GWAS Uncovers Differential Genetic Bases for Drought and Salt Tolerances in Sesame at the Germination Stage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Salt and Polyethylene Glycol Stress Treatment
2.3. Statistical Analysis
2.4. Phenotype-Genotype Association Analysis
2.5. Mining of Potential Candidate Genes
3. Results
3.1. Identification of Suitable Concentrations for NaCl and Polyethylene Glycol Stress Induction
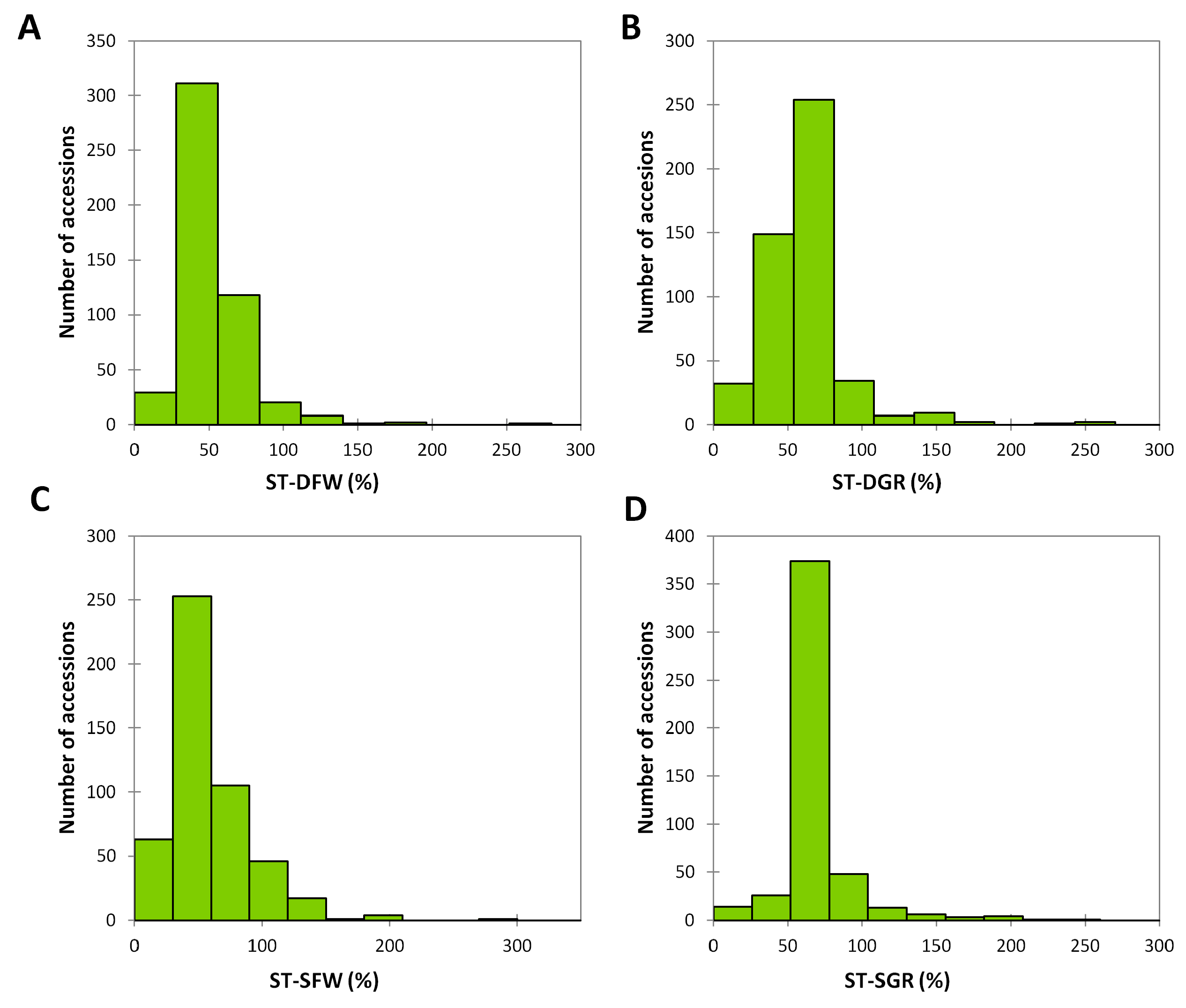
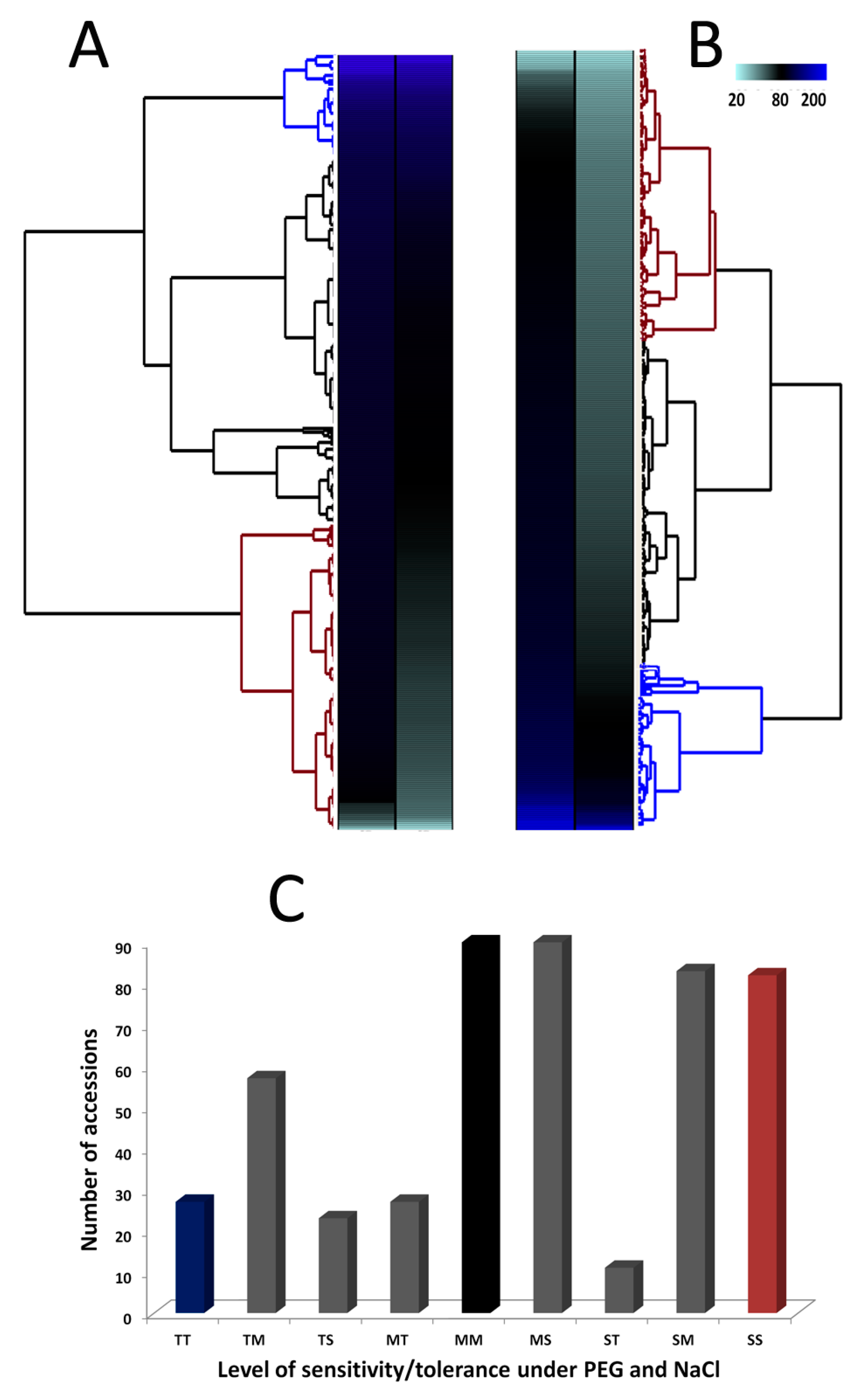
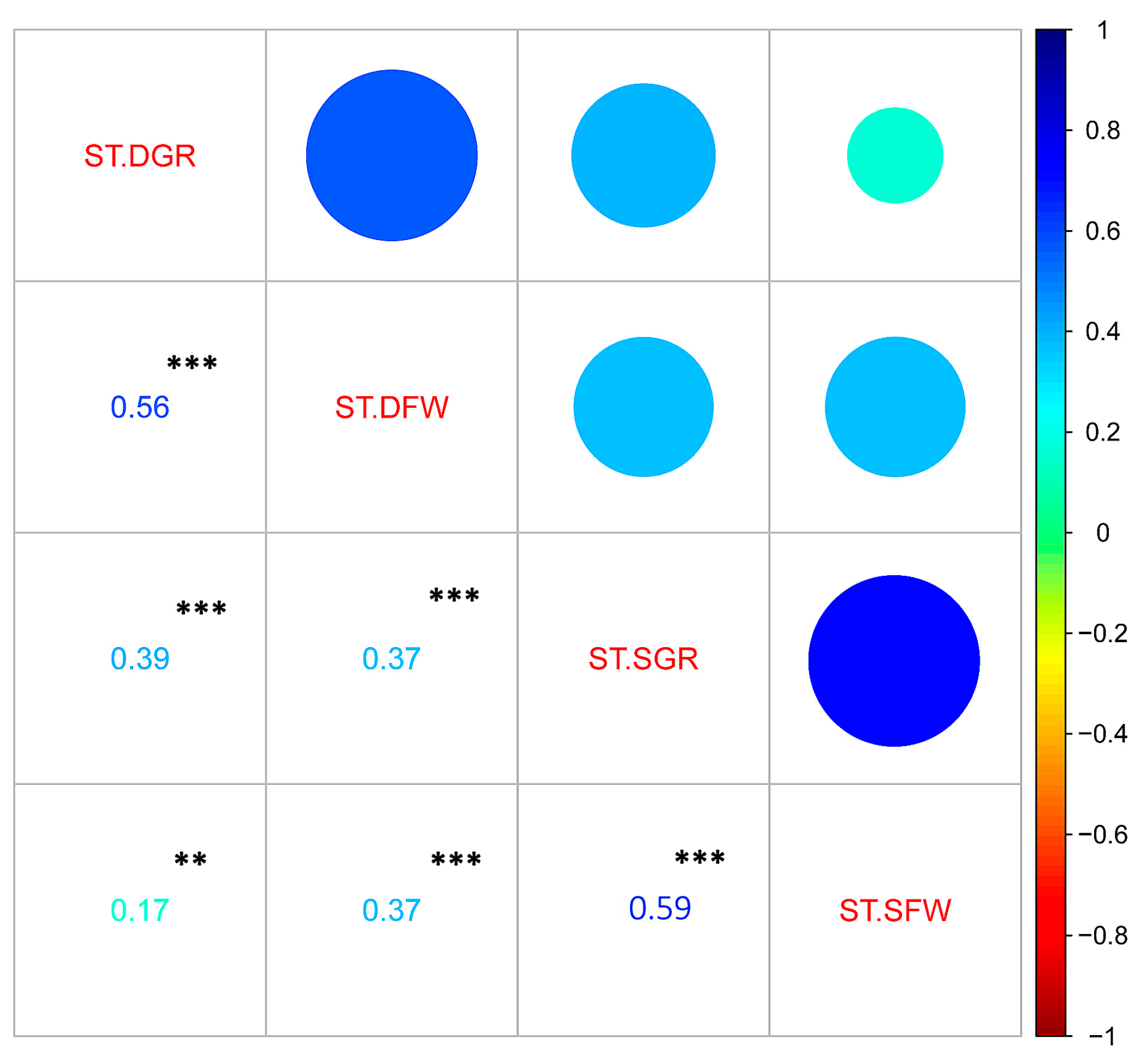
3.2. Phenotypic Variation for Salt and Drought Tolerances in the Sesame Germplasm
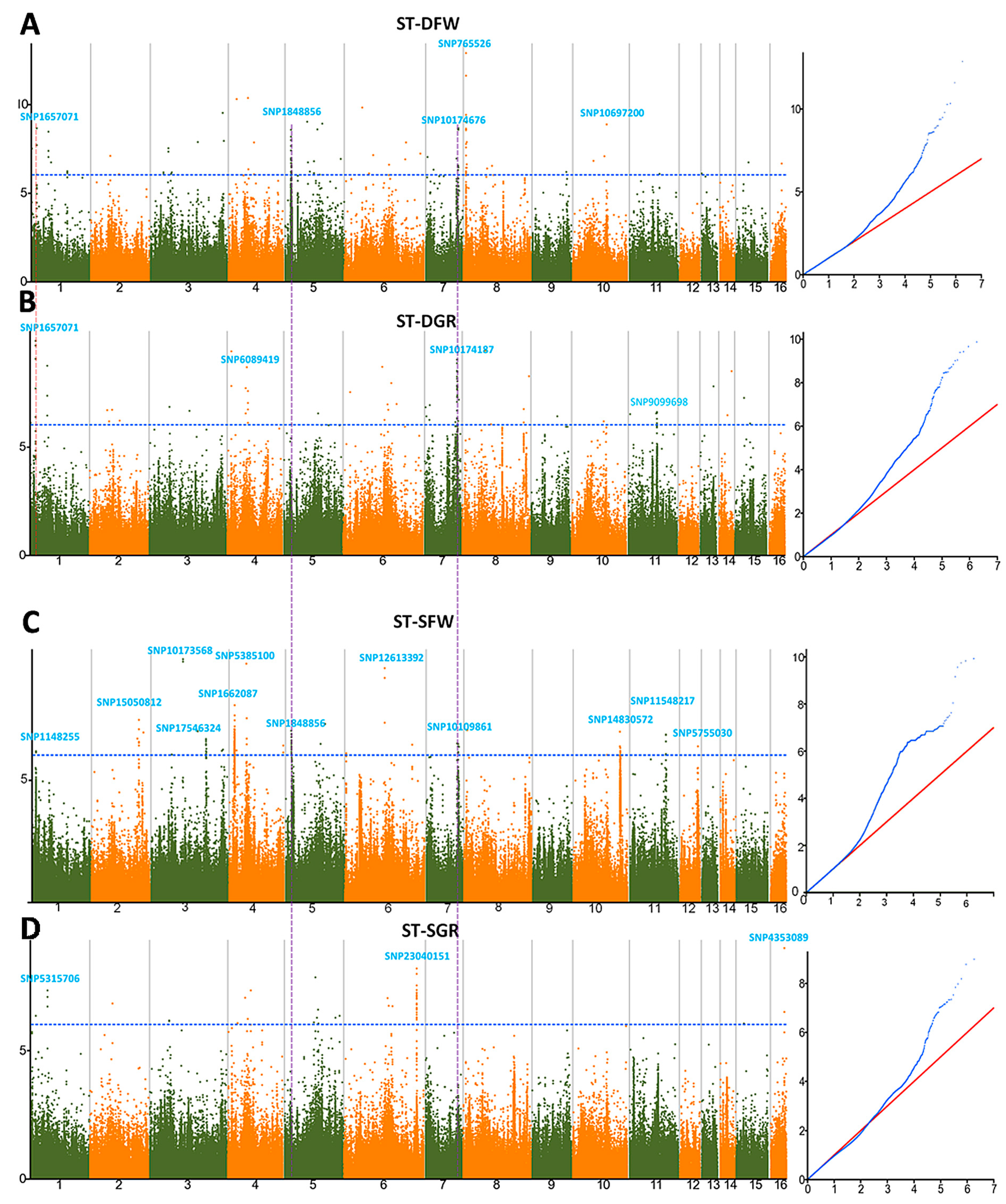
3.3. Genome-Wide Association Studies for Drought and Salt Tolerance Indexes
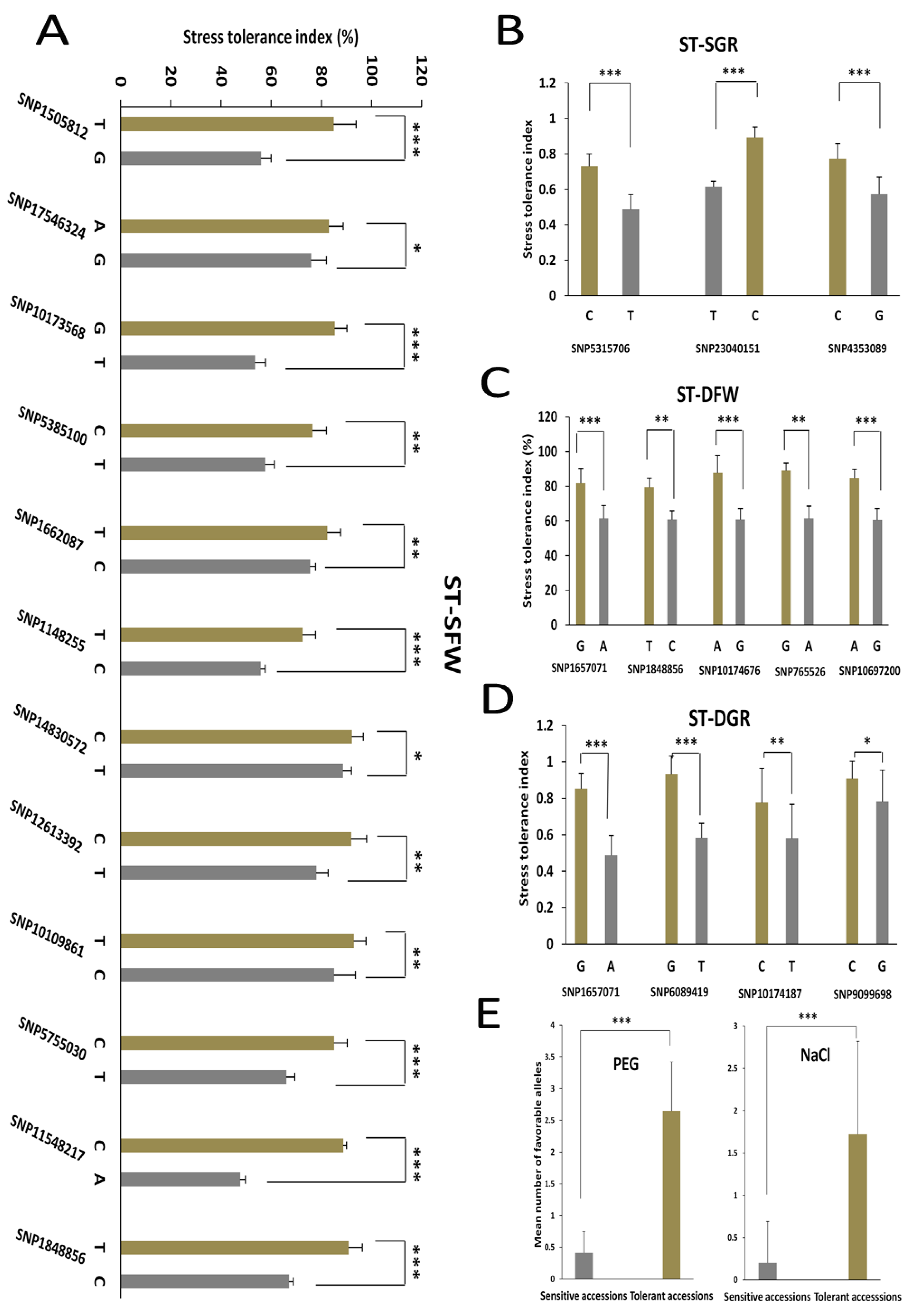
3.4. Allelic Effects of the Associated SNPs on the Salt and Drought Tolerance Indexes in Sesame
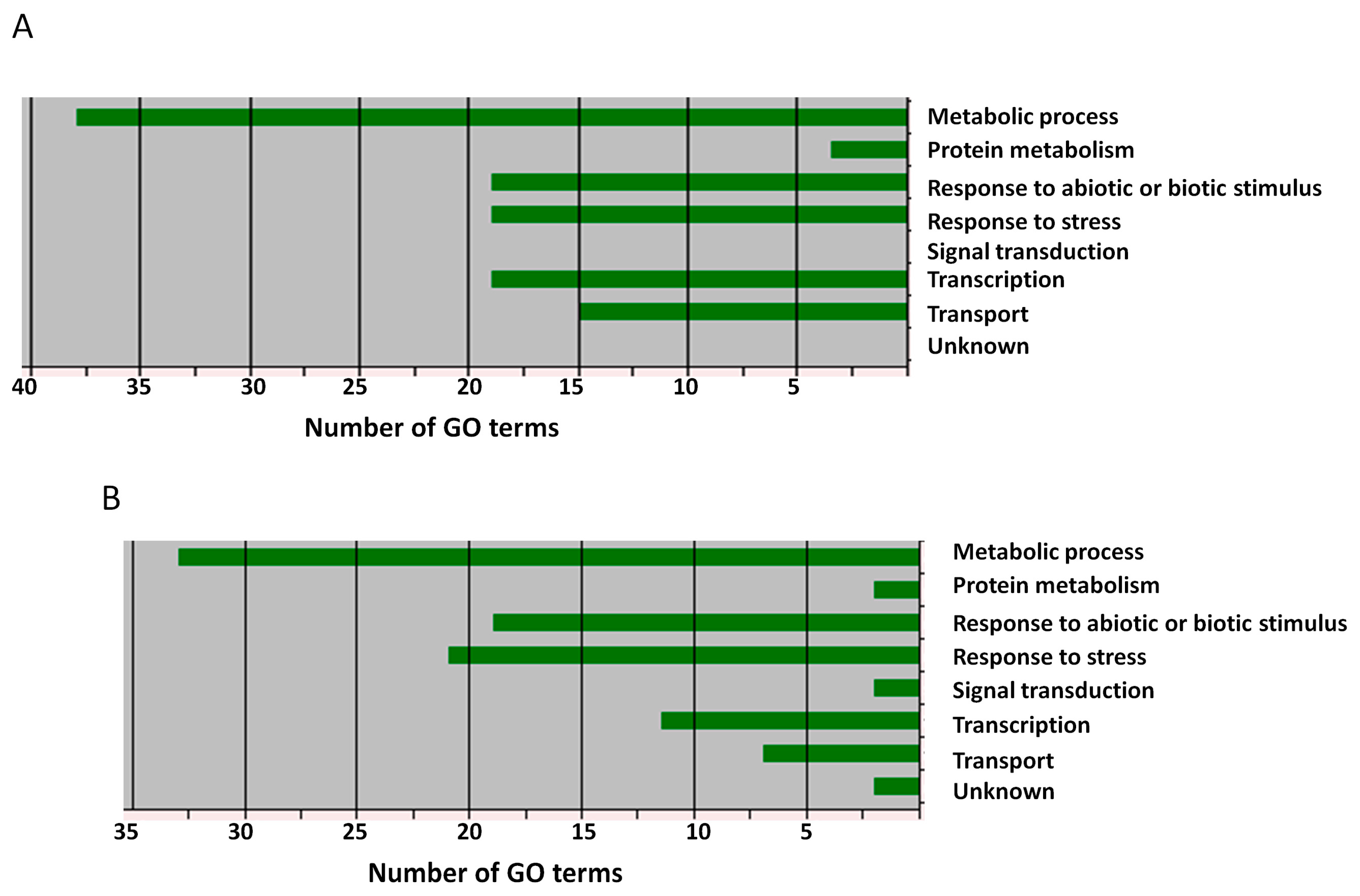
3.5. Assigning Significant SNPs Associated with Drought and Salt Tolerance to Potential Candidate Genes
4. Discussion
4.1. Drought and Salt Responses Are Governed by Different Genetic Components in Sesame
4.2. GWAS Is an Effective Approach to Identify Functional SNPs and Candidate Genes for Drought and Salt Tolerances in Sesame
4.3. Discovering New Functional Genes for the Enhancement of Drought and Salt Tolerances in Sesame
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ashri, A. Sesame Breeding. Plant Breed. Rev. 1998, 16, 179–228. [Google Scholar]
- Dossa, K.; Diouf, D.; Wang, L.; Wei, X.; Zhang, Y.; Niang, M.; Fonceka, D.; Yu, J.; Mmadi, M.A.; Yehouessi, L.W.; et al. The emerging oilseed crop Sesamum indicum enters the “Omics” era. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, R.; Gutierrez, D.; Villafane, R.; Lizaso, J.I. Salt tolerance of sesame genotypes at germination, vegetative and maturity stages. Commun. Soil Sci. Plant Anal. 2005, 36, 2405–2419. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Lee, I.-J. Penicillium–sesame interactions: A remedy for mitigating high salinity stress effects on primary and defense metabolites in plants. Environ. Exp. Bot. 2015, 116, 47–60. [Google Scholar] [CrossRef]
- Islam, F.; Gill, R.A.; Ali, B.; Farooq, M.A.; Xu, L.; Najeeb, U.; Zhou, W. Sesame. In Breeding Oilseed Crop for Sustainable Production: Opportunities and Constraints; Gupta, S.K., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 135–147. [Google Scholar]
- Yousif, H.Y.; Bingham, F.T.; Yermason, D.M. Growth, mineral composition and seed oil of sesame (Sesamum indicum L.) as affected by NaCl. Soil Sci. Soc. Am. Proc. 1972, 36, 450–453. [Google Scholar] [CrossRef]
