Dynamics and Functional Potential of Stormwater Microorganisms Colonizing Sand Filters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Site and Protocol
2.2. Column Description, Set-Up, and Operation
2.3. Media and Culture Conditions
2.4. DNA Extraction and 16S rRNA Gene Library Protocol
2.5. Sequence Analysis and Quality Control
2.6. Statistical Analysis
2.7. Quantitative Polymerase Chain Reaction (qPCR)
3. Results
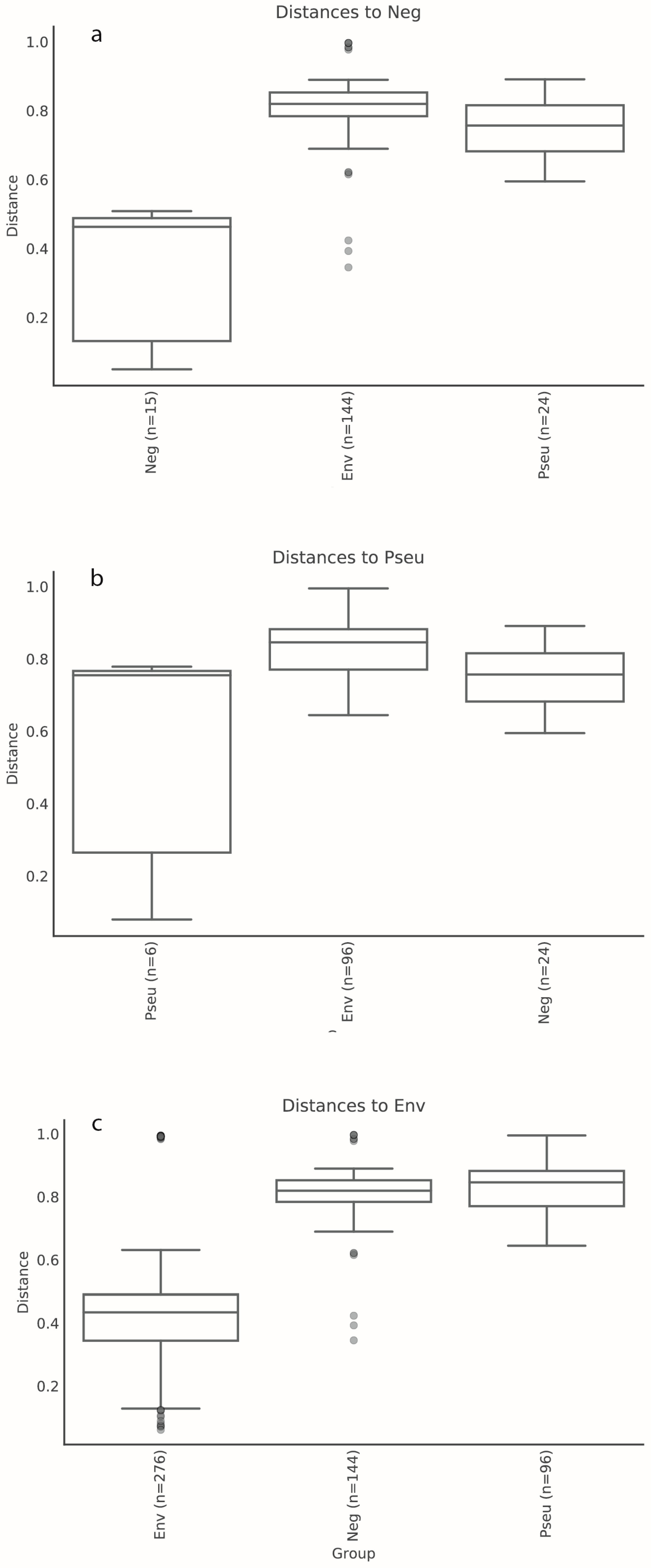
3.1. Stormwater-Inoculated Column Communities Are Distinct from Positive and Non-Inoculated Control Column Communities
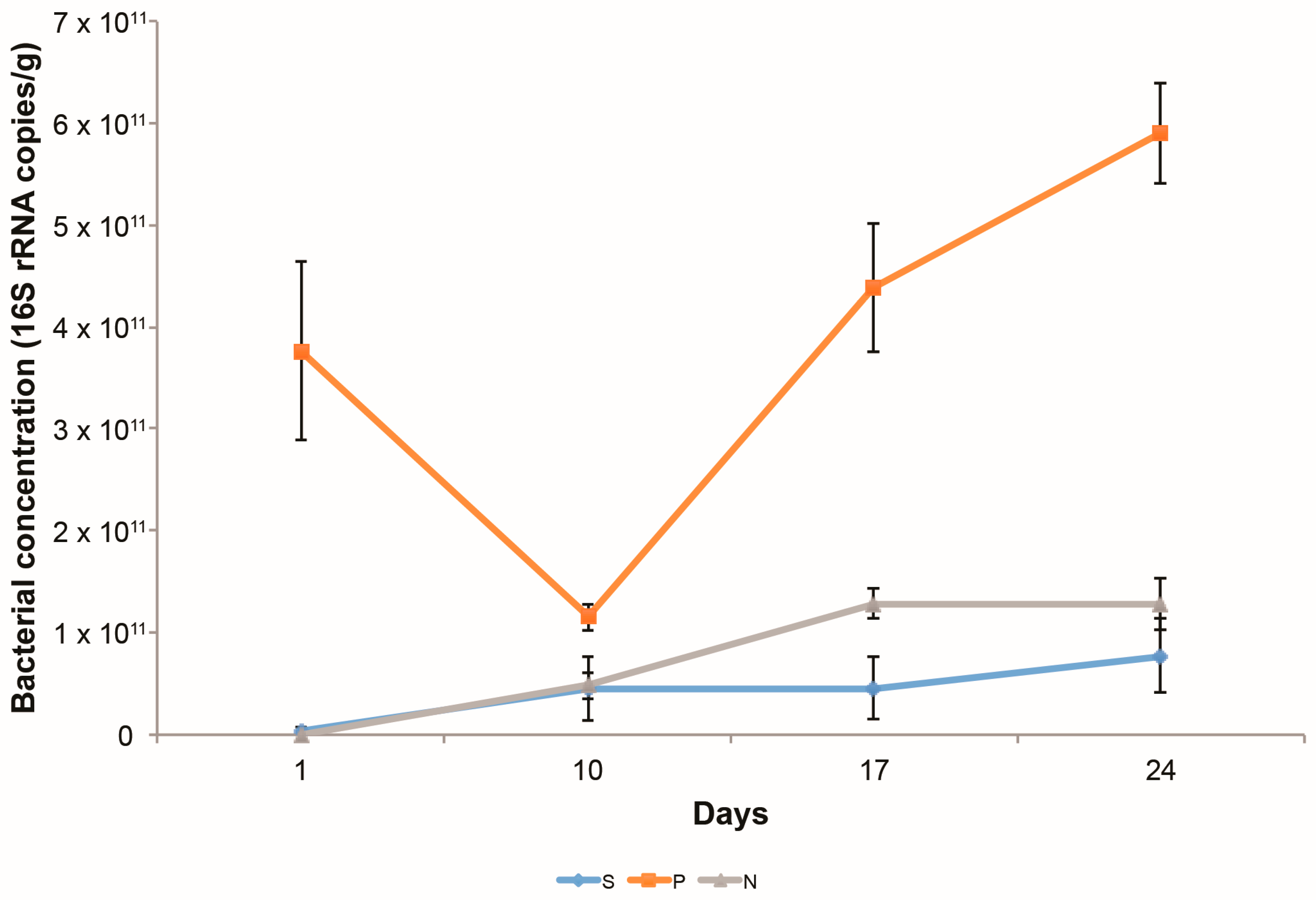
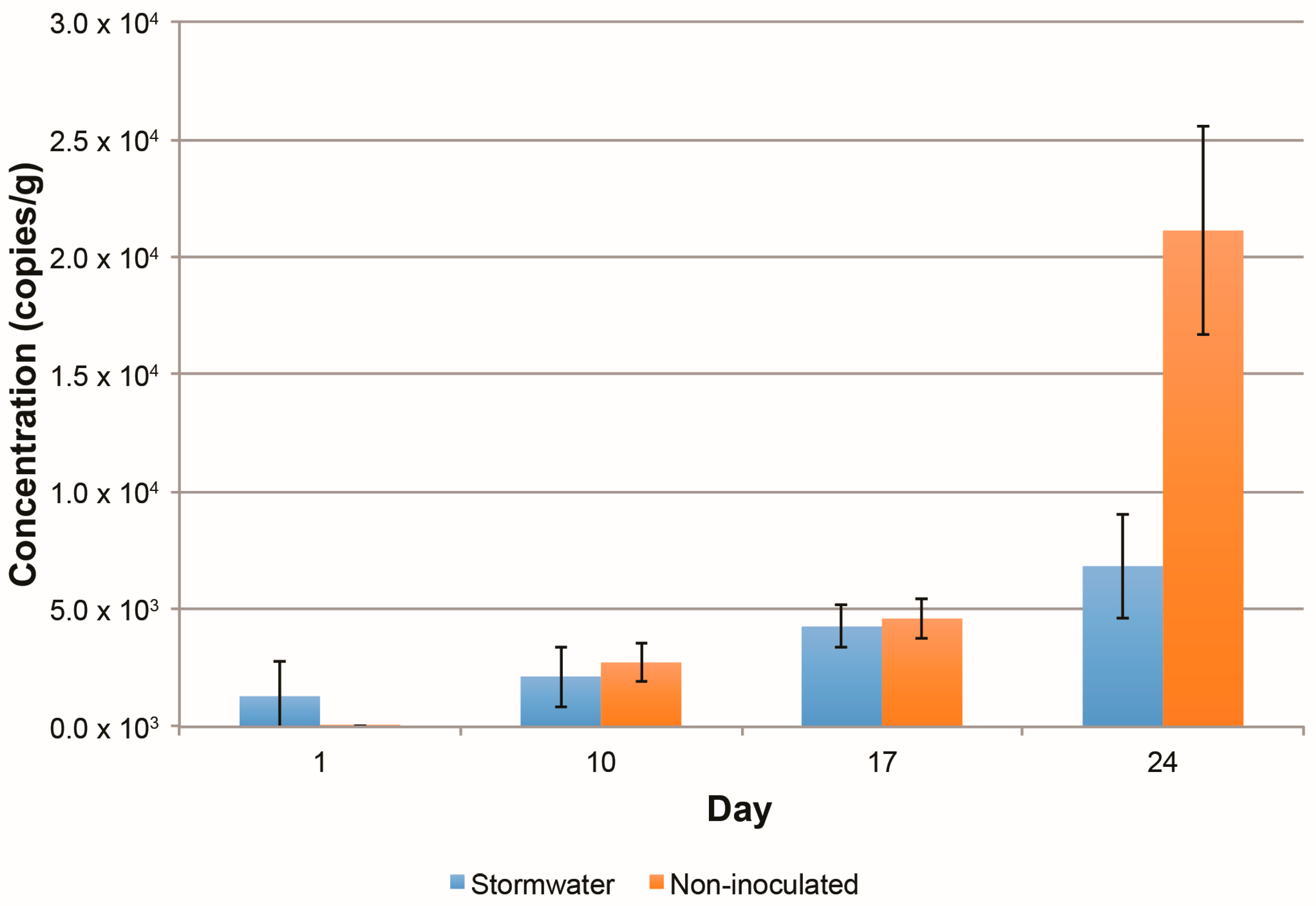
3.1.1. Bacterial Growth on Columns
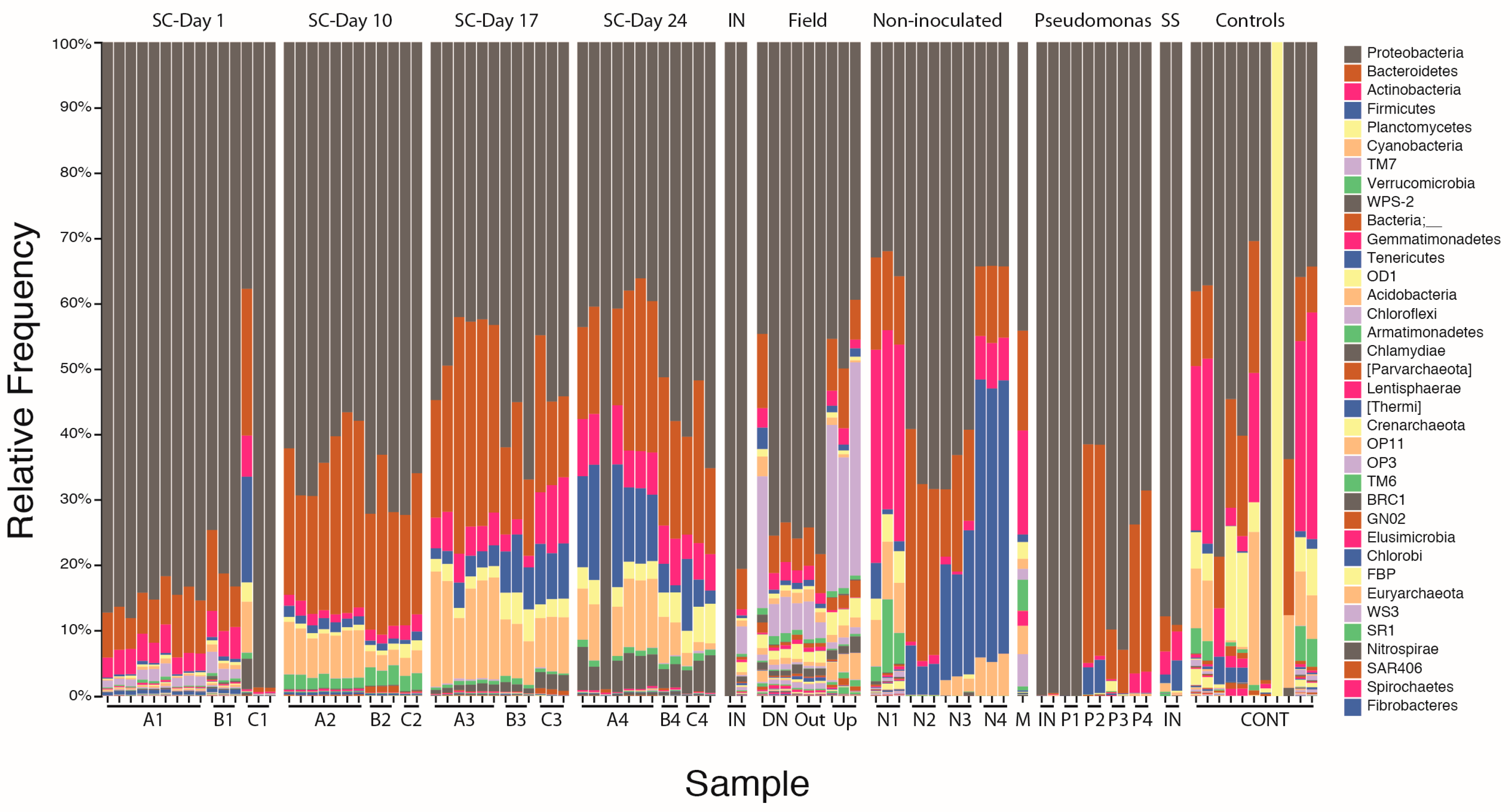
3.1.2. Bacterial Community Composition on Columns
3.2. Microbial Community Succession on Stormwater Columns
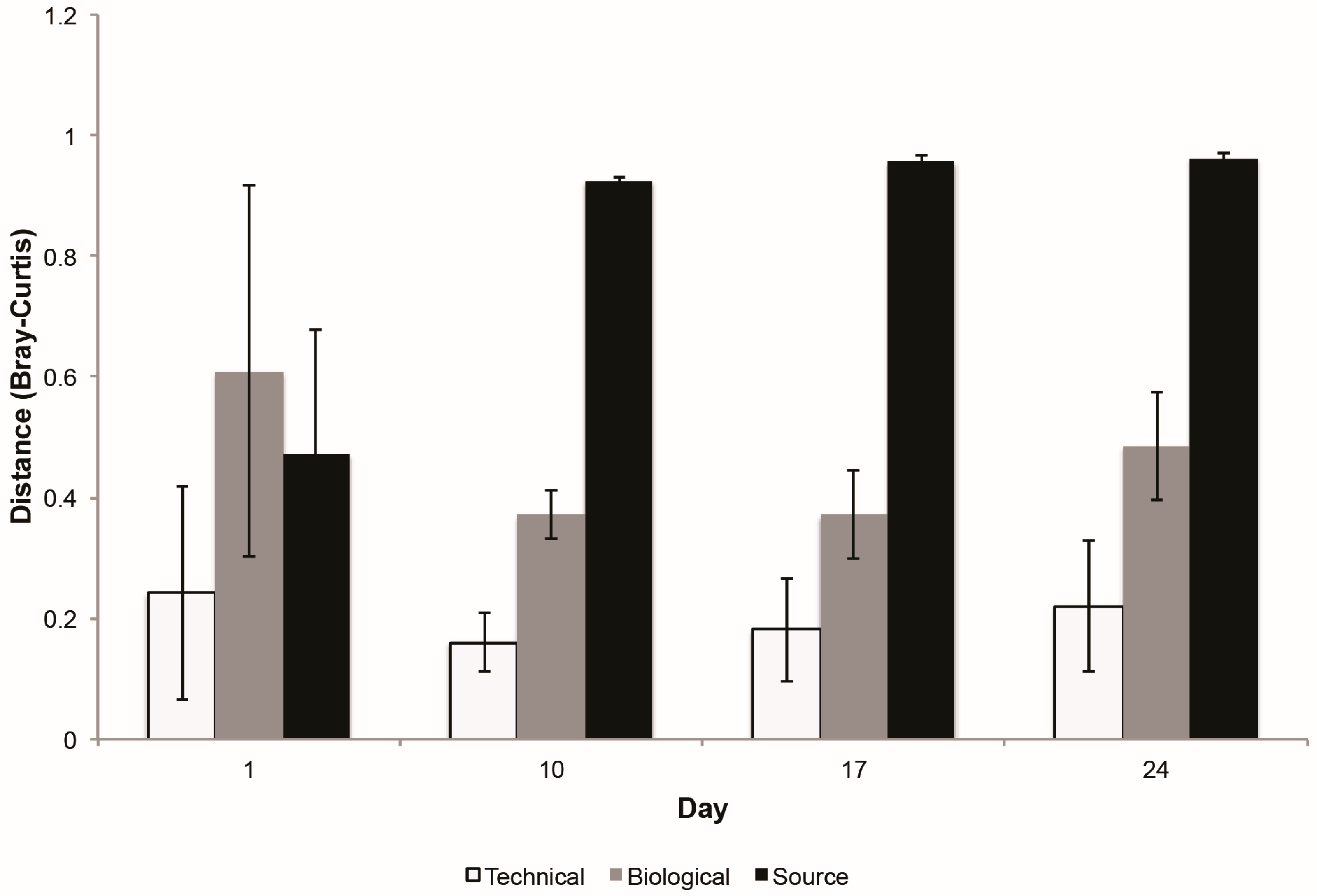
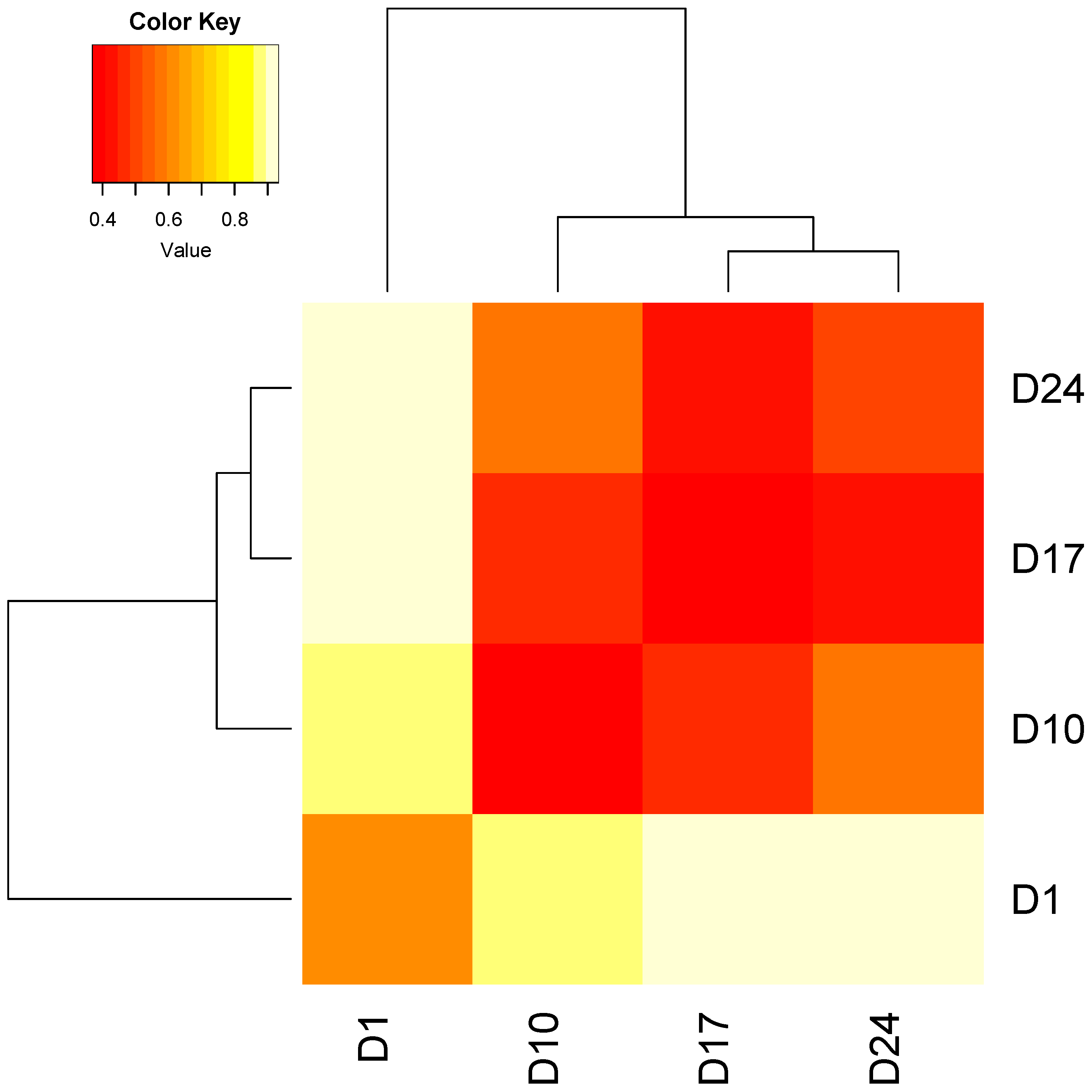
3.2.1. Technical Variability Is Less Than Biological Variability between Replicate Columns
3.2.2. Stability of Stormwater-Inoculated Columns over Time
3.2.3. Dynamics of Potential Pathogens and Denitrifying Bacteria
3.2.4. Changes in the Abundance of Denitrification Potential over Time
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Council, N.R. Urban Stormwater Management in the United States; The National Academies Press: Washington, DC, USA, 2009; p. 610. [Google Scholar] [CrossRef]
