | 1.91 × 10−5 | 95 | |
| Pathway | R-HSA-453274 | Mitotic G2-G2/M phases [R-HSA-453274] | 3.59 × 10−3 | 28 | |
| Pathway | R-HSA-1266738 | Developmental Biology [R-HSA-1266738] | 6.80 × 10−3 | 102 | |
| Pathway | R-HSA-69275 | G2/M Transition [R-HSA-69275] | 8.10 × 10−3 | 27 | |
| Pathway | vegfr1_2_pathway | Signaling events mediated by VEGFR1 and VEGFR2 [vegfr1_2_pathway] | 1.13 × 10−2 | 20 | |
| Pathway | R-HSA-68877 | Mitotic Prometaphase [R-HSA-68877] | 1.27 × 10−2 | 26 | |
| Pathway | hsa04520 | Adherens junction [hsa04520] | 1.43 × 10−2 | 20 | |
| GO BP | GO:0000278 | mitotic cell cycle [GO:0000278] | 4.75 × 10−13 | 153 | |
| GO BP | GO:0007049 | cell cycle [GO:0007049] | 1.61 × 10−9 | 199 | |
| GO BP | GO:0022402 | cell cycle process [GO:0022402] | 2.19 × 10−9 | 170 | |
| GO BP | GO:1903047 | mitotic cell cycle process [GO:1903047] | 6.39 × 10−9 | 119 | |
| GO BP | GO:0090304 | nucleic acid metabolic process [GO:0090304] | 2.52 × 10−7 | 454 | |
| GO CC | GO:0005622 | intracellular [GO:0005622] | 1.99 × 10−18 | 1144 | |
| GO CC | GO:0044424 | intracellular part [GO:0044424] | 1.31 × 10−17 | 1124 | |
| GO CC | GO:0031981 | nuclear lumen [GO:0031981] | 1.42 × 10−17 | 448 | |
| GO CC | GO:0005654 | nucleoplasm [GO:0005654] | 2.48 × 10−17 | 392 | |
| GO CC | GO:0044428 | nuclear part [GO:0044428] | 4.85 × 10−17 | 472 | |
| GO MF | GO:0005515 | protein binding [GO:0005515] | 1.75 × 10−12 | 1042 | |
| GO MF | GO:0005488 | binding [GO:0005488] | 1.61 × 10−10 | 1141 | |
| GO MF | GO:0003723 | RNA binding [GO:0003723] | 3.90 × 10−7 | 198 | |
| GO MF | GO:0003676 | nucleic acid binding [GO:0003676] | 8.01 × 10−7 | 310 | |
| GO MF | GO:0044822 | poly(A) RNA binding [GO:0044822] | 3.15 × 10−6 | 172 | |
| Down | Pathway | hsa04144 | Endocytosis [hsa04144] | 4.00 × 10−4 | 52 |
| Pathway | hdac_classi_pathway | Signaling events mediated by HDAC Class I [hdac_classi_pathway] | 4.83 × 10−3 | 20 | |
| Pathway | hsa04666 | Fc gamma R-mediated phagocytosis [hsa04666] | 9.38 × 10−3 | 24 | |
| GO BP | GO:0016192 | vesicle-mediated transport [GO:0016192] | 7.26 × 10−5 | 154 | |
| GO BP | GO:0051179 | localization [GO:0051179] | 9.44 × 10−5 | 511 | |
| GO BP | GO:0006810 | transport [GO:0006810] | 5.10 × 10−4 | 419 | |
| GO BP | GO:0051234 | establishment of localization [GO:0051234] | 3.12 × 10−3 | 423 | |
| GO BP | GO:0048518 | positive regulation of biological process [GO:0048518] | 3.96 × 10−3 | 490 | |
| GO CC | GO:0044424 | intracellular part [GO:0044424] | 3.47 × 10−10 | 1156 | |
| GO CC | GO:0005622 | intracellular [GO:0005622] | 2.75 × 10−8 | 1167 | |
| GO CC | GO:0005737 | cytoplasm [GO:0005737] | 4.37 × 10−6 | 901 | |
| GO CC | GO:0030054 | cell junction [GO:0030054] | 3.72 × 10−5 | 104 | |
| GO CC | GO:0012505 | endomembrane system [GO:0012505] | 1.57 × 10−3 | 327 | |
| GO MF | GO:0016773 | phosphotransferase activity, alcohol group as acceptor [GO:0016773] | 1.58 × 10−3 | 90 | |
| GO MF | GO:0016772 | transferase activity, transferring phosphorus-containing groups [GO:0016772] | 2.30 × 10−3 | 108 | |
| GO MF | GO:0016301 | kinase activity [GO:0016301] | 2.52 × 10−3 | 96 | |
| GO MF | GO:0000166 | nucleotide binding [GO:0000166] | 1.29 × 10−2 | 78 | |
| GO MF | GO:1901265 | nucleoside phosphate binding [GO:1901265] | 1.40 × 10−2 | 78 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Qin, T.; Hu, W.; Chen, B.; Dai, M.; Xu, G. Genome-Wide Methylation Patterns in Androgen-Independent Prostate Cancer Cells: A Comprehensive Analysis Combining MeDIP-Bisulfite, RNA, and microRNA Sequencing Data. Genes 2018, 9, 32. https://doi.org/10.3390/genes9010032
Wang Y, Qin T, Hu W, Chen B, Dai M, Xu G. Genome-Wide Methylation Patterns in Androgen-Independent Prostate Cancer Cells: A Comprehensive Analysis Combining MeDIP-Bisulfite, RNA, and microRNA Sequencing Data. Genes. 2018; 9(1):32. https://doi.org/10.3390/genes9010032
Chicago/Turabian StyleWang, Yumin, Tingting Qin, Wangqiang Hu, Binghua Chen, Meijie Dai, and Gang Xu. 2018. "Genome-Wide Methylation Patterns in Androgen-Independent Prostate Cancer Cells: A Comprehensive Analysis Combining MeDIP-Bisulfite, RNA, and microRNA Sequencing Data" Genes 9, no. 1: 32. https://doi.org/10.3390/genes9010032