- Hassanzadeh, M.; Asghari, A.; Jamaati-e-Somarin, S.; Saeidi, M.; Zabihi-e-Mahmoodabad, R.; Hokmalipour, S. Effects of water deficit on drought tolerance indices of sesame (Sesamum indicum L.) genotypes in Moghan Region. Res. J. Environ. Sci. 2009, 3, 116–121. [Google Scholar] [CrossRef]
- Sun, J.; Rao, Y.; Le, M.; Yan, T.; Yan, X.; Zhou, H. Effects of drought stress on sesame growth and yield characteristics and comprehensive evaluation of drought tolerance. Chin. J. Oil Crop Sci. 2010, 32, 525–533. (In Chinese) [Google Scholar]
- Boureima, S.; Eyletters, M.; Diouf, M.; Diop, T.A.; Van Damme, P. Sensitivity of seed germination and seedling radicle growth to drought stress in sesame (Sesamum indicum L.). Res. J. Environ. Sci. 2011, 5, 557–564. [Google Scholar] [CrossRef]
- Dossa, K.; Niang, M.; Assogbadjo, A.E.; Cisse, N.; Diouf, D. Whole genome homology-based identification of candidate genes for drought resistance in (Sesamum indicum L.). Afr. J. Biotechnol. 2016, 15, 1464–1475. [Google Scholar]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshou, L.; Diouf, D.; Cissé, N.; et al. Insight into the AP2/ERF transcription factor superfamily in sesame (Sesamum indicum) and expression profiling of the DREB subfamily under drought stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Diouf, D.; Cissé, N. Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 2016, 7, 1522. [Google Scholar] [CrossRef] [PubMed]
- Cerda, A.; Bingham, F.T.; Hoffman, G.J. Interactive effect of salinity and phosphorus on sesame. Soil Sci. Soc. Am. J. 1977, 41, 915–918. [Google Scholar] [CrossRef]
- Koca, H.; Bor, M.; Ozdemir, F.; Turkan, I. The effect of salt stress on lipid peroxidation, antioxidative enzymes and proline content of sesame cultivars. Environ. Exp. Bot. 2007, 60, 344–351. [Google Scholar] [CrossRef]
- Yahya, A. Selectivity and partitioning of potassium and sodium in sesame. J. Plant Nutr. 2010, 33, 670–683. [Google Scholar] [CrossRef]
- Bazrafshan, A.H.; Ehsanzadeh, P. Growth, photosynthesis and ion balance of sesame (Sesamum indicum L.) genotypes in response to NaCl concentration in hydroponic solutions. Photosynthetica 2014, 52, 134–147. [Google Scholar] [CrossRef]
- Weiss, E.A. Oil Seed Crop, 2nd ed.; Black Well Science: Malden, MA, USA, 2000. [Google Scholar]
- Betram, K.; Janssens, M.J.J.; Abdalwahab, A. Breeding for drought tolerance in sesame (Sesamum indicum). In Proceedings of the Conference on Technological and Institutional Innovations for Sustainable Rural Development, Gottingen, Germany, 8–10 October 2003; p. 135. [Google Scholar]
- Gehlot, H.S.; Purohit, A.; Shekhawat, N.S. Metabolic changes and protein patterns associated with adaptation to salinity in Sesamum indicum cultivars. J. Cell. Mol. Biol. 2005, 4, 31–39. [Google Scholar]
