Abstract
Heavy metals (HMs) are compounds that can be hazardous and impair growth of living organisms. Bacteria have evolved the capability not only to cope with heavy metals but also to detoxify polluted environments. Three heavy metal-resistant strains of Mucilaginibacer rubeus and one of Mucilaginibacter kameinonensis were isolated from the gold/copper Zijin mining site, Longyan, Fujian, China. These strains were shown to exhibit high resistance to heavy metals with minimal inhibitory concentration reaching up to 3.5 mM Cu(II), 21 mM Zn(II), 1.2 mM Cd(II), and 10.0 mM As(III). Genomes of the four strains were sequenced by Illumina. Sequence analyses revealed the presence of a high abundance of heavy metal resistance (HMR) determinants. One of the strain, M. rubeus P2, carried genes encoding 6 putative PIB-1-ATPase, 5 putative PIB-3-ATPase, 4 putative Zn(II)/Cd(II) PIB-4 type ATPase, and 16 putative resistance-nodulation-division (RND)-type metal transporter systems. Moreover, the four genomes contained a high abundance of genes coding for putative metal binding chaperones. Analysis of the close vicinity of these HMR determinants uncovered the presence of clusters of genes potentially associated with mobile genetic elements. These loci included genes coding for tyrosine recombinases (integrases) and subunits of mating pore (type 4 secretion system), respectively allowing integration/excision and conjugative transfer of numerous genomic islands. Further in silico analyses revealed that their genetic organization and gene products resemble the Bacteroides integrative and conjugative element CTnDOT. These results highlight the pivotal role of genomic islands in the acquisition and dissemination of adaptive traits, allowing for rapid adaption of bacteria and colonization of hostile environments.
1. Introduction
Heavy metals (HMs) have a dualistic impact on living organisms. On the one hand, metal ions are essential for numerous biological processes mandatory for cellular activity, including homeostasis, enzyme activity, and protein functionality [1]. On the other hand, when present in excess in the environment, HM can have toxic effect hindering diverse cellular processes and thus cellular life.
Heavy metal pollution has been part of Earth’s history as it can originate from natural processes such as volcanic eruption. Recent (over)industrialization and exploitation of Earth resources worldwide has accelerated HM release into the environment and led to high levels of water, air, and soil pollution. Especially, mine exploitation for metal extraction is one of the most important sources of heavy metal pollution [2]. This comes not only from excavating deep-buried HMs to be exposed to the surface, but also from extraction protocols that often rely on the use of other contaminants, including HMs [2].
Beyond its effects on people, HM toxicity was shown to have profound impacts on microbial communities, including fungi and bacteria [2]. Heavy metals were shown to have critical consequences on bacterial viability due to their pleiotropic effect on cellular processes. Excess of HM can disrupt the cell membrane, damage nucleic acids and proteins, impair enzymatic activities, and inhibit key processes such as transcription [1]. The presence of HM pollution exerts a high selective pressure on microbial communities, reducing their diversity, biomass, and activity, thus strongly impacting the biological activity of polluted environments [3].
In order to cope with the presence of elevated concentration of HMs, a myriad of bacterial genetic programs has been selected encoding functions that allow efflux and/or sequestration of HMs, and modification to inactivate or reduce reactivity of certain metal ions. The main mechanism to resist toxicity of HMs is efflux [1]. Important classes of HM transporters include PIB-type ATPases and cation diffusion facilitators (CDF). Both types of transporters translocate HM ions from the cytoplasm across the cytoplasmic membrane into the periplasm [4]. In the context described here with microbes having to handle very high external concentrations of HMs, P-type ATPases are much more relevant since they are much more powerful using ATP to pump HMs against their concentration gradient out of the cytoplasm [5]. In addition, HMs are translocated from the periplasm across the outer membrane into the extracellular space by resistance-nodulation-division (RND)-type transport systems. These multicomponent transporters of the RND type contain 3 RND transport proteins, 6 membrane fusion proteins (MFPs), and 3 outer membrane factor (OMF) proteins. The fascinating transport mechanism of the RND-type transport complex has been described in detail [4]. PIB-type ATPases and RND-type transport systems were described as being the most important systems to confer a high HM resistance (HMR).
