The Influences of O2 Availability on the Microbial Activities and Fe Transformations in the Iron Formation Caves of the Southern Espinhaço Range, Brazil
Abstract
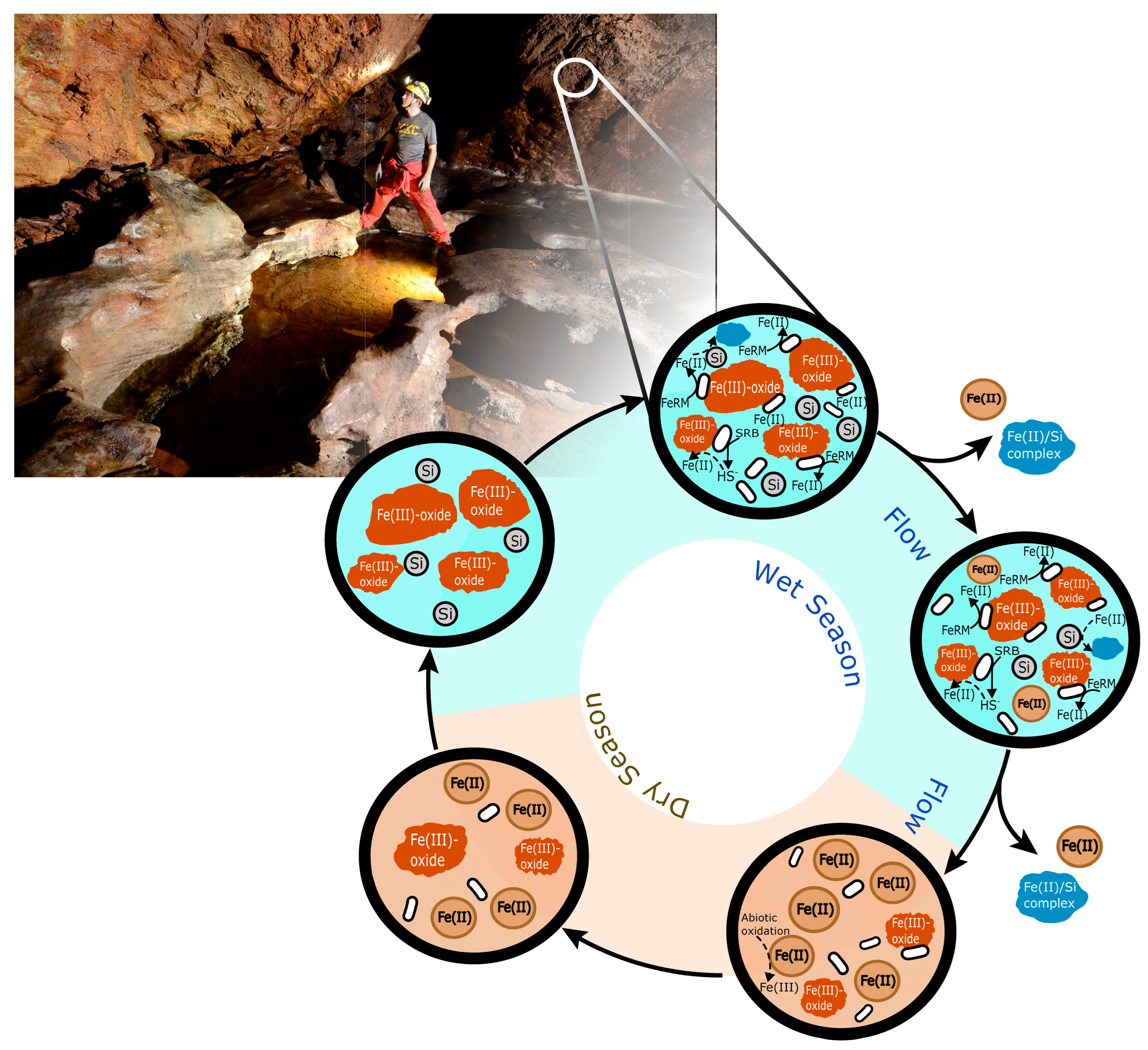
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation for Incubations
2.2. Batch Experiments and Fe(II) Measurement
2.3. 16S rRNA Gene Sequencing and Analysis
2.4. Shotgun Metagenomic Sequencing and Analysis
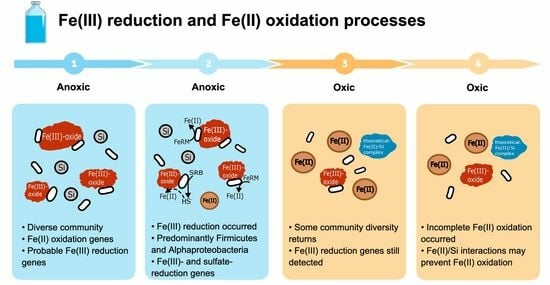
3. Results and Discussion
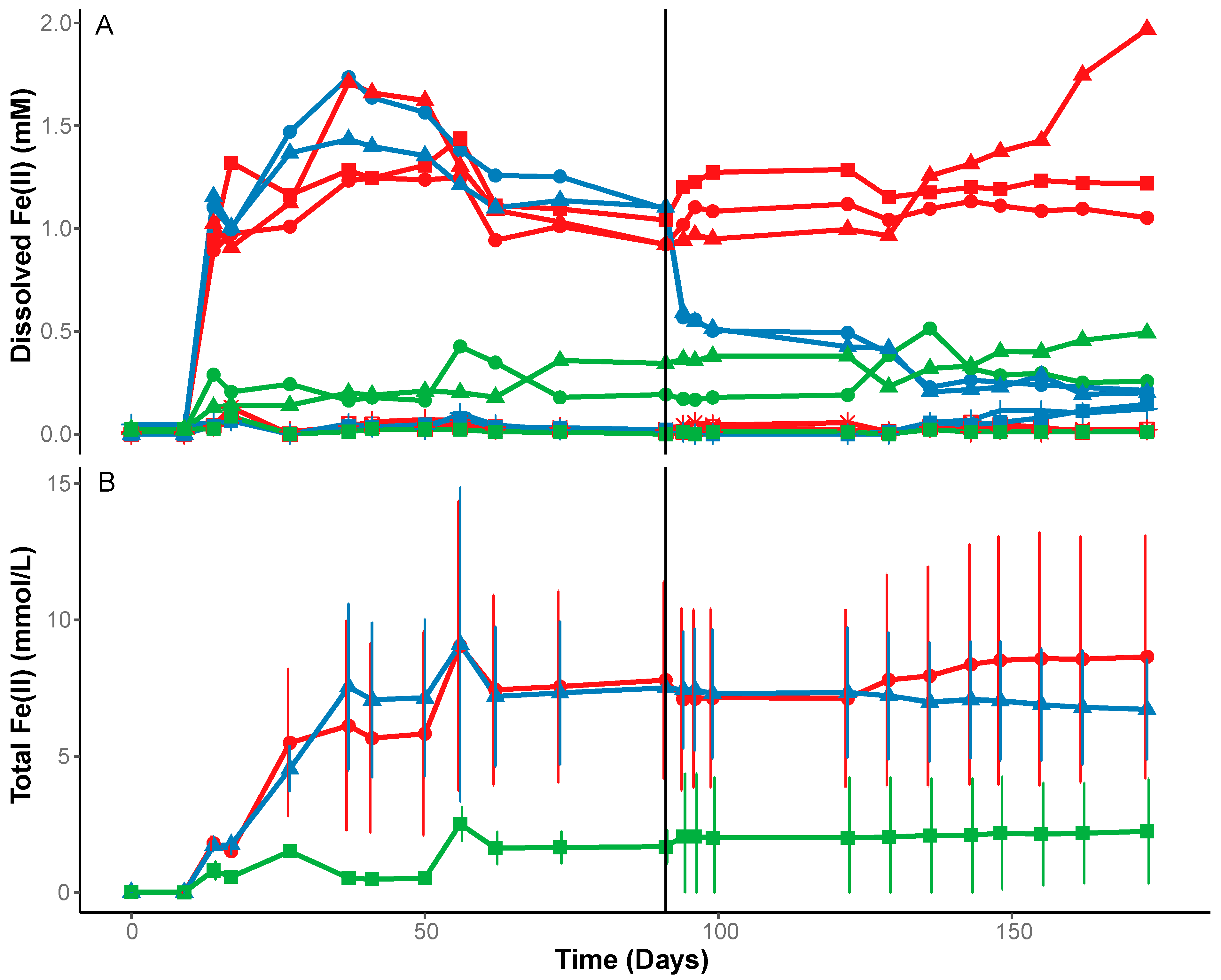
3.1. Fe(III) Reduction under Anoxic Conditions
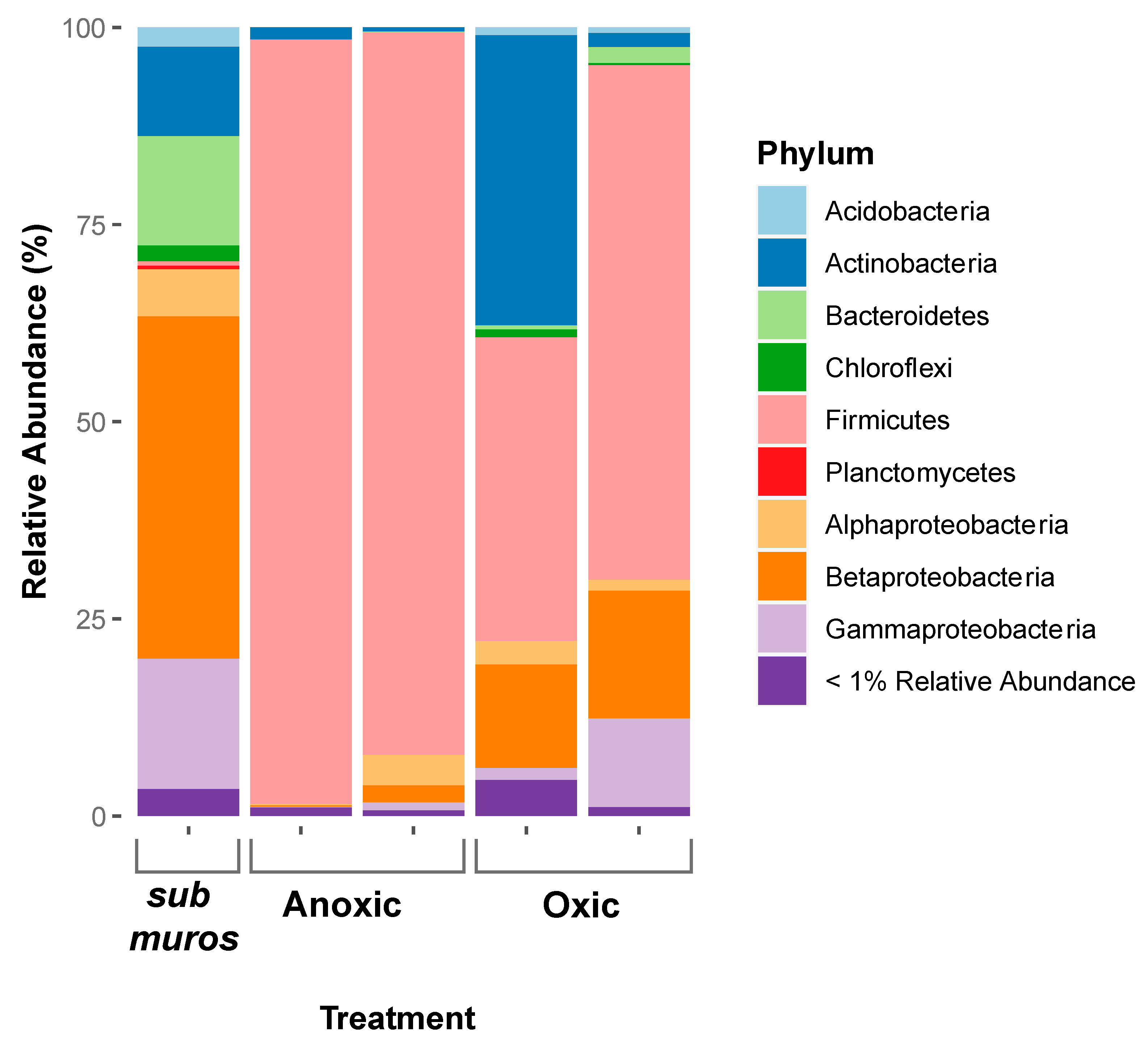
3.2. Changes in the Microbial Community Composition
3.3. Metabolic Potential of Anoxic and Oxic Treatments
3.4. Metagenome Assembled Genome Analyses
3.5. Fe(III)-Reducing Potential of Acidobacteria
3.6. Uncharacterized Fe(III)-Reducing Potential of Alphaproteobacteria
3.7. The Effects of Aeration on the Microbial Community
3.8. Other Metabolic Characteristics of MAGs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parker, C.W.; Wolf, J.A.; Auler, A.S.; Barton, H.A.; Senko, J.M. Microbial Reducibility of Fe(III) Phases Associated with the Genesis of Iron Ore Caves in the Iron Quadrangle, Minas Gerais, Brazil. Minerals 2013, 3, 395–411. [Google Scholar] [CrossRef]