- Boureima, S.; Oukarroum, A.; Diouf, M.; Cissé, N.; Van Damme, P. Screening for drought tolerance in mutant germplasm of sesame (Sesamum indicum) probing by chlorophyll a fluorescence. Environ. Exp. Bot. 2012, 81, 37–43. [Google Scholar] [CrossRef]
- Boureima, S.; Diouf, M.; Amoukou, A.I.; Damme, V.P. Screening for sources of tolerance to drought in sesame induced mutants: Assessment of indirect selection criteria for seed yield. Int. J. Pure Appl. Biosci. 2016, 4, 45–60. [Google Scholar] [CrossRef]
- Bahrami, H.; Razmjoo, J.; Jafari, A.O. Effect of drought stress on germination and seedling growth of sesame cultivars (Sesamum indicum L.). Int. J. AgriSci. 2012, 2, 423–428. [Google Scholar]
- Kadkhodaie, A.; Zahedi, M.; Razmjoo, J.; Pessarakli, M. Changes in some anti-oxidative enzymes and physiological indices among sesame genotypes (Sesamum indicum L.) in response to soil water deficits under field conditions. Acta Physiol. Plant. 2013, 36, 641–650. [Google Scholar] [CrossRef]
- Kadkhodaie, A.; Razmjoo, J.; Zahedi, M.; Pessarakli, M. Selecting sesame genotypes for drought tolerance based on some physiochemical traits. Agron. J. 2014, 106, 111–118. [Google Scholar] [CrossRef]
- Bazrafshan, A.H.; Ehsanzadeh, P. Evidence for differential lipid peroxidation and antioxidant enzyme activities in Sesamum indicum L. genotypes under NaCl salinity. J. Agric. Sci. Technol. 2016, 18, 207–222. [Google Scholar]
- Hussein, Y.; Amin, G.; Azab, A.; Gahin, H. Induction of drought stress resistance in (Sesamum indicum L.) plant by salicylic acid and kinetin. J. Plant Sci. 2015, 10, 128–141. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thuillet, A.C.; Yu, J.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005, 44, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, A.; Mithra, S.V.A.; Krishnamurthy, S.L.; Parida, S.K.; Jain, S.; Tiwari, K.K.; Kumar, P.; Rao, A.R.; Sharma, S.K.; et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 2015, 22, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yu, L.; Zheng, P.; Li, Y.; Rivera, M.; Main, D.; Greene, S.L. Identification of loci associated with drought resistance traits in heterozygous autotetraploid Alfalfa (Medicago sativa L.) using genome-wide association studies with genotyping by sequencing. PLoS ONE 2015, 10, e0138931. [Google Scholar] [CrossRef] [PubMed]
- Clauw, P.; Coppens, F.; Korte, A.; Herman, D.; Slabbinck, B.; Dhondt, S.; Van Daele, T.; De Milde, L.; Vermeersch, M.; Maleux, K.; et al. Leaf growth response to mild drought: Natural variation in Arabidopsis sheds light on trait architecture. Plant Cell 2016, 28, 2417–2434. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Klei, K.; Fokkens, L.; Haring, M.A.; Schranz, E.M.; Testerink, C. Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J. Exp. Bot. 2016, 67, 2127–2138. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hu, K.; Zhang, Z.; Guan, C.; Chen, S.; Hua, W.; Li, J.; Wen, J.; Yi, B.; Shen, J.; et al. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res. 2016, 23, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.; Chen, P.; Korth, K.; Hancock, F.; Pereira, A.; Brye, K.; Wu, C.; Shi, A. Genome-wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Mol. Breed. 2017, 37, 30. [Google Scholar] [CrossRef]
- Patishtan, J.; Hartley, T.N.; de Carvalho, R.F.F.; Maathuis, J.M. Genome wide association studies to identify rice salt-tolerance markers. Plant Cell Environ. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Liao, F.; Hou, L.; Wang, J.; Wei, L.; Jian, H.; Xu, X.; Li, J.; Liu, L. Genome-wide association analysis of seed germination percentage and germination index in Brassica napus L. under salt and drought stresses. Euphytica 2017, 213, 40. [Google Scholar] [CrossRef]
- Liu, X.; Yu, L. Genome-wide association mapping of loci associated with plant growth and forage production under salt stress in Alfalfa (Medicago sativa L.). Front. Plant Sci. 2017, 8, 853. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef] [PubMed]
- Long, N.V.; Dolstra, O.; Malosetti, M.; Kilian, B.; Graner, A.; Visser, R.G.F.; van der Linden, C.G. Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2013, 126, 2335–2351. [Google Scholar] [CrossRef] [PubMed]
- Dray, S.; Dufour, A.B.; Chessel, D. The ade4 package-II: Two-table and K-table methods. R News 2007, 7, 47–52. [Google Scholar]
- De Mendiburu, F. agricolae: Statistical Procedures for Agricultural Research, R Package Version 1.2-1. 2014. Available online: http://CRAN.R-project.org/package=agricolae (accessed on 15 August 2017).
- Wei, T.; Simko, V. corrplot: Visualization of a Correlation Matrix, R Package Version 0.77. 2016. Available online: http://CRAN.R-project.org/package=corrplot, (accessed on 15 August 2017).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Hardy, O.J.; Vekemans, X. SPAGEDi: A versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]
- Dossa, K.; Wei, X.; Zhang, Y.; Fonceka, D.; Yang, W.; Diouf, D.; Liao, B.; Cissé, N.; Zhang, X. Analysis of genetic diversity and population structure of sesame accessions from Africa and Asia as major centers of its cultivation. Genes 2016, 7, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Sul, J.; Service, S.; Zaitlen, N.; Kong, S.; Freimer, N.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, H.; Wang, X.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Gong, H.; Yu, J.; Liu, P.; Wang, L.; Zhang, Y.; Zhang, X. Sesame FG: An integrated database for the functional genomics of sesame. Sci. Rep. 2017, 7, 2342. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Mondini, L.; Pagnotta, M.A. Drought and salt stress in cereal. In Sustainable Agriculture Reviews; Lichtfouse, E., Goyal, A., Eds.; Springer: Cham, Switzerland, 2015; p. 16. [Google Scholar]