- Fraley-McNeal, L.; Schueler, T.; Winer, R. National Pollutant Removal Performance Database, Version 3; Center for Watershed Protection: Ellicott City, MD, USA, 2007; pp. 1–10. [Google Scholar]
- Grebel, J.E.; Mohanty, S.K.; Torkelson, A.A.; Boehm, A.B.; Higgins, C.P.; Maxwell, R.M.; Nelson, K.L.; Sedlak, D.L. Engineered Infiltration Systems for Urban Stormwater Reclamation. Environ. Eng. Sci. 2013, 30, 437–454. [Google Scholar] [CrossRef]
- Bekele, E.; Page, D.; Vanderzalm, J.; Kaksonen, A.; Gonzalez, D. Water Recycling via Aquifers for Sustainable Urban Water Quality Management: Current Status, Challenges and Opportunities. Water 2018, 10, 457. [Google Scholar] [CrossRef]
- Haig, S.J.; Schirmer, M.; D’Amore, R.; Gibbs, J.; Davies, R.L.; Collins, G.; Quince, C. Stable-isotope probing and metagenomics reveal predation by protozoa drives E-coli removal in slow sand filters. ISME J. 2015, 9, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Afrooz, A.R.M.N.; Boehm, A.B. Escherichia coli Removal in Biochar-Modified Biofilters: Effects of Biofilm. PLoS ONE 2016, 11, e0167489. [Google Scholar] [CrossRef] [PubMed]
- Waller, L.J.; Evanylo, G.K.; Krometis, L.A.H.; Strickland, M.S.; Wynn-Thompson, T.; Badgley, B.D. Engineered and Environmental Controls of Microbial Denitrification in Established Bioretention Cells. Environ. Sci. Technol. 2018, 52, 5358–5366. [Google Scholar] [CrossRef] [PubMed]