- Shuster, D.L.; Farley, K.A.; Vasconcelos, P.M.; Balco, G.; Monteiro, H.S.; Waltenberg, K.; Stone, J.O. Cosmogenic 3He in Hematite and Goethite from Brazilian “Canga” Duricrust Demonstrates the Extreme Stability of These Surfaces. Earth Planet. Sci. Lett. 2012, 329–330, 41–50. [Google Scholar] [CrossRef]
- Dorr, J.V.N. Supergene Iron Ores of Minas Gerais, Brasil. Econ. Geol. 1964, 59, 1203–1240. [Google Scholar] [CrossRef]
- Parker, C.W.; Auler, A.S.; Barton, M.D.; Sasowsky, I.D.; Senko, J.M.; Barton, H.A. Fe(III) Reducing Microorganisms from Iron Ore Caves Demonstrate Fermentative Fe(III) Reduction and Promote Cave Formation. Geomicrobiol. J. 2018, 35, 311–322. [Google Scholar] [CrossRef]
- Piló, L.B.; Auler, A. Geoespeleologia das Cavernas em Rochas Ferriferas da Regiao de Carajas, PA. In Proceedings of the ANAIS do XXX Congresso Brasileiro de Espeleologia, Montes Carlos, Brazil, 9–12 July 2009; pp. 181–186. [Google Scholar]
- Auler, A.S.; Parker, C.W.; Barton, H.A.; Soares, G.A. Iron Formation Caves: Genesis and Ecology. In Encyclopedia of Caves; Academic Press: Cambridge, MA, USA, 2019; pp. 559–566. [Google Scholar] [CrossRef]
- Auler, A.S.; Piló, L.B.; Parker, C.W.; Senko, J.M.; Sasowsky, I.D.; Barton, H.A. Hypogene Cave Patterns in Iron Ore Caves: Convergence of Forms or Processes; Karst Waters Institute Special Publication: Leesburg, VA, USA, 2014; pp. 15–19. [Google Scholar]
- Auler, A.S.; Barton, H.A.; Zambelli, B.; Senko, J.; Parker, C.W.; Sasowsky, I.D.; Souza, T.A.R.; Pujoni, D.; Peñaranda, J.; Davis, R. Silica and Iron Mobilization, Cave Development and Landscape Evolution in Iron Formations in Brazil. Geomorphology 2022, 398, 108068. [Google Scholar] [CrossRef]
- Gagen, E.J.; Zaugg, J.; Tyson, G.W.; Southam, G. Goethite Reduction by a Neutrophilic Member of the Alphaproteobacterial Genus Telmatospirillum. Front. Microbiol. 2019, 10, 02938. [Google Scholar] [CrossRef]
- Levett, A.; Gagen, E.; Shuster, J.; Rintoul, L.; Tobin, M.; Vongsvivut, J.; Bambery, K.; Vasconcelos, P.; Southam, G. Evidence of Biogeochemical Processes in Iron Duricrust Formation. J. S. Am. Earth Sci. 2016, 71, 131–142. [Google Scholar] [CrossRef]
- Gagen, E.J.; Levett, A.; Paz, A.; Gastauer, M.; Caldeira, C.F.; Valadares, R.B.d.S.; Bitencourt, J.A.P.; Alves, R.; Oliveira, G.; Siqueira, J.O.; et al. Biogeochemical Processes in Canga Ecosystems: Armoring of Iron Ore against Erosion and Importance in Iron Duricrust Restoration in Brazil. Ore Geol. Rev. 2019, 107, 573–586. [Google Scholar] [CrossRef]