- Zhang, Y.; Wang, L.; Li, D.; Gao, Y.; Lu, H.; Zhang, X. Mapping of sesame waterlogging tolerance QTL and identification of excellent waterlogging tolerant germplasm. Sci. Agric. Sin. 2014, 47, 422–430. (In Chinese) [Google Scholar]
- Wang, L.; Li, D.; Zhang, Y.; Gao, Y.; Yu, J.; Wei, X.; Zhang, X. Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. PLoS ONE 2016, 11, e0149912. [Google Scholar] [CrossRef] [PubMed]
- Patade, V.Y.; Bhargava, S.; Suprasanna, P. Salt and drought tolerance of sugarcane and antioxidant defense. J. Plant Interact. 2011, 6, 275–282. [Google Scholar] [CrossRef]
- Patade, V.Y.; Bhargava, S.; Suprasanna, P. Effects of NaCl and iso-osmotic PEG stress on growth, osmolytes accumulation and antioxidant defense in cultured sugarcane cells. Plant Cell Tissue Organ. Cult. 2012, 108, 279–286. [Google Scholar] [CrossRef]
- Lokhande, V.H.; Nikam, T.D.; Penna, S. Differential osmotic adjustment to iso-osmotic NaCl and PEG stress in the in vitro cultures of Sesuvium portulacastrum (L.) L. J. Crop Sci. Biotechnol. 2010, 13, 251–256. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- De Olivera, A.B.; Alencar, N.L.M.; Gomes-Filho, E. Comparison between the Water and Salt Stress Effects on Plant Growth and Development; Intech: Rijeka, Croatia, 2013; pp. 67–94. [Google Scholar]
- Kreps, J.A.; Wu, Y.; Chang, H.S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic and cold stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xia, Q.; Zhang, Y.; Zhu, X.; Zhu, X.; Li, D.; Ni, X.; Gao, Y.; Xiang, H.; Wei, X.; et al. Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genom. 2016, 17, 31. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Liu, H.; Yang, M.; Tao, Y.; Ma, H.; Wu, W.; Zuo, Y.; Zhao, Y. High-density genetic map construction and QTLs analysis of grain yield-related traits in sesame (Sesamum indicum L.) based on RAD-Seq technology. BMC Plant Biol. 2014, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Sacco, A.; Di, M.A.; Lombardi, N.; Trotta, N.; Punzo, B.; Mari, A.; Barone, A. Quantitative trait loci pyramiding for fruit quality traits in tomato. Mol. Breed. 2013, 31, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, W.; Chang, X.; Li, R.; Jing, R. Effects of favorable alleles for water soluble carbohydrates at grain filling on grain weight under drought and heat stresses in wheat. PLoS ONE 2014, 29, e102917. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Li, D.; Wang, L.; Zheng, X.; Yua, J.; Wei, X.; Fonceka, D.; Diouf, D.; Liao, B.; Cisse, N.; et al. Dynamic transcriptome landscape of Sesame (Sesamum indicum L.) under progressive drought and after rewatering. Genom. Data 2017, 11, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Li, D.; Wang, L.; Zheng, X.; Liu, A.; Yu, J.; Wei, X.; Zhou, R.; Fonceka, D.; Diouf, D.; et al. Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- De Ollas, C.; Arbona, V.; Gomez-Cadenas, A. Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell Environ. 2015, 38, 2157–2170. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Muszynska, A.; Gruszka, D. The role of strigolactones in nutrient-stress responses in plants. Int. J. Mol. Sci. 2013, 14, 9286–9304. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Belghazi, M.; Huguet, R.; Robin, C.; Moreau, A.; Job, C.; Job, D. Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 2006, 141, 910–923. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Arisz, S.A.; Dekker, H.L.; Kramer, G.; de Koster, C.G.; Haring, M.A.; Munnik, T.; Testerink, C. Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 2013, 450, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ke, T.; Tehrim, S.; Sun, F.; Liao, B.; Hua, W. PTGBase: An integrated database to study tandem duplicated genes in plants. Database 2015, 2015, bav017. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Gong, Q.; Bohnert, H.J. Dissecting salt stress pathways. J. Exp. Bot. 2006, 57, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hsu, F.; Li, J.; Wang, N.; Shih, M. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011, 156, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wang, Y.; Liu, G.; Wang, C.; Jiang, J.; Yang, C. Cloning of ten peroxidase (POD) genes from Tamrix hispida and characterization of their responses to abiotic stress. Plant Mol. Biol. Rep. 2010, 28, 77. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Shkolnik-Inbar, D.; Adler, G.; Bar-Zvi, D. ABI4 down regulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 2013, 73, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Park, H.C.; Aman, R.; Ali, Z.; Yun, D. Role of HKT1 in Thellungiella salsuginea, a model extremophile plant. Plant Signal. Behav. 2013, 8, e25196. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Raddatz, N.; Aman, R.; Kim, S.; Park, H.C.; Jan, M.; Baek, D.; Khan, I.U.; Oh, D.; Lee, S.Y.; et al. A single amino-acid substitution in the sodium transporter HKT1 associated with plant salt tolerance. Plant Physiol. 2016, 171, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Sakuma, Y.; Tran, L.P.; Maruyama, K.; Kidokoro, S.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.; Tanokura, M.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Mieulet, D.; Hubberten, H.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fernie, A.R.; Fisahn, J.; San Segundo, B.; Guiderdoni, E.; et al. SALT-RESPONSIVE ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 2013, 25, 2115–2131. [Google Scholar] [CrossRef] [PubMed]

| Trait | Treatment (T) | Mean | SD | Range | Variance | Skewness | Kurtosis | A | T | A × T |
|---|---|---|---|---|---|---|---|---|---|---|
| GR | Distilled water | 40 | 9.40 | 33–50 | 88.40 | −1.75 | 3.65 | *** | *** | *** |
| PEG stress | 25.24 | 4.60 | 0–50 | 112.42 | −1.46 | 1.94 | ||||
| Salt stress | 24.23 | 3.46 | 0–50 | 109.51 | −1.41 | 1.66 | ||||
| FW (g) | Distilled water | 1.49 | 0.52 | 1.08–2.93 | 0.26 | −0.28 | −0.30 | *** | *** | *** |
| PEG stress | 0.66 | 0.18 | 0.0.5–1.61 | 0.08 | −0.27 | 0.00 | ||||
| Salt stress | 0.7 | 0.16 | 0.05–1.92 | 0.22 | 0.23 | 0.05 |
| Stress | Tolerance Levels | Number of Accessions | Percentage (%) | Mean ST-GR | Mean ST-FW |
|---|---|---|---|---|---|
| Sensitive | 195 | 40 | 0.76 | 66.28 | |
| Salt | Moderate | 230 | 47 | 0.87 | 81.83 |
| Tolerant | 65 | 13 | 1.17 | 100.62 | |
| Sensitive | 176 | 36 | 0.78 | 50.31 | |
| PEG | Moderate | 207 | 42 | 0.94 | 56.75 |
| Tolerant | 107 | 22 | 1.06 | 91.14 |
| Trait | LG | QTL | Peak SNP | –log10 (p) | Ref Base | SNP Base | MAF | Genes in LD | PVE (%) | Candidate Gene ID | Gene Name |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ST-DFW | LG1 | qDFW1.1 | SNP1657071 | 8.63 | A | G | 0.06 | 24 | 7.23 | SIN_1021558 | SiCCD8 |
| SIN_1021566 | SiEMF1 | ||||||||||
| LG5 | qDFW5.1 | SNP1848856 | 8.55 | C | T | 0.07 | 24 | 6.89 | SIN_1007701 | SiGPAT3 | |
| SIN_1007708 | SiAGL37 | ||||||||||
| SIN_1007698 | SiHKT1 | ||||||||||
| LG7 | qDFW7.1 | SNP10174676 | 6.54 | G | A | 0.04 | 27 | 4.93 | SIN_1008841 | SiGDH2 | |
| SIN_1008842 | SiCYP76C7 | ||||||||||
| SIN_1009337 | SiDREB | ||||||||||
| LG8 | qDFW8.1 | SNP765526 | 12.88 | A | G | 0.03 | 33 | 10.45 | SIN_1019660 | SiRABA1D | |
| SIN_1019661 | SiWRKY69 | ||||||||||
| LG10 | qDFW10.1 | SNP10697200 | 8.85 | G | A | 0.06 | 7 | 7.67 | NA | NA | |
| ST-DGR | LG1 | qDGR1.1 | SNP1657071 | 8.63 | A | G | 0.06 | 24 | 7.23 | SIN_1021566 | SiEMF1 |
| SIN_1021558 | SiCCD8 | ||||||||||
| LG4 | qDGR4.1 | SNP6089419 | 8.66 | T | G | 0.07 | 10 | 7.22 | SIN_1001572 | SiGRV2 | |
| LG7 | qDGR7.1 | SNP10174187 | 9.01 | T | C | 0.03 | 26 | 6.61 | SIN_1008842 | SiCYP76C7 | |
| SIN_1009337 | SiDREB | ||||||||||
| SIN_1008841 | SiGDH2 | ||||||||||
| LG11 | qDGR11.1 | SNP9099698 | 6.59 | G | C | 0.06 | 25 | 5.3 | SIN_1024695 | SiGRF5 | |
| SIN_1024693 | SiOPR3 |
| Trait | LG | QTL | Peak SNP | –log10 (p) | REF BASE | SNP Base | MAF | Genes in LD | PVE (%) | Candidate Gene ID | Gene Name |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ST-SFW | LG1 | qSFW1.1 | SNP1148255 | 6.16 | C | T | 0.05 | 20 | 3.93 | SIN_1021624 | SiLHCB6 |
| LG2 | qSFW2.1 | SNP15050812 | 7.45 | G | T | 0.05 | 21 | 8.03 | SIN_1021337 | SiMLP31 | |
| SIN_1021330 | SiANTH | ||||||||||
| SIN_1021327 | SiPOD | ||||||||||
| SIN_1021326 | SiPOD | ||||||||||
| SIN_1021325 | SiPOD | ||||||||||
| SIN_1021324 | SiPOD | ||||||||||
| SIN_1021323 | SiPOD | ||||||||||
| SIN_1021322 | SiPOD | ||||||||||
| LG3 | qSFW3.1 | SNP17546324 | 6.66 | G | A | 0.03 | 16 | 4.04 | SIN_1015378 | SiHSFA1 | |
| LG3 | qSFW3.2 | SNP10173568 | 9.92 | T | G | 0.08 | 15 | 8.32 | SIN_1017475 | SiDUF538 | |
| LG4 | qSFW4.1 | SNP5385100 | 9.74 | T | C | 0.05 | 6 | 7.45 | SIN_1018894 | SiCC-NBS-LRR | |
| LG4 | qSFW4.2 | SNP1662087 | 8.04 | C | T | 0.06 | 8 | 6.52 | SIN_1008463 | SiUDG | |
| LG5 | qSFW5.1 | SNP1848856 | 8.55 | C | T | 0.07 | 24 | 2.92 | SIN_1007701 | SiGPAT3 | |
| SIN_1007708 | SiAGL37 | ||||||||||
| SIN_1007698 | SiHKT1 | ||||||||||
| LG6 | qSFW6.1 | SNP12613392 | 9.5 | T | C | 0.06 | 6 | 6.52 | SIN_1018616 | SiNAC43 | |
| LG7 | qSFW7.1 | SNP10109861 | 6.45 | C | T | 0.04 | 9 | 5.88 | SIN_1008841 | SiGDH2 | |
| SIN_1008842 | SiCYP76C7 | ||||||||||
| SIN_1009337 | SiDREB | ||||||||||
| LG10 | qSFW10.1 | SNP14830572 | 6.95 | T | C | 0.03 | 23 | 4.91 | SIN_1026087 | SiCP24 | |
| LG11 | qSFW11.1 | SNP11548217 | 6.83 | A | C | 0.04 | 32 | 7.7 | SIN_1013032 | NA | |
| LG12 | qSFW12.1 | SNP5755030 | 6.37 | T | C | 0.06 | 16 | 6.68 | SIN_1006749 | SiWRKY14 | |
| SIN_1006753 | SiLSD1 | ||||||||||
| ST-SGR | LG1 | qSGR1.1 | SNP5315706 | 7.33 | T | C | 0.06 | 10 | 5.91 | SIN_1026318 | SiXXT5 |
| LG6 | qSGR6.1 | SNP23040151 | 8.18 | C | T | 0.03 | 24 | 8.45 | SIN_1022410 | SiXTH15 | |
| LG16 | qSGR16.1 | SNP4353089 | 8.98 | G | C | 0.07 | 10 | 7.45 | SIN_1003799 | SiG6PD1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Dossa, K.; Zhang, Y.; Wei, X.; Wang, L.; Zhang, Y.; Liu, A.; Zhou, R.; Zhang, X. GWAS Uncovers Differential Genetic Bases for Drought and Salt Tolerances in Sesame at the Germination Stage. Genes 2018, 9, 87. https://doi.org/10.3390/genes9020087
Li D, Dossa K, Zhang Y, Wei X, Wang L, Zhang Y, Liu A, Zhou R, Zhang X. GWAS Uncovers Differential Genetic Bases for Drought and Salt Tolerances in Sesame at the Germination Stage. Genes. 2018; 9(2):87. https://doi.org/10.3390/genes9020087
Chicago/Turabian StyleLi, Donghua, Komivi Dossa, Yanxin Zhang, Xin Wei, Linhai Wang, Yujuan Zhang, Aili Liu, Rong Zhou, and Xiurong Zhang. 2018. "GWAS Uncovers Differential Genetic Bases for Drought and Salt Tolerances in Sesame at the Germination Stage" Genes 9, no. 2: 87. https://doi.org/10.3390/genes9020087
APA StyleLi, D., Dossa, K., Zhang, Y., Wei, X., Wang, L., Zhang, Y., Liu, A., Zhou, R., & Zhang, X. (2018). GWAS Uncovers Differential Genetic Bases for Drought and Salt Tolerances in Sesame at the Germination Stage. Genes, 9(2), 87. https://doi.org/10.3390/genes9020087