- Morse, N.R.; McPhillips, L.E.; Shapleigh, J.P.; Walter, M.T. The Role of Denitrification in Stormwater Detention Basin Treatment of Nitrogen. Environ. Sci. Technol. 2017, 51, 7928–7935. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, S.; Bradford, S.A.; Torkzaban, S.; Ye, X.Y.; Vanderzalm, J.; Du, X.Q.; Page, D. Unraveling the complexities of the velocity dependency of E-coli retention and release parameters in saturated porous media. Sci. Total Environ. 2017, 603, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Bozorg, A.; Gates, I.D.; Sen, A. Impact of biofilm on bacterial transport and deposition in porous media. J. Contam. Hydrol. 2015, 183, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Seagren, E.A.; Davis, A.P.; Karns, J.S. Long-Term Sustainability of Escherichia Coli Removal in Conventional Bioretention Media. J. Environ. Eng.-ASCE 2011, 137, 669–677. [Google Scholar] [CrossRef]
- Wennekes, P.L.; Rosindell, J.; Etienne, R.S. The Neutral-Niche Debate: A Philosophical Perspective. Acta Biotheor. 2012, 60, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teles, F.R.; Teles, R.P.; Uzel, N.G.; Song, X.Q.; Torresyap, G.; Socransky, S.S.; Haffajee, A.D. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J. Periodontal Res. 2012, 47, 95–104. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Maurice, C.; Carmody, R.; Gootenberg, D.; Button, J.; Wolfe, B.; Ling, A.; Devlin, A.; Varma, Y.; Fischbach, M.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannette, R.L.; Fukami, T. Historical contingency in species interactions: Towards niche-based predictions. Ecol. Lett. 2014, 17, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Vignola, M.; Werner, D.; Wade, M.J.; Meynet, P.; Davenport, R.J. Medium shapes the microbial community of water filters with implications for effluent quality. Water Res. 2018, 129, 499–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albers, C.N.; Ellegaard-Jensen, L.; Harder, C.B.; Rosendahl, S.; Knudsen, B.E.; Ekelund, F.; Aamand, J. Groundwater Chemistry Determines the Prokaryotic Community Structure of Waterworks Sand Filters. Environ. Sci. Technol. 2015, 49, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Siripattanakul, S.; Wirojanagud, W.; McEvoy, J.M.; Casey, F.X.M.; Khan, E. Atrazine removal in agricultural infiltrate by bioaugmented polyvinyl alcohol immobilized and free Agrobacterium radiobacter J14a: A sand column study. Chemosphere 2009, 74, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, G.M.; Martin, D.; Bingzhang, L.; Parzybok, T.; Yekta, M.; Riley, D. Precipitation-Frequency Atlas of the United States, Volume 2, Version 3.0; National Weather Service: Silver Spring, MD, USA, 2004. [Google Scholar]
- Department of Environmental Resources. Design Manual for Use of Bioretention in Stormwater Management; Department of Environmental Protection, Watershed Protection Branch: Prince George’s County, MD, USA, 1993.
- Mohanty, S.K.; Torkelson, A.A.; Dodd, H.; Nelson, K.L.; Boehm, A.B. Engineering Solutions to Improve the Removal of Fecal Indicator Bacteria by Bioinfiltration Systems during Intermittent Flow of Stormwater. Environ. Sci. Technol. 2013, 47, 10791–10798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, D.; Vanderzalm, J.; Miotlinski, K.; Barry, K.; Dillon, P.; Lawrie, K.; Brodie, R.S. Determining treatment requirements for turbid river water to avoid clogging of aquifer storage and recovery wells in siliceous alluvium. Water Res. 2014, 66, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: Chicheester, UL, USA, 1991; pp. 115–147. [Google Scholar]
- Preheim, S.P.; Olesen, S.W.; Spencer, S.J.; Materna, A.; Varadharajan, C.; Blackburn, M.; Friedman, J.; Rodríguez, J.; Hemond, H.; Alm, E.J. Surveys, simulation and single-cell assays relate function and phylogeny in a lake ecosystem. In Nature Microbiology; Macmillan Publishers Limited: Basingstoke, UK, 2016; Volume 1, p. 16130. [Google Scholar]
- Preheim, S.P.; Perrotta, A.R.; Martin-Platero, A.M.; Gupta, A.; Alm, E.J. Distribution-Based Clustering: Using Ecology To Refine the Operational Taxonomic Unit. Appl. Environ. Microbiol. 2013, 79, 6593–6603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Olesen, S.W.; Duvallet, C.; Alm, E.J. dbOTU3: A new implementation of distribution-based OTU calling. PLoS ONE 2017, 12, e0176335. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Assih, E.A.; Ouattara, A.S.; Thierry, S.; Cayol, J.L.; Labat, M.; Macarie, H. Stenotrophomonas acidaminiphila sp. nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int. J. Syst. Evol. Microbiol. 2002, 52, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Denton, M.; Kerr, K.G. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 1998, 11, 57–80. [Google Scholar] [PubMed]
- Drancourt, M.; Bollet, C.; Raoult, D. Stenotrophomonas africana sp nov, an opportunistic human pathogen in Africa. Int. J. Syst. Bacteriol. 1997, 47, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Dalsgaard, A.; Olsen, J.E. Phenotypic characterization and antibiotic resistance of Acinetobacter spp. isolated from aquatic sources. J. Appl. Microbiol. 1999, 87, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Seifert, H.; Dijkshoorn, L.; GernerSmidt, P.; Pelzer, N.; Tjernberg, I.; Vaneechoutte, M. Distribution of Acinetobacter species on human skin: Comparison of phenotypic and genotypic identification methods. J. Clin. Microbiol. 1997, 35, 2819–2825. [Google Scholar] [PubMed]
- Rodriguez, C.H.; Nastro, M.; Dabos, L.; Barberis, C.; Vay, C.; Famiglietti, A. First isolation of Acinetobacter johnsonii co-producing PER-2 and OXA-58 beta-lactamases. Diagn. Microbiol. Infect. Dis. 2014, 80, 341–342. [Google Scholar] [CrossRef] [PubMed]
- Seifert, H.; Strate, A.; Schulze, A.; Pulverer, G. Vascular catheter-related blood-stream infection due to Acinetobacter-johnsonii (formerly Acinetobacter calcoaceticus var iwoffii)—Report of 13 cases. Clin. Infect. Dis. 1993, 17, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Haig, S.J.; Quince, C.; Davies, R.L.; Dorea, C.C.; Collins, G. Replicating the microbial community and water quality performance of full-scale slow sand filters in laboratory-scale filters. Water Res. 2014, 61, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Albers, C.N.; Ellegaard-Jensen, L.; Hansen, L.H.; Sorensen, S.R. Bioaugmentation of rapid sand filters by microbiome priming with a nitrifying consortium will optimize production of drinking water from groundwater. Water Res. 2018, 129, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bowen, J.L.; Babbin, A.R.; Kearns, P.J.; Ward, B.B. Connecting the dots: Linking nitrogen cycle gene expression to nitrogen fluxes in marine sediment mesocosms. Front. Microbiol. 2014, 5, 429. [Google Scholar] [CrossRef] [PubMed]
- Tomasek, A.; Kozarek, J.L.; Hondzo, M.; Lurndahl, N.; Sadowsky, M.J.; Wang, P.; Staley, C. Environmental drivers of denitrification rates and denitrifying gene abundances in channels and riparian areas. Water Resour. Res. 2017, 53, 6523–6538. [Google Scholar] [CrossRef]
- Martiny, A.C.; Treseder, K.; Pusch, G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013, 7, 830–838. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fraser, A.N.; Zhang, Y.; Sakowski, E.G.; Preheim, S.P. Dynamics and Functional Potential of Stormwater Microorganisms Colonizing Sand Filters. Water 2018, 10, 1065. https://doi.org/10.3390/w10081065
Fraser AN, Zhang Y, Sakowski EG, Preheim SP. Dynamics and Functional Potential of Stormwater Microorganisms Colonizing Sand Filters. Water. 2018; 10(8):1065. https://doi.org/10.3390/w10081065
Chicago/Turabian StyleFraser, Andrea Naimah, Yue Zhang, Eric Gregory Sakowski, and Sarah Pacocha Preheim. 2018. "Dynamics and Functional Potential of Stormwater Microorganisms Colonizing Sand Filters" Water 10, no. 8: 1065. https://doi.org/10.3390/w10081065
APA StyleFraser, A. N., Zhang, Y., Sakowski, E. G., & Preheim, S. P. (2018). Dynamics and Functional Potential of Stormwater Microorganisms Colonizing Sand Filters. Water, 10(8), 1065. https://doi.org/10.3390/w10081065