- Levett, A.; Vasconcelos, P.M.; Gagen, E.J.; Rintoul, L.; Spier, C.; Guagliardo, P.; Southam, G. Microbial Weathering Signatures in Lateritic Ferruginous Duricrusts. Earth Planet. Sci. Lett. 2020, 538, 116209. [Google Scholar] [CrossRef]
- Calapa, K.A.; Mulford, M.K.; Rieman, T.D.; Senko, J.M.; Auler, A.S.; Parker, C.W.; Barton, H.A. Hydrologic Alteration and Enhanced Microbial Reductive Dissolution of Fe(III) (Hydr)Oxides Under Flow Conditions in Fe(III)-Rich Rocks: Contribution to Cave-Forming Processes. Front. Microbiol. 2021, 12, 696534. [Google Scholar] [CrossRef]
- Parker, C.W.; Senko, J.M.; Auler, A.S.; Sasowsky, I.D.; Schulz, F.; Woyke, T.; Barton, H.A. Enhanced Terrestrial Fe(II) Mobilization Identified through a Novel Mechanism of Microbially Driven Cave Formation in Fe(III)-Rich Rocks. Sci. Rep. 2022, 12, 17062. [Google Scholar] [CrossRef] [PubMed]
- Wolthoorn, A.; Temminghoff, E.J.M.; Weng, L.; Van Riemsdijk, W.H. Colloid Formation in Groundwater: Effect of Phosphate, Manganese, Silicate and Dissolved Organic Matter on the Dynamic Heterogeneous Oxidation of Ferrous Iron. Appl. Geochem. 2004, 19, 611–622. [Google Scholar] [CrossRef]
- Monteiro, H.S.; Vasconcelos, P.M.P.; Farley, K.A. A Combined (U-Th)/He and Cosmogenic 3He Record of Landscape Armoring by Biogeochemical Iron Cycling. J. Geophys. Res. Earth Surf. 2018, 123, 298–323. [Google Scholar] [CrossRef]