Bacteria also show an astonishing capability to spread HMR genes within bacterial communities via horizontal gene transfer. Dissemination of genetic material conferring HMR is frequently associated with conjugative plasmids, genomic islands, and transposons [6]. Conjugative plasmids are extrachromosomal replicative entities able to transfer from a donor cell toward a recipient cell by conjugation [7]. Conjugative plasmids have been recognized as major contributors for the spread of adaptive traits such as antibiotic resistance, new metabolic capacities, and HMR [8]. Conjugative plasmid-borne HMR is associated with occurrence of large clusters of HMR genes that can span over several kb [9,10,11,12,13,14,15]. Portions of genomic DNA called genomic islands (GIs) were also shown to play a pivotal role into the horizontal dissemination of genetic material [16]. Although the mechanisms underlying the mobility of some GIs remain obscure, current knowledge describes different strategies that ultimately rely on conjugative transfer [17,18]. GI-associated HMR was described in Enterobacteriaceae and Shewanellaceae [19], Listeria monocytogenes [20], and Acinetobacter baumannii [21]. Also, HMR was shown to be conferred by an IncC-dependent mobilizable genomic island SGI1 variant called SGI1-K in Salmonella enterica [22,23,24]. Transposons are genetic entities able to move intra-molecularly (on the same replicon) or inter-molecularly (between different replicons) [25]. Most transposons can hitchhike by integrating into a conjugative plasmid or a GI for intercellular mobility. Transposons conferring HMR were described to be in association with other mobile genetic elements [13,22].
In this study, we describe the isolation and characterization of four heavy metal-resistant Mucilaginibacter strains isolated from a gold/copper mine in China. Genomes of these strains were sequenced and further in silico analysis revealed a high number of heavy metal resistance determinants. Moreover, at least part of these HMR gene clusters were shown to be potentially mobile as they are in the close vicinity of the core region of putative integrative and conjugative elements (ICEs).
2. Materials and Methods
2.1. Bacterial Isolation
Strains Mucilaginibacter rubeus P1, P2, and P3 were isolated from samples collected at 5–10 cm below the surface of a soil located near a waste water treatment dam of a copper-gold mine, and Mucilaginibacter kameinonensis P4, was isolated from a hillside with little human activity within the gold and copper mine (Zijin mining) in Longyan city of Fujian province, China (Table 1). After serial dilutions with 0.85% NaCl, the soil sample was spread on R2A (DSM medium 830) agar plates containing 2 mM CuSO4·5H2O. After incubation at 28 °C for 1 week, the strains were isolated and later stored at −80 °C in 20% glycerol (w/v).

Table 1.
Characteristics of the heavy metal (HM)-contaminated soil from where the strains were isolated.
2.2. Taxonomic Analysis
Strains were incubated at 28 °C for 24 h on R2A agar plates. As described in Brosius et al. [26], the universal primer pair 27F/1492R was used to amplify 16S sequences and the amplified PCR product was subsequently sequenced [26]. PCR products were sequenced by Biosune Company (Fuzhou, China) using the Sanger method. Based on the EzTaxon database (http://eztaxon-e.ezbiocloud.net) [27], pairwise sequence similarity and phylogenetic neighbors of the sequences of each individual strain (1382–1432 bp) were obtained through BLAST searches. In total, 19 Mucilaginibacter strains with publicly available 16S ribosomal RNA (rRNA) gene sequences were selected, with Pedobacter africanus DSM 12126T (AJ438171) as an out-group, to do the alignment via Mega 7.0 software [28]. A Neighbor-joining (NJ) tree was generated and the Kimura’s two-parameter model was used to calculate evolutionary distances [29], and bootstrap analysis with 1000 replications was conducted to obtain confidence levels of the branches [30].
2.3. Determination of the Minimal Inhibitory Concentration
To determine the level of resistance to various metals of all strains, M. rubeus P1, P2, and P3 and M. kameinonensis P4 were grown on Cu, As, Cd, and Zn agar plates containing different Cu(II), Zn(II), As(III), and Cd(II) concentrations to determine the minimal inhibitory concentration (MIC). The different R2A plates contained 0–10.0 mM of copper or arsenic, with 0.5 mM increments, 0–30.0 mM with 1.5 mM increments in case of zinc, and 0–2.0 mM cadmium with the increments being 0.2 mM. 1M CuSO4·5H2O, ZnCl2, NaAsO2, and CdCl2·5H2O stock solutions were prepared and stored after filtration through a 0.22 µm filter.
2.4. Cell Morphology and Flagella Observation
Overnight cultures of strains M. rubeus P1, P2, and P3 and M. kameinonensis P4 were inoculated into 50 mL of R2A medium at 28 °C with 180 rpm shaking. After 24 h of growth with shaking, cells were centrifuged (1000× g, 10 min, 4 °C) and observed under scanning electron microscopy (SEM). Cells were harvested and washed three times with cold (4 °C) phosphate buffered saline (0.2 M PBS, pH 7.2). Fixation was performed with 2.5% glutaraldehyde (24 h, 4 °C). Fixed cells were dehydrated through a series of alcohol dehydration steps (30%, 50%, 70%, 85%, 95%, and 100%) and finally freeze dried and sputter coated. The samples were then viewed using a scanning electron microscope JSM-6390 SEM (JEOL, Tokyo, Japan).
2.5. Growth Conditions Optimization
To optimize NaCl concentration and pH of the medium for growth of the Mucilaginibacter strains, 50 μL precultures were added to 5 mL R2A liquid medium supplemented with 0–3% NaCl at pH 7, or to R2A without any NaCl and with pH set to the range between pH 2–11. Cultures were incubated at 28 °C for 7 days, after which culture turbidities optical density (OD) at 600nm were evaluated. Anaerobic growth was tested by incubating R2A plates in an anaerobic chamber at 28 °C for 1 week. Optimal growth temperature was tested in the incubator on R2A agar plates at temperatures between 4 to 40 °C for 1 week.
2.6. Genomic DNA Extraction
Genomic DNA (gDNA) was extracted by using a TIANamp Bacteria DNA Kit (Tiangen Biotech, Beijing, China) from cultures grown on R2A. The quantity and purity of gDNA were assessed using an UV spectrophotometry (Nanodrop ND-1000, J & H Technology Co., Ltd. Wilmington, USA). Genomic DNA with OD260/280 value higher than 1.80 was selected and examined on agarose gel electrophoresis (0.8%). Samples containing more than 25 μg of intact gDNA (fragment size > 20 kb) were sent out for whole-genome sequencing.
2.7. Whole-Genome Sequencing
Whole-genome shotgun sequencing was preformed using an Illumina HiSeq X Ten System provided by Vazyme Biotech Co., Ltd. (Nanjing, China). The DNA library was constructed using the Illumina V3 VAHTS Universal DNA Library Prep Kit according to the VAHTS Universal DNA sample preparation protocol (Illumina, Santiago, USA). The insert size was 300 bp for all strains, and 16,980,768, 18,531,104, 18,306,636, and 20,005,292 read-pairs and 2.86, 3.12, 3.09, and 3.37 Gb of raw data were obtained for strains M. kameinonensis P4 and M. rubeus P1, P2, and P3, respectively.
2.8. De novo Genome Assembly and Annotation
Illumina reads were quality-filtered, trimmed, and de novo assembled with default settings using CLC Genomic Workbench 11.0 (QIAGEN, Hilden, Germany). The draft genome sequences were annotated by NCBI PGAP, and are accessible under GenBank numbers QEYR0000000, QFKW0000000, QFKV0000000, and QFKU0000000 for M. kameinonensis P4 and M. rubeus P1, P2, and P3, respectively. M. kameinonensis P4 generated 78 contigs with an n50 value of 350.607 bp. M rubeus P1 generated 158 contigs with an n50 value of 139.339 bp. M rubeus P2 generated 118 contigs with an n50 value of 132.524 bp. M rubeus P3 generated 107 contigs with an n50 value of 148.541 bp.
2.9. TraG Proteins Phylogenetic Analyses
Molecular phylogenetic analysis of TraG proteins was performed using MEGA6 [28]. The 807- to 850-amino acid sequences of TraG proteins were recovered from genome sequences of Mucilaginibacter isolated in this study. The corresponding sequence in CTnDOT (TraGDOT accession number: AAG17832.1) was added to the dataset as an outgroup. Analyses were computed using an amino acid alignment generated by MUSCLE [31]. The evolutionary history was inferred by using the Maximum Likelihood method based on the Jone, Taylor and Thornton (JTT) matrix-based model [32]. Initial tree(s) for the heuristic search were obtained by applying the NJ method to a matrix of pairwise distances estimated using a JTT model. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 2.9848)). The analysis involved 18 amino acid sequences. All positions with less than 95% site coverage were eliminated, providing a total of 716 positions in the final dataset.
3. Results and Discussion
3.1. Isolation of Four Heavy Metal-Resistant Mucilaginibacter
We intended to isolate heavy metal resistant strains from the ZiJin copper-gold mine to gain insights into how bacterial strains adapt to high concentrations of HMs. We recovered four strains that were morphologically similar with a high tolerance to a number of HMs.
Based on phylogenetic analysis (NJ) of the 16S rRNA gene three strains (P1, P2, and P3) were closely related to M. rubeus EF23T (98.34–99.93%) and M. gossypiicola Gh-67T (98.12–99.01 %). The fourth strain (P4) grouped closely with M. kameinonensis SCKT (98.8 %) (Figure 1). All strains belonged to the Sphingobacteriaceae family in the class Sphingobacteriia.
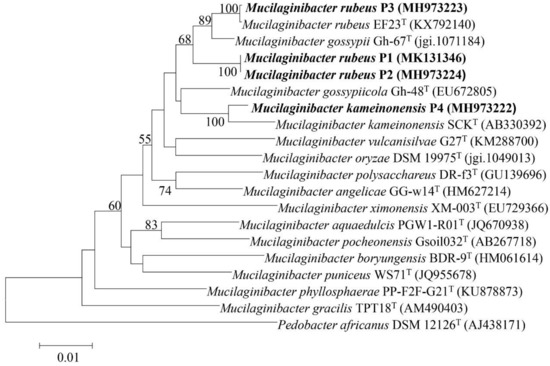
Figure 1.
Neighbour-joining phylogenetic tree constructed based on the 16S ribosomal RNA (rRNA) gene sequences from the draft genome sequence showing the phylogenetic relationships between strains Mucilaginibacter rubeus P1, P2 and P3 and Mucilaginibacter kameinonensis P4 and other species in the genus Mucilaginibacter. Values indicate percentages of identical branching in 1000 bootstrappings. The sequence of Pedobacter koreensis WPCB189T was used as an out-group. Bar, 0.01 substitutions per nucleotide position.
3.2. Phenotypic Characterization of Mucilaginibacter Strains Uncovered Multiple Heavy Metal Resistances
The HM concentration of the soil is extremely high, even the lowest concentration of total Zn, As, Cd, and Cu was found to be 49.27, 1.43, 1.19, and 18.37 mg·kg−1, respectively. The MICs of the four strains reached up to 3.5 mM Cu(II), 21 mM Zn(II), 1.2 mM Cd(II), and 10.0 mM As(III). Strain M. kameinonensis P4 displayed higher Cd resistance compared to strains M. rubeus P1, P2, and P3 (Table 2). Related, not heavy metal resistant Mucilaginibacter pedocola sp. TBZ30T, cultured under similar conditions displayed MICs of 0.4 mM Cu(II), 3 mM Zn(II), 0.2 mM Cd(II), and 0.2 mM As(III) [33]. Such high resistance to multiple HMs as reported here has therefore not been observed before in the genus Mucilaginibacter [33].

Table 2.
Minimal inhibitory concentration (MIC) of strains to Zn(II), As(III), Cd(II), and Cu(II) and respective concentrations of the HM in the soil where the strains were isolated from.
Strains M. rubeus P1, P2, and P3 and M. kameinonensis P4 formed a light orange or pink, moist, circular, and convex colony with smooth margins on R2A agar plates. All strains were Gram-negative and aerobic. Growth of strains was observed at 4–30 °C (optimum, 28 °C). Optimal growth occurred in absence of further NaCl, but the strains could still grow in R2A with up to 1.5% NaCl added. These characteristics are consistent with description of the genus Mucilaginibacter [34,35,36]. Medium pH for optimal growth (~pH 5.0) and pH tolerance (pH 5.0–9.0) varied slightly between the four strains (Table 3).

Table 3.
General features of strains M. rubeus P1, P2, and P3 and M. kameinonensis P4.
3.3. Mucilaginobacter Strains Exhibit an Arsenal of Genetic Determinants to Deal with High Concentrations of Heavy Metals
To gain insight in the genetic basis of how the four strains were able to deal with these high HM concentrations, we determined draft genome sequences. Draft genomes were automatically annotated through RAST (Rapid Annotation using Subsystem Technology) database (http://rast.nmpdr.org/). Based on inferred protein homologies, between 6 and 16 putative P1B type-ATPase [37] were encoded in the four genomes (Table 4). All strains further encoded a variety of putative RND type metal transporter systems of the CzcCBAD type. Three strains further encoded putative CusCBA Cu(I) translocating RND-type transport systems, except M. kameinonensis P4 (Table 3). Multiple genes for putative multicopper oxidases were found on the different genomes, which may constitute the basis for the observed copper resistance (Table 3). Genes for putative multicopper oxidases were only taken into account if they were located adjacent to genes encoding P1B type Cu(I) translocating P-type ATPase. Finally, between 2 and 4 putative ars operons (arsNCR, acr3, arsMCR) were among the Mucilaginibacter genomes (Table 3). The higher number of ars operons in strain P1 and P2 genomes correlated to their high MICs on As(III) (10.0 and 9.0 mM, respectively).

Table 4.
Heavy metal related genes in the analyzed Mucilaginibacter strains in comparison to Cupriavidus metallidurans CH34.
The number of HMR determinants in Mucilaginibacter genomes was unusually high, even in comparison to the well-known HM-resistant strain Cupriavidus metallidurans CH34 [5,38,39] (Table 4), suggesting a strong selection for HM resistance in their natural living environment. The HM resistance determinants are often clustered together, and often located adjacent to tra genes. They could be identified on many different contigs.
3.4. Heavy Metal Resistance Is Associated with CTnDOT-Related Genomics Islands
Tolerance to HMs is frequently acquired by horizontal transmission among and between bacterial populations. Given the important size of HMR clusters identified in Mucilaginibacter genomes (up to 150 kb), we wondered whether some might be encompassed by GIs (Figure 2). We examined the close vicinity of the HMR clusters for the hallmark of conjugative systems, also known as Type 4 secretion systems (T4SSs) [40]. T4SSs have been classified based on their VirB4 protein, a ubiquitous constituent of conjugative systems [41]. A total of 17 genes encoding VirB4 proteins (traG) exhibiting sizes between 807 to 850 amino acids were identified on the 4 genomes, most of them in the close proximity of HMR clusters: 6 in M. rubeus P1 (contigs 1, 4, 24, 29, 42, 55), 6 in M. rubeus P2 (contigs 24, 26, 29, 32, 34, 42), 2 in M. rubeus P3 (contigs 5 and 11), and 3 in M. kameinonensis P4 (contigs 6, 9, and 35).
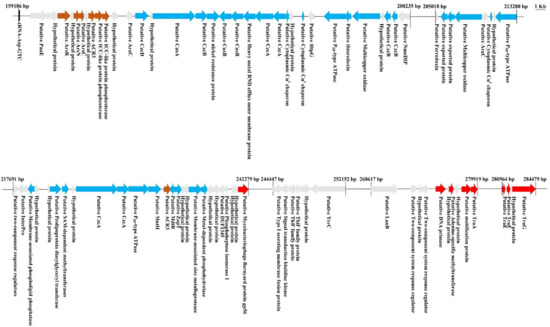
Figure 2.
Representative putative genomic island carrying genes encoding HM determinants in contig 26 of M. Rubeus P2. Genomic analysis was performed via RAST (http://rast.nmpdr.org/). Genes encoding determinants related to metalloid (Arsenical and Antimony) resistance are highlighted in maroon, metal (Copper/Cobalt/Cadmium/Zinc/Lead/Mercury) resistance in blue, and the genes encoding putative transfer functions in red. Genes encoding hypothetical proteins and unknown functions are highlighted in gray.
TraG proteins of the putative conjugative GIs identified in Mucilaginibacter genomes were compared to the 838-amino acid TraG of CTnDOT (TraGDOT) and showed 32 to 53% of identity over 94 to 98% of their amino acid sequence. The evolutionary history of TraG proteins was inferred using TraGDOT as an outgroup (Figure 3). Two strongly supported clades were delineated, suggesting that they belong to two distinct lineages (Figure 3, green and red boxes). As expected, each one of the TraG proteins of strain M. rubeus P1 grouped with one TraG protein from strain M. rubeus P2, confirming that these identical strains contain the same 6 elements. More interestingly, TraGP1-1, TraGP2-26, TraGP3-5, and TraGP4-9 grouped together, and their gene sequences were identical.
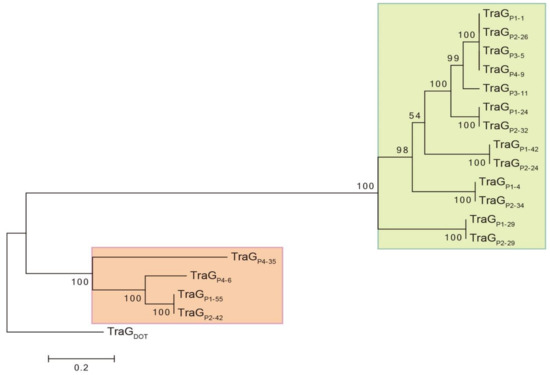
Figure 3.
Molecular phylogenetic analysis of TraG proteins of putative conjugative genomic islands (GIs) of Mucilaginibacter. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model [32]. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA6 [28]. Initial alignment of sequences was performed using Muscle for the presented tree. An identical tree with minor changes in bootstrap values was obtained using ClustalW for alignment. The VirB4 subunit of MPFB T4SS is named TraG [41]. For convenience and consistence in TraG protein identification, nomenclature is as follows: TraGPX-Y, where X is the strain number and Y the contig carrying the gene coding for TraG. TraG CTnDOT (TraGDOT) accession number: AAG17832.1.
Closer analysis revealed the presence of genes coding for other T4SS subunits adjacent to each one of the traG genes. This grouping of tra genes may be regarded as a conjugation module, i.e., genes and sequences implicated in the same biological process [42]. In particular the regions including traGP1-1, traGP2-26, traGP3-5, and traGP4-9 were 100% identical across a circa 150-kb region, including an about 75-kb cluster coding for multiple HMR. The organization of these putative conjugation modules resembles the one encoded in CTnDOT, a protypical ICE of Bacteroides [41,43]. As in CTnDOT, putative conjugative modules encoded by GIs of Mucilaginibacter thus belong to the mating pair formation (MPF) category B (MPFB) [41]. Also, most of the putative conjugative modules identified in Mucilaginibacter strains are in the proximity of genes encoding a putative tyrosine recombinase related to IntDOT, the integrase of CTnDOT [44]. Moreover, the Mucilaginibacter GIs carried a gene predicted to encode a RteC-like protein reminiscent of the CTnDOT regulation system [45,46]. Mucilaginibacter GIs are thus likely to be ICEs, whose maintenance relies on integration into the chromosome and dissemination depends on its excision from the chromosome as a circular element that would transfer by conjugation [17,47,48]. The presence of at least one identical contiguous region over 150 kb (represented by the traGP1-1 gene) in the four different Mucilaginibacter recovered strains suggest active mobility and recent transfer of this GI.
Since the draft genomes were not completely curated to a single contiguous scaffold, we could not confidently delimit the boundaries of the putative conjugative GIs. As a matter of fact, a single GI might be spread over multiple contigs, or could be a defective element lacking flanking or internal parts of the original GI. Also, IntDOT was reported to not require strict homology between the recombining sites in contrast to the majority of tyrosine recombinases [46,49]. The integration/excision is, in that case, site-selective rather than site-specific, strongly impairing the precise identification of the right and left attachment sites (attR and attL, respectively).
4. Concluding Remark
This work allowed the isolation and characterization of four heavy metal-resistant Mucilaginibacter strains recovered from polluted soil of gold mines. Sequencing of genomic content allowed inspection of HMR loci into the chromosome of these strains and their close association with loci coding for conjugation of CTnDOT-related GIs. Further genome closure and experimental investigation should allow testing the functionality of such putative ICEs found in the Mucilaginibacter strains. Notably, phenotypes such as their ability to excise from the chromosome and their capability to transfer toward a new host by conjugation will be monitored. In particular the 150-kb (at least) conserved putative ICE present in the four strains is an interesting candidate, likely to be functional given its complete identity among all four strains. Protein BLAST using TraG of this conserved element and search Mucilaginibacter genomes did not give other perfect hits, suggesting that presence of this GI is restricted to sampling locations of this study (Table 1). Increasing availability of fully sequenced genomes should allow further data meaning in order to evaluate the abundance of specific GIs in Mucilaginibacter and predict their functionality.
The presence of putative CTnDOT-related ICEs into genomes of Mucilaginibacter strains does not seem uncommon. Protein BLAST analysis using TraGDOT as query and searching Mucilaginibacter genomes gave multiple hits with identity ranging from 70% over 99% of aa sequence, down to 30% over 79% of aa sequence (considering 96 hits with more than 75% of coverage). This observation highlights the presence of multiple putative CTnDOT-related GIs populating Mucilaginibacter genomes, most likely playing a key role in their genome evolution and adaptation. One can speculate that such GIs may be involved in conferring specific capabilities such as the ability to degrade pectin, xylan, and laminarin of M. paludis and M. gracilis [50] plant growth promotion capacity conferred by M. gossypii and M. gossypiicola [51], or yet to be discovered adaptive functions that may be conferred by such GIs. Further in silico analyses may reveal interesting features of such putative ICEs considering accessory functions they could confer, or functionality of their recombination, conjugation, or regulatory systems.
This exploratory work on HMR GIs of Mucilaginibacter together with other research done on Mucilaginibacter species so far constitutes a solid ground for future experimental research aiming at developing molecular tools. Such tools would greatly facilitate further investigation of M. rubeus and M. kameinonensis biology and could likely be extended to other Mucilaginibacter species.
Author Contributions
Y.P.L. isolated strains from mine and generated the phylogenetic tree of 16SrDNA, annotated the genome, mapped the gene cluster, and partially drafted the manuscript. N.C. contributed to the general concept of the study, performed gene sequence prediction, molecular phylogenetic analysis of TraG proteins, and partially drafted the manuscript. N.Y. contributed to the MICs determination and partially drafted the manuscript. B.L. performed growth conditions optimization and genomic DNA extraction. X.X. assembled the genomes. Q.S. and H.A.Al-W. contributed to data analysis. R.F. contributed to the general concept of the study and manuscript verification. J.R.v.d.M. contributed to the general concept of the study and manuscript verification. C.R. contributed to the general concept of the study, design of experiments and manuscript preparation.
Funding
This research was funded by National Natural Science Foundation of China (NSFC) grant number 31770123, State administration of Foreign Expert Project numbers GDT20173600005 and 110000214620170006, and Swiss National Science Foundation (SNSF) grant number 31003A_175638, Twasol Research Excellence Program (TRE), King Saud University, Saudi Arabia for support.
Acknowledgments
We like to thank engineers Huaiguo Huang, Hongwen Li, and Xianzheng Chen of Zijin Mining Group Co., Ltd. for the technical support in the sampling.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chandrangsu, P.; Rensing, C.; Helmann, J.D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017, 15, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Viet, H.; Gilbert, D.; Mitchell, E.A.D.; Badot, P.M.; Bernard, N. Effects of experimental lead pollution on the microbial communities associated with Sphagnum fallax (Bryophyta). Microb. Ecol. 2007, 54, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.N.; Kuang, J.L.; Shu, W.S. Microbial ecology and evolution in the acid mine drainage model system. Trends Microbiol. 2016, 24, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Nies, D.H. The biological chemistry of the transition metal “transportome” of Cupriavidus metallidurans. Metallomics 2016, 8, 481–507. [Google Scholar] [CrossRef] [PubMed]
- Reva, O.; Bezuidt, O. Distribution of horizontally transferred heavy metal resistance operons in recent outbreak bacteria. Mob. Genet. Elem. 2012, 2, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Llosa, M.; Gomis-Rüth, F.X.; Coll, M.; De la Cruz, F. Bacterial conjugation: A two-step mechanism for DNA transport. Mol. Microbiol. 2010, 45, 1–8. [Google Scholar] [CrossRef]
- Cabezón, E.; Ripoll-Rozada, J.; Peña, A.; De la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2014, 39, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Monchy, S.; Benotmane, M.A.; Janssen, P.; Vallaeys, T.; Taghavi, S.; Van der Lelie, D.; Mergeay, M. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 2007, 189, 7417–7425. [Google Scholar] [CrossRef] [PubMed]
- Mergeay, M.; Monchy, S.; Vallaeys, T.; Auquier, V.; Benotmane, A.; Bertin, P.; Taghavi, S.; Dunn, J.; van der Lelie, D.; Wattiez, R. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: Towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 2010, 27, 385–410. [Google Scholar] [CrossRef]
- Hernández-Ramírez, K.C.; Reyes-Gallegos, R.I.; Chávez-Jacobo, V.M.; Díaz-Magaña, A.; Meza-Carmen, V.; Ramírez-Díaz, M.I. A plasmid-encoded mobile genetic element from Pseudomonas aeruginosa that confers heavy metal resistance and virulence. Plasmid 2018, 98, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Bezuidt, O.; Pierneef, R.; Mncube, K.; Limamendez, G.; Reva, O.N. Mainstreams of horizontal gene exchange in enterobacteria: Consideration of the outbreak of enterohemorrhagic E. coli O104:H4 in Germany in 2011. PLoS ONE 2012, 2, 96–100. [Google Scholar]
- Schneiker, S.; Keller, M.; Dröge, M.; Lanka, E.; Pühler, A.; Selbitschka, W. The genetic organization and evolution of the broad host range mercury resistance plasmid pSB102 isolated from a microbial population residing in the rhizosphere of alfalfa. Nucl. Acids Res. 2001, 29, 5169–5181. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, M.W.; Thomson, N.R.; Sanders, M.; Parkhill, J.; Taylor, D.E. The complete nucleotide sequence of the resistance plasmid R478: Defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 2004, 52, 182–202. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, K.; Sota, M.; Tamai, Y.; Nagata, N.; Konda, T.; Inoue, T.; Top, E.M.; Arakawa, Y. Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP-1β plasmids without accessory mobile elements. Microbiology 2006, 152, 3477–3484. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Quintais, L.; Garcillán-Barcia, M.P.; De la Cruz, F.; Rocha, E.P.C. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011, 7, e1002222. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Burrus, V. The dualistic nature of integrative and conjugative elements. Mob. Genet. Elem. 2015, 5, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Rivard, N.; Burrus, V.; Ceccarelli, D. Mobilizable genomic islands, different strategies for the dissemination of multidrug resistance and other adaptive traits. Mob. Genet. Elem. 2017, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Staehlin, B.M.; Gibbons, J.G.; Rokas, A.; O’Halloran, T.V.; Slot, J.C. Evolution of a heavy metal homeostasis/resistance island reflects increasing copper stress in enterobacteria. Genome Boil. Evol. 2016, 8, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ward, T.J.; Jima, D.D.; Parsons, C.; Kathariou, S. The arsenic resistance Listeria genomic island LGI2 exhibits sequence and integration site diversity and propensity for three Listeria monocytogenes clones with enhanced virulence. Appl. Environ. Microbiol. 2017, 83, 1189–11117. [Google Scholar] [CrossRef] [PubMed]
- Al-Jabri, Z.; Zamudio, R.; Horvath-Papp, E.; Ralph, J.; Al-Muharrami, Z.; Rajakumar, K.; Oggioni, M. Integrase-controlled excision of metal-resistance genomic islands in Acinetobacter baumannii. Genes 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Levings, R.S.; Partridge, S.R.; Djordjevic, S.P.; Hall, R.M. SGI1-K, a variant of the SGI1 genomic island carrying a mercury resistance region, in Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 2007, 51, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Matteau, D.; Luo, P.; Rodrigue, S.; Burrus, V. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 2014, 10, e1004714. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Durand, R.; Rivard, N.; Anquetil, C.; Barrette, C.; Humbert, M.; Burrus, V. Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet. 2017, 13, e1006705. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.; Chandler, M.; Courvalin, P.; Guédon, G.; Mullany, P.; Pembroke, T.; Rood, J.I.; Smith, C.J.; Summers, A.O.; Tsuda, M.; et al. Revised nomenclature for transposable genetic elements. Plasmid 2008, 60, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J.; Arfsten, U. Primary structure of protein L19 from the large subunit of Escherichia coli ribosomes. Biochemistry 1978, 17, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Lee, J.H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K.; Lim, Y.W. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limit on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Tang, J. Identification and Genome Analysis of Mucilaginibacter pedocola sp. nov. Ph.D. Thesis, Huazhong Agriculture University, Wuhan, China, 2017. [Google Scholar]
- Liu, Q.; Siddiqi, M.Z.; Kim, M.S.; Kim, S.Y.; Im, W.T. Mucilaginibacter hankyongensis, sp. nov. isolated from soil of ginseng field Baekdu mountain. J. Microbiol. 2017, 55, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.M.; Siddiqi, M.Z.; Im, W.T. Mucilaginibacter ginsenosidivorans, sp. nov. isolated from soil of ginseng field. Curr. Microbiol. 2017, 74, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, J.; Qiao, Z.; Wang, R.; Wang, G. Mucilaginibacter pedocola sp. nov. isolated from a heavy-metal-contaminated paddy field. Int. J. Syst. Evol. Microbiol. 2016, 66, 4033–4038. [Google Scholar] [PubMed]
- Purohit, R.; Ross, M.O.; Batelu, S.; Kusowski, A.; Stemmler, T.L.; Hoffman, B.M.; Rosenzweig, A.C. Cu+-specific CopB transporter: Revising P1B-type ATPase classification. Proc. Natl. Acad. Sci. USA 2018, 115, 2108–2113. [Google Scholar] [CrossRef] [PubMed]
- Mergeay, M.; Nies, D.; Schlegel, H.G.; Gerits, J.; Charles, P.; Van Gijsegem, F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 1985, 162, 328–334. [Google Scholar] [PubMed]
- Janssen, P.J.; Van Houdt, R.; Moors, H.; Monsieurs, P.; Morin, N.; Michaux, A.; Benotmane, M.A.; Leys, N.; Vallaeys, T.; Lapidus, A.; et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE 2010, 5, e10433. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J. The mosaic type IV secretion systems. EcoSal Plus 2016, 7, 1–22. [Google Scholar]
- Guglielmini, J.; Néron, B.; Abby, S.S.; Garcillánbarcia, M.P.; Cruz, F.D.L.; Rocha, E.P.C. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucl. Acids Res. 2014, 42, 5715–5727. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, A.; Merlin, C. Mobile elements as a combination of functional modules. Plasmid 2002, 47, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Grossman, A.D. Integrative and conjugative elements (ICEs): What they do and how they work. Annu. Rev. Genet. 2015, 49, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, N.B.; Wang, G.R.; Salyers, A.A. NBU1, a mobilizable site-specific integrated element from Bacteroides spp. can integrate non-specifically in Escherichia coli. J. Bacteriol. 1996, 178, 3601–3607. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Salyers, A.A. Characterization of the Bacteroides CTnDOT regulatory protein RteC. J. Bacteriol. 2011, 193, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Sutanto, Y.; Shoemaker, N.B.; Gardner, J.F.; Salyers, A.A. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 2001, 41, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Delavat, F.; Miyazaki, R.; Carraro, N.; Pradervand, N.; Van der Meer, J.R. The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 2017, 41, 512–537. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Burrus, V. Biology of three ICE families: SXT/R391, ICEBs1, and ICESt1/ICESt3. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Wood, M.M.; Gardner, J.F. The integration and excision of CTnDOT. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Pankratov, T.A.; Tindall, B.W.; Dedysh, S.N. Mucilaginibacter paludis gen. nov. sp. nov. and Mucilaginibacter gracilis sp. nov. pectin-, xylan- and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 2007, 57, 2349–2354. [Google Scholar] [CrossRef] [PubMed]
- Madhaiyan, M.; Poonguzhali, S.; Jungsook, L.; Senthilkumar, M.; Lee, K.C.; Sundaram, S. Mucilaginibacter gossypii sp. nov. and Mucilaginibacter gossypiicola sp. nov. plant-growth-promoting bacteria isolated from cotton rhizosphere soils. Int. J. Syst. Evol. Microbiol. 2010, 60, 2451–2457. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).