Archaeal Viruses from High-Temperature Environments
Abstract
:1. Introduction
2. Challenges Associated with Archaeal Virology
3. Comparison of Gene Content of Bacteriophage, Archaeal, and Eukaryotic Viruses
4. Archaeal Virus Life Style and Gene Functions
5. Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Colman, D.R.; Poudel, S.; Hamilton, T.L.; Havig, J.R.; Selensky, M.J.; Shock, E.L.; Boyd, E.S. Geobiological feedbacks and the evolution of thermoacidophiles. ISME J. 2017, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Spang, A.; Saw, J.H.; Jørgensen, S.L.; Zaremba-Niedzwiedzka, K.; Martijn, J.; Lind, A.E.; van Eijk, R.; Schleper, C.; Guy, L.; Ettema, T.J.G. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 2015, 521, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Zaremba-Niedzwiedzka, K.; Caceres, E.F.; Saw, J.H.; Bäckström, D.; Juzokaite, L.; Vancaester, E.; Seitz, K.W.; Anantharaman, K.; Starnawski, P.; Kjeldsen, K.U.; et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017, 541, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, B.M.; Kim, M.; Lai-Hoe, A.; Shukor, N.A.A.; Rahim, R.A.; Go, R.; Adams, J.M. pH dominates variation in tropical soil archaeal diversity and community structure. FEMS Microbiol. Ecol. 2013, 86, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Karner, M.B.; Delong, E.F.; Karl, D.M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 2001, 409, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Vila-Costa, M.; Barberan, A.; Auguet, J.C.; Sharma, S.; Moran, M.A.; Casamayor, E.O. Bacterial and archaeal community structure in the surface microlayer of high mountain lakes examined under two atmospheric aerosol loading scenarios. FEMS Microbiol. Ecol. 2013, 84, 387–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348, e1261359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munson-McGee, J.H.; Field, E.K.; Bateson, M.; Rooney, C.; Stepanauskas, R.; Young, M.J. Nanoarchaeota, their sulfolobales host, and Nanoarchaeota virus distribution across Yellowstone National Park Hot Springs. Appl. Environ. Microbiol. 2015, 81, 7860–7868. [Google Scholar] [CrossRef] [PubMed]
- Inskeep, W.P.; Jay, Z.J.; Tringe, S.G.; Herrgård, M.J.; Rusch, D.B. The YNP Metagenome Project: Environmental parameters responsible for microbial distribution in the Yellowstone Geothermal Ecosystem. Front. Microbiol. 2013, 4, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jay, Z.J.; Rusch, D.B.; Tringe, S.G.; Bailey, C.; Jennings, R.M.; Inskeep, W.P. Predominant acidilobus-like populations from geothermal environments in yellowstone national park exhibit similar metabolic potential in different hypoxic microbial communities. Appl. Environ. Microbiol. 2014, 80, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Valentine, D. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 2007, 5, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Inskeep, W.P.; Rusch, D.B.; Jay, Z.J.; Herrgard, M.J.; Kozubal, M.A.; Richardson, T.H.; Macur, R.E.; Hamamura, N.; Jennings, R.D.; Fouke, B.W.; et al. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS ONE 2010, 5, e9773. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, B.; Wirth, J.F.; Mazurie, A.; Young, M.J. Viral assemblage composition in Yellowstone acidic hot springs assessed by network analysis. ISME J. 2015, 9, 2162–2177. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, T.; Patterson, M.; Richardson, P.M.; Wommack, K.E.; Young, M.; Mead, D. Assembly of viral metagenomes from Yellowstone hot springs. Appl. Environ. Microbiol. 2008, 74, 4164–4174. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, B.; Shaughnessy, D.P.; Wolf, Y.I.; Koonin, E.V.; Roberto, F.F.; Young, M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J. Virol. 2012, 86, 5562–5573. [Google Scholar] [CrossRef] [PubMed]
- Dellas, N.; Snyder, J.C.; Bolduc, B.; Young, M.J. Archaeal viruses: Diversity, replication, and structure. Annu. Rev. Virol. 2014, 1, 399–426. [Google Scholar] [CrossRef] [PubMed]
- Rachel, R.; Bettstetter, M.; Hedlund, B.P.; Häring, M.; Kessler, A.; Stetter, K.O.; Prangishvili, D. Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Arch. Virol. 2002, 147, 2419–2429. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D.; Bamford, D.H.; Forterre, P.; Iranzo, J.; Koonin, E.V.; Krupovic, M. The enigmatic archaeal virosphere. Nat. Rev. Microbiol. 2017, 15, 724–739. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses. Available online: http://www.ictvonline.org (accessed on 6 December 2017).
- NCBI Taxonomy Browser. Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi (accessed on 9 February 2018).
- Nigro, O.D.; Jungbluth, S.P.; Lin, H.T.; Hsieh, C.C.; Miranda, J.A.; Schvarcz, C.R.; Rappé, M.S.; Steward, G.F. Viruses in the oceanic basement. M. Bio. 2017, 8, e02129-16. [Google Scholar] [CrossRef] [PubMed]
- Philosof, A.; Yutin, N.; Flores-Uribe, J.; Sharon, I.; Koonin, E.V.; Béjà, O. Novel abundant oceanic viruses of uncultured marine group II Euryarchaeota. Curr. Biol. 2017, 27, 1362–1368. [Google Scholar] [CrossRef] [PubMed]
- Vik, D.R.; Roux, S.; Brum, J.R.; Bolduc, B.; Emerson, J.B.; Padilla, C.C.; Stewart, F.J.; Sullivan, M.B. Putative archaeal viruses from the mesopelagic ocean. PeerJ 2017, 5, e3428. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, M.; Santos, F.; Moreno-Paz, M.; Parro, V.; Antón, J. Unveiling viral–host interactions within the “microbial dark matter”. Nat. Commun. 2014, 5, 1–8. [Google Scholar] [CrossRef]
- Labonté, J.M.; Field, E.K.; Lau, M.; Chivian, D.; Van Heerden, E.; Wommack, K.E.; Kieft, T.L.; Onstott, T.C.; Stepanauskas, R. Single cell genomics indicates horizontal gene transfer and viral infections in a deep subsurface Firmicutes population. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and biogeochemical impacts of uncultivated globally abundant ocean viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef] [PubMed]
- NCBI Viral Genomes. Available online: https://www.ncbi.nlm.nih.gov/genome/viruses/ (accessed on 6 December 2017).
- Krupovic, M.; Cvirkaite-Krupovic, V.; Iranzo, J.; Prangishvili, D.; Koonin, E.V. Viruses of archaea: Structural, functional, environmental and evolutionary genomics. Virus Res. 2018, 244, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Bize, A.; Karlsson, E.A.; Ekefjard, K.; Quax, T.E.F.; Pina, M.; Prevost, M.; Forterre, P.; Tenaillon, O.; Bernander, R.; Prangishvili, D. A unique virus release mechanism in the Archaea. Proc. Natl. Acad. Sci. USA 2009, 106, 11306–11311. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.C.; Brumfield, S.K.; Peng, N.; She, Q.; Young, M.J. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J. Virol. 2011, 85, 6287–6292. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.C.; Young, M.J. Potential role of cellular ESCRT proteins in the STIV life cycle. Biochem. Soc. Trans. 2011, 39, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.C.; Samson, R.Y.; Brumfield, S.K.; Bell, S.D.; Young, M.J. Functional interplay between a virus and the ESCRT machinery in archaea. Proc. Natl. Acad. Sci. USA 2013, 110, 10783–10787. [Google Scholar] [CrossRef] [PubMed]
- Fouqueau, T.; Blombach, F.; Hartman, R.; Cheung, A.C.M.; Young, M.J.; Werner, F. The transcript cleavage factor paralogue TFS4 is a potent RNA polymerase inhibitor. Nat. Commun. 2017, 8, 1914. [Google Scholar] [CrossRef] [PubMed]
- Campos-Olivas, R.; Louis, J.M.; Clerot, D.; Gronenborn, B.; Gronenborn, A.M. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. USA 2002, 99, 10310–10315. [Google Scholar] [CrossRef] [PubMed]
- Oke, M.; Kerou, M.; Liu, H.; Peng, X.; Garrett, R.A.; Prangishvili, D.; Naismith, J.H.; White, M.F. A dimeric rep protein initiates replication of a linear Archaeal virus genome: Implications for the rep mechanism and viral replication. J. Virol. 2011, 85, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Pietila, M.K.; Laurinmaki, P.; Russell, D.A.; Ko, C.-C.; Jacobs-Sera, D.; Hendrix, R.W.; Bamford, D.H.; Butcher, S.J. Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Proc. Natl. Acad. Sci. USA 2013, 110, 10604–10609. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.; Tang, L.; Stedman, K.; Roberto, F.; Spuhler, J.; Gillitzer, E.; Johnson, J.E.; Douglas, T.; Young, M. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl. Acad. Sci. USA 2004, 101, 7716–7720. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, S.; Tschitschko, B.; Zhong, L.; Raftery, M.J.; Cavicchioli, R. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat. Microbiol. 2017, 2, 1446–1455. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P.; Da Cunha, V.; Catchpole, R. Plasmid vesicles mimicking virions. Nat. Microbiol. 2017, 2, 1340–1341. [Google Scholar] [CrossRef] [PubMed]
- Pietila, M.K.; Atanasova, N.S.; Manole, V.; Liljeroos, L.; Butcher, S.J.; Oksanen, H.M.; Bamford, D.H. Virion architecture unifies globally distributed pleolipoviruses infecting halophilic Archaea. J. Virol. 2012, 86, 5067–5079. [Google Scholar] [CrossRef] [PubMed]
- Bettstetter, M.; Peng, X.; Garrett, R.A.; Prangishvili, D. AFV1, a novel virus infecting hyperthermophilic archaea of the genus Acidianus. Virology 2003, 315, 68–79. [Google Scholar] [CrossRef]
- Mochizuki, T.; Yoshida, T.; Tanaka, R.; Forterre, P.; Sako, Y.; Prangishvili, D. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology 2010, 402, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Hochstein, R.; Amenabar, M.J.; Munson-McGee, J.H.; Boyd, E.S.; Young, M.J. Acidianus tailed spindle virus: a new archaeal large tailed spindle virus discovered by culture-independent methods. J. Virol. 2016, 90, 3458–3468. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D.; Vestergaard, G.; Häring, M.; Aramayo, R.; Basta, T.; Rachel, R.; Garrett, R.A. Structural and genomic properties of the hyperthermophilic Archaeal Virus ATV with an extracellular stage of the reproductive cycle. J. Mol. Biol. 2006, 359, 1203–1216. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, N.S.; Roine, E.; Oren, A.; Bamford, D.H.; Oksanen, H.M. Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. 2012, 14, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Weidenbach, K.; Nickel, L.; Neve, H.; Alkhnbashi, O.S.; Künzel, S.; Kupczok, A.; Bauersachs, T.; Cassidy, L.; Tholey, A.; Backofen, R.; et al. Methanosarcina Spherical Virus, a novel archaeal lytic virus targeting Methanosarcina strains. J. Virol. 2017, 91, e00955-17. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.; Reddy, V.; Asturias, F.; Khoshouei, M.; Johnson, J.E.; Manrique, P.; Munson-McGee, J.; Baumeister, W.; Lawrence, C.M.; Young, M.J. Isolation and characterization of metallosphaera turreted icosahedral virus, a founding member of a new family of archaeal viruses. J. Virol. 2017, 91, e00925-17. [Google Scholar] [CrossRef] [PubMed]
- Witte, A.; Baranyi, U.; Klein, R.; Sulzner, M.; Luo, C.; Wanner, G.; Krüger, D.H.; Lubitz, W. Characterization of Natronobacterium magadii phage phi Ch1, a unique archaeal phage containing DNA and RNA. Mol. Microbiol. Microbiol. 1997, 23, 603–616. [Google Scholar] [CrossRef]
- Reisser, W.; Burbank, D.E.; Meints, S.M.; Meints, R.H.; Becker, B.; Van Etten, J.L. A comparison of viruses infecting two different Chlorella-like green Algae. Virology 1988, 167, 143–149. [Google Scholar] [CrossRef]
- Olsen, R.H.; Siak, J.S.; Gray, R.H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J. Virol. 1974, 14, 689–699. [Google Scholar] [PubMed]
- Prangishvili, D.; Arnold, H.P.; Götz, D.; Ziese, U.; Holz, I.; Kristjansson, J.K.; Zillig, W. A novel virus family, the Rudiviridae: Structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics 1999, 152, 1387–1396. [Google Scholar] [PubMed]
- Palm, P.; Schleper, C.; Grampp, B.; Yeats, S.; McWilliam, P.; Reiter, W.D.; Zillig, W. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 1991, 185, 242–250. [Google Scholar] [CrossRef]
- Janekovic, D.; Wunderl, S.; Holz, I.; Zillig, W.; Gierl, A.; Neumann, H. TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic, anaerobic, sulfur reducing archaebacterium Thermoproteus tenax. MGG Mol. Gen. Genet. 1983, 192, 39–45. [Google Scholar] [CrossRef]
- Grazziotin, A.L.; Koonin, E.V.; Kristensen, D.M. Prokaryotic Virus Orthologous Groups (pVOGs): A resource for comparative genomics and protein family annotation. Nucleic Acids Res. 2017, 45, D491–D498. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, B.L.; Brum, J.R.; Sullivan, M.B. Depth-stratified functional and taxonomic niche specialization in the “core” and “flexible” Pacific Ocean Virome. ISME J. 2014, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Ignacio-espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S. Patterns and ecological drivers of ocean viral communities. Science 2015, 348, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, S.; Chan, L.K.; Egan, R.; Malmstrom, R.R.; McMahon, K.D.; Sullivan, M.B. Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Hallam, S.J.; Woyke, T.; Sullivan, M.B. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. eLife 2015, 4, e08490. [Google Scholar] [CrossRef] [PubMed]
- Dimaio, F.; Yu, X.; Rensen, E.; Krupovic, M.; Prangishvili, D.; Egelman, E.H. A virus that infects a hyperthermophile encapsidates A-form DNA. Science 2015, 348, 914–917. [Google Scholar] [CrossRef] [PubMed]
- Frenkiel-Krispin, D.; Sack, R.; Englander, J.; Shimoni, E.; Eisenstein, M.; Bullitt, E.; Horowitz-Scherer, R.; Hayes, C.S.; Setlow, P.; Minsky, A.; et al. Structure of the DNA-SspC Complex: Implications for DNA Packaging, Protection, and Repair in Bacterial Spores. J. Bacteriol. 2004, 186, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Ptchelkine, D.; Gillum, A.; Mochizuki, T.; Lucas-Staat, S.; Liu, Y.; Krupovic, M.; Phillips, S.E.V.; Prangishvili, D.; Huiskonen, J.T. Unique architecture of thermophilic archaeal virus APBV1 and its genome packaging. Nat. Commun. 2017, 8, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.C.; Brumfield, S.K.; Kerchner, K.M.; Quax, T.E.F.; Prangishvili, D.; Young, M.J. Insights into a viral lytic pathway from an archaeal virus-host system. J. Virol. 2013, 87, 2186–2192. [Google Scholar] [CrossRef] [PubMed]
- Quemin, E.R.J.; Chlanda, P.; Sachse, M.; Forterre, P.; Prangishvili, D.; Krupovic, M. Eukaryotic-like virus budding in archaea. mBio 2016, 7, e01439-16. [Google Scholar] [CrossRef] [PubMed]
- Hartman, R.; Bollschweiler, D.; Young, M.; Lawrence, M.C. Insights into first attachment of an archaeal virus using cryo-electron tomography. Unpublished work. 2018. [Google Scholar]
- Häring, M.; Vestergaard, G.; Rachel, R.; Chen, L.; Garrett, R.; Prangishvili, D. Independent virus development outside a host. Nature 2005, 436, 1102. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D.; Forterre, P.; Garrett, R. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 2006, 4, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, N.S.; Bamford, D.H.; Oksanen, H.M. Virus-host interplay in high salt environments. Environ. Microbiol. Rep. 2016, 8, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Demina, T.A.; Pietilä, M.K.; Svirskaitė, J.; Ravantti, J.J.; Atanasova, N.S.; Bamford, D.H.; Oksanen, H.M. HCIV-1 and other tailless icosahedral internal membrane-containing viruses of the family Sphaerolipoviridae. Viruses 2017, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Liu, Y.; Wang, Y.; Zhang, Z.; Oksanen, H.M.; Bamford, D.H.; Chen, X. Identification and characterization of SNJ2, the first temperate pleolipovirus integrating into the genome of the SNJ1-lysogenic archaeal strain. Mol. Microbiol. 2015, 98, 1002–1020. [Google Scholar] [CrossRef] [PubMed]
- Challberg, M.D.; Kelly, T.J. Adenovirus DNA replication in vitro: Origin and direction of daughter strand synthesis. J. Mol. Biol. 1979, 135, 999–1012. [Google Scholar] [CrossRef]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef] [PubMed]
- Saren, A.M.; Ravantti, J.J.; Benson, S.D.; Burnett, R.M.; Paulin, L.; Bamford, D.H.; Bamford, J.K.H. A snapshot of viral evolution from genome analysis of the Tectiviridae family. J. Mol. Biol. 2005, 350, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Qiu, J. Human parvovirus B19: A mechanistic overview of infection and DNA replication. Future Virol. 2005, 10, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Quemin, E.R.J.; Lucas, S.; Daum, B.; Quax, T.E.F.; Kuhlbrandt, W.; Forterre, P.; Albers, S.-V.; Prangishvili, D.; Krupovic, M. First insights into the entry process of hyperthermophilic archaeal viruses. J. Virol. 2013, 87, 13379–13385. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, S.; Le, S.; Bauer, M.; Garrett, R.A. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 2014, 91, 900–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birkenbihl, R.P.; Neef, K.; Prangishvili, D.; Kemper, B. Holliday junction resolving enzymes of archaeal viruses SIRV1 and SIRV2. J. Mol. Biol. 2001, 309, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Peng, N.; Han, W.; Li, Y.; Liang, Y.; She, Q. Genetic technologies for extremely thermophilic microorganisms of Sulfolobus, the only genetically tractable genus of crenarchaea. Sci. China Life Sci. 2017, 60, 370–385. [Google Scholar] [CrossRef] [PubMed]
- Stedman, K.M.; Schleper, C.; Rumpf, E.; Zillig, W. Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: Construction and testing of viral shuttle vectors. Genetics 1999, 152, 1397–1405. [Google Scholar] [PubMed]
- Iverson, E.A.; Goodman, D.A.; Gorchels, M.E.; Stedman, K.M. Genetic analysis of the major capsid protein of the archaeal Fusellovirus SSV1: Mutational flexibility and conformational change. Genes 2017, 8, 373. [Google Scholar] [CrossRef] [PubMed]
- Iverson, E.A.; Goodman, D.A.; Gorchels, M.E.; Stedman, K.M. Extreme mutation tolerance: Nearly half of the archaeal Fusellovirus Sulfolobus spindle-shaped Virus 1 genes are not required for virus function, Including the Minor Capsid Protein Gene vp3. J. Virol. 2017, 91, e02406-16. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.C.; Bolduc, B.; Bateson, M.M.; Young, M.J. The prevalence of STIV c92-like proteins in acidic thermal environments. Adv. Virol. 2011, 2011, 650930. [Google Scholar] [CrossRef] [PubMed]
- Selb, R.; Derntl, C.; Klein, R.; Alte, B.; Hofbauer, C.; Kaufmann, M.; Beraha, J.; Schöner, L.; Witte, A. The viral gene ORF79 encodes a repressor regulating induction of the lytic life cycle in the haloalkaliphilic virus ϕCh1. J. Virol. 2017, 91, e00206-17. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.T.; Reiter, D.; Young, M.; Lawrence, C.M. Structure of A197 from Sulfolobus turreted icosahedral virus: A crenarchaeal viral glycosyltransferase exhibiting the GT-A fold. J. Virol. 2006, 80, 7636–7644. [Google Scholar] [CrossRef] [PubMed]
- Franco, O.L.; Rigden, D.J. Fold recognition analysis of glycosyltransferase families: Further members of structural superfamilies. Glycobiology 2003, 13, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Dellas, N.; Snyder, J.C.; Dills, M.; Nicolay, S.J.; Kerchner, K.M.; Brumfield, S.K.; Lawrence, C.M.; Young, M. Structure-based mutagenesis of sulfolobus turreted icosahedral virus B204 reveals essential residues in the virion-associated DNA-packaging ATPase. J. Virol. 2016, 90, 2729–2739. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.T.; Eilers, B.; Menon, S.; Reiter, D.; Ortmann, A.; Young, M.J.; Lawrence, C.M. A winged-helix protein from sulfolobus turreted icosahedral virus points toward stabilizing disulfide bonds in the intracellular proteins of a hyperthermophilic virus. Virology 2007, 368, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.T.; Eilers, B.J.; Reiter, D.; Ortmann, A.C.; Young, M.J.; Lawrence, C.M. A new DNA binding protein highly conserved in diverse crenarchaeal viruses. Virology 2007, 363, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Lintner, N.G. Crenarchaeal virus-host systems: Structure-function studies of crenarchaeal viruses and prokaryotic adaptive immunity. Ph.D. Thesis, Montana State University, Bozeman MT, USA, March 2011. [Google Scholar]
- Veesler, D.; Ng, T. Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc. Natl. Acad. Sci. USA 2013, 110, 5504–5509. [Google Scholar] [CrossRef] [PubMed]
- Hochstein, R.; Bollschweiler, D.; Engelhardt, H.; Lawrence, C.M.; Young, M. Large tailed spindle viruses of Archaea: a new way of doing viral business. J. Virol. 2015, 89, 9146–9149. [Google Scholar] [CrossRef] [PubMed]
- Hochstein, R.; Bollschweiler, D.; Dharmavaram, S.; Lintner, N.G.; Plitzko, J.M.; Bruinsma, R.; Engelhardt, H.; Young, M.J.; Klug, W.S.; Lawrence, C.M. Structural studies of Acidianus tailed spindle virus reveal a structural paradigm used in the assembly of spindle-shaped viruses. Proc. Natl. Acad. Sci. USA 2018, 201719180. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Gregory, A.; Yilmaz, S. Contrasting life strategies of viruses that infect photo- and heterotrophic bacteria, as revealed by viral tagging. mBio 2013, 3, e00373-12. [Google Scholar] [CrossRef]
- Deng, L.; Ignacio-Espinoza, J.C.; Gregory, A.C.; Poulos, B.T.; Weitz, J.S.; Hugenholtz, P.; Sullivan, M.B. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 2014, 513, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Allers, E.; Moraru, C.; Duhaime, M.B.; Beneze, E.; Solonenko, N.; Barrero-Canosa, J.; Amann, R.; Sullivan, M.B. Single-cell and population level viral infection dynamics revealed by phage FISH, a method to visualize intracellular and free viruses. Env. Microbiol 2013, 15, 2306–2318. [Google Scholar] [CrossRef] [PubMed]
- Labonté, J.M.; Swan, B.K.; Poulos, B.; Luo, H.; Koren, S.; Hallam, S.J.; Sullivan, M.B.; Woyke, T.; Eric Wommack, K.; Stepanauskas, R. Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J. 2015, 9, 2386–2399. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Hawley, A.K.; Torres Beltran, M.; Scofield, M.; Schwientek, P.; Stepanauskas, R.; Woyke, T.; Hallam, S.J.; Sullivan, M.B. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta- genomics. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: mining viral signal from microbial genomic data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Ahlgren, N.A.; Ren, J.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. Alignment-free d2∗ oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 2017, 45, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, S.; Hasegawa, T.; Imachi, H.; Yamaguchi, T.; Harada, H.; Ohashi, A.; Kubota, K. Detection of single-copy functional genes in prokaryotic cells by two-pass TSA-FISH with polynucleotide probes. J. Microbiol. Methods 2012, 88, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Barrero-Canosa, J.; Moraru, C.; Zeugner, L.; Fuchs, B.M.; Amann, R. Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environ. Microbiol. 2017, 19, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Kim, E.-H.; Trubl, G.; Jones, R.M.; Roux, S.; VerBerkmoes, N.C.; Rich, V.I.; Sullivan, M.B. Illuminating structural proteins in viral “dark matter” with metaproteomics. Proc. Natl. Acad. Sci. USA 2016, 113, 201525139. [Google Scholar] [CrossRef] [PubMed]
- Auguet, J.C.; Barberan, A.; Casamayor, E.O. Global ecological patterns in uncultured Archaea. ISME J. 2010, 4, 182–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
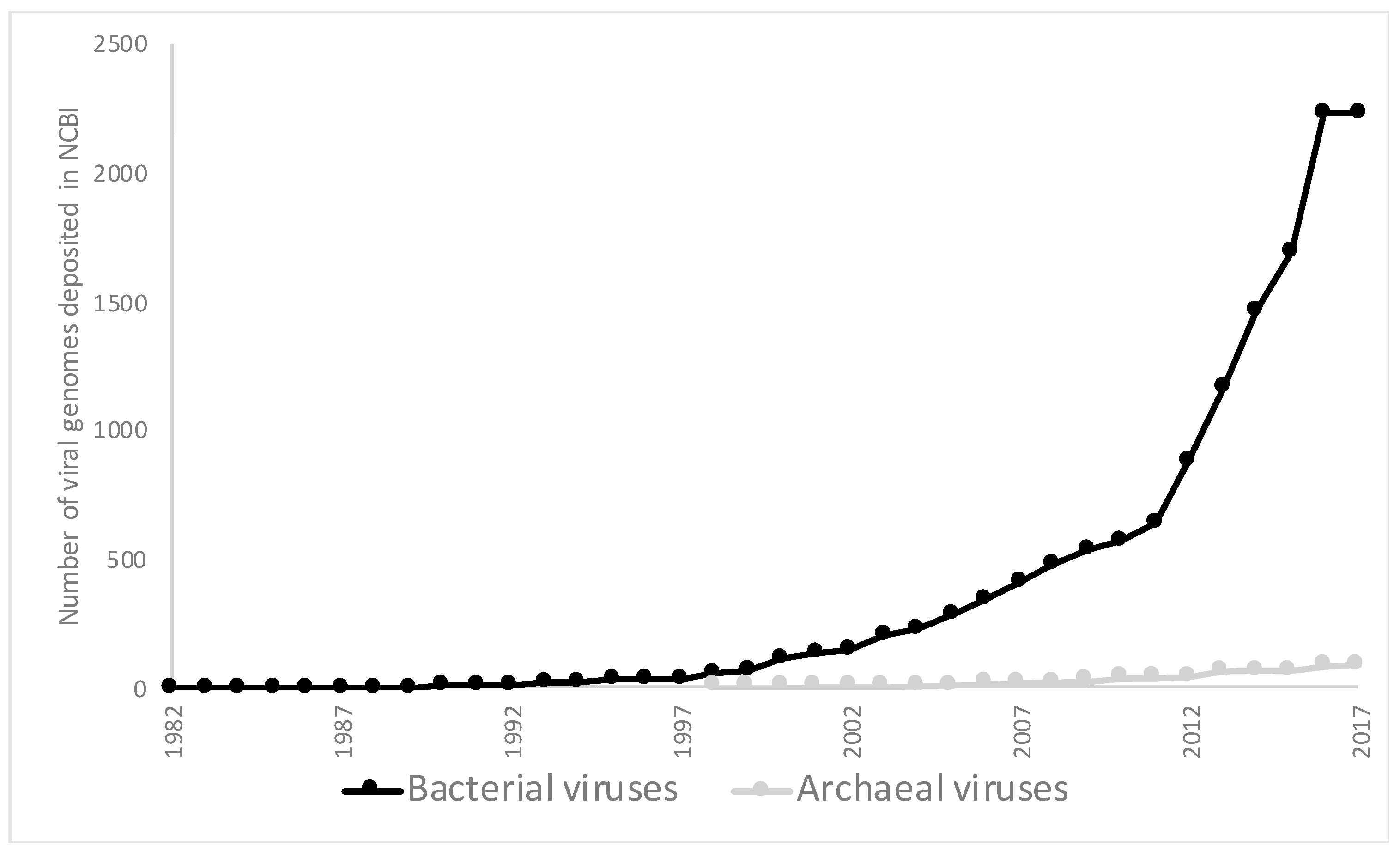
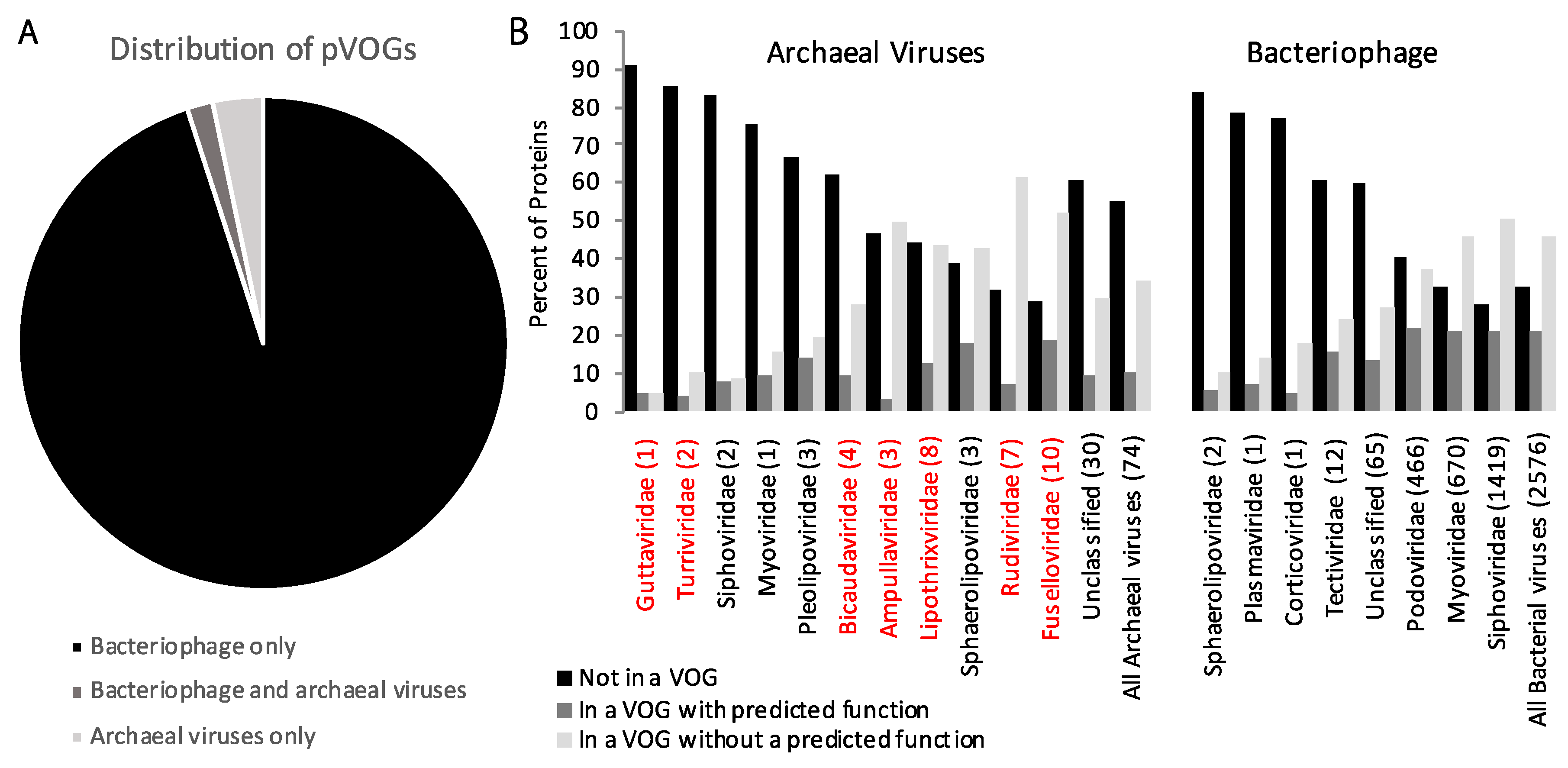
| Phyla | Number of Viruses in NCBI | Metagenomic Viruses |
|---|---|---|
| Candiditus Aenigmarchaeota | 0 | No |
| Candiditus Bathyarchaeota | 0 | Yes [21] |
| Crenarchaeota | 55 | Yes [13] |
| Candiditus Diapherotrites | 0 | No |
| Euryarchaeota | 32 | Yes [21,22,23] |
| Candiditus Geothermarchaeota | 0 | No |
| Candiditus Heimdallarchaeota | 0 | No |
| Candiditus Korarchaeota | 0 | No |
| Candiditus Lokiarchaeota | 0 | No |
| Candiditus Micrarchaeota | 0 | No |
| Nanoarchaeota | 0 | Yes [8] |
| Candiditus Nanohaloarchaeota | 0 | Yes [24] |
| Candiditus Odinarchaeota | 0 | No |
| Candiditus Pacearchaeota | 0 | No |
| Candiditus Parvarchaeota | 0 | No |
| Thaumarchaeota | 0 | Yes [23,25,26] |
| Candiditus Thorarchaeota | 0 | No |
| Candiditus Woesearchaeota | 0 | No |
| Abbreviation | Virus Name | Reference |
|---|---|---|
| AFV1 | Acidianus filamentous virus 1 | Bettstetter et al., 2003 [41] |
| APBV1 | Aeropyrum pernix bacilliform virus 1 | Mochizuki et al., 2010 [42] |
| ATSV | Acidianus tailed spindle virus | Hochstein et al., 2015 [43] |
| ATV | Acidianus two-tailed virus | Prangishvili et al., 2006 [44] |
| HHTV-2 | Haloarcula hispanica tailed virus 2 | Atanasova et al., 2012 [45] |
| HSTV-1 | Haloarcula sinaiiensis tailed virus 1 | Atanasova et al., 2012 [45] |
| MetSV | Methanosarcina spherical virus | Weidenbach et al., [46] |
| MTIV | Metallosphaera turreted icosahedral virus | Wagner et al., 2017 [47] |
| ϕCh1 | Natrialba magadii phi Ch1 | Witte et al., 1997 [48] |
| PBCV-1 | Paramecium bursaria chlorella virus 1 | Reisser et al., 1988 [49] |
| PRD1 | Phage PRD1 | Olsen et al., 1974 [50] |
| SIRV1 | Sulfolobus islandicus rod-shaped virus 1 | Prangishvili et al., 1999 [51] |
| SIRV2 | Sulfolobus islandicus rod-shaped virus 2 | Prangishvili et al., 1999 [51] |
| SSV1 | Sulfolobus spindle virus 1 | Palm et al., 1991 [52] |
| STIV | Sulfolobus turreted icosahedral virus | Rice et al., 2004 [37] |
| TTV-1 | Thermoproteus tenax virus 1 | Janekovic et al., 1983 [53] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munson-McGee, J.H.; Snyder, J.C.; Young, M.J. Archaeal Viruses from High-Temperature Environments. Genes 2018, 9, 128. https://doi.org/10.3390/genes9030128
Munson-McGee JH, Snyder JC, Young MJ. Archaeal Viruses from High-Temperature Environments. Genes. 2018; 9(3):128. https://doi.org/10.3390/genes9030128
Chicago/Turabian StyleMunson-McGee, Jacob H., Jamie C. Snyder, and Mark J. Young. 2018. "Archaeal Viruses from High-Temperature Environments" Genes 9, no. 3: 128. https://doi.org/10.3390/genes9030128
APA StyleMunson-McGee, J. H., Snyder, J. C., & Young, M. J. (2018). Archaeal Viruses from High-Temperature Environments. Genes, 9(3), 128. https://doi.org/10.3390/genes9030128