- Levett, A.; Gagen, E.J.; Vasconcelos, P.M.; Zhao, Y.; Paz, A.; Southam, G. Biogeochemical Cycling of Iron: Implications for Biocementation and Slope Stabilisation. Sci. Total Environ. 2020, 707, 136128. [Google Scholar] [CrossRef] [PubMed]
- Kinsela, A.S.; Jones, A.M.; Bligh, M.W.; Pham, A.N.; Collins, R.N.; Harrison, J.J.; Wilsher, K.L.; Payne, T.E.; Waite, T.D. Influence of Dissolved Silicate on Rates of Fe(II) Oxidation. Environ. Sci. Technol. 2016, 50, 11663–11671. [Google Scholar] [CrossRef] [PubMed]
- Urrutia, M.M.; Roden, E.E.; Zachara, J.M. Influence of Aqueous and Solid-Phase Fe(II) Complexants on Microbial Reduction of Crystalline Iron(II) Oxides. Environ. Sci. Technol. 1999, 33, 4022–4028. [Google Scholar] [CrossRef]
- Stookey, L.L. Ferrozine—A New Spectrophotometric Reagent for Iron. Anal. Chem. 1970, 42, 779–781. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S RRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Amir, A.; Daniel, M.; Navas-Molina, J.; Kopylova, E.; Morton, J.; Xu, Z.Z.; Eric, K.; Thompson, L.; Hyde, E.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. Am. Soc. Microbiol. 2017, 2, 10–1128. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-Filtering Vastly Improves Diversity Estimates from Illumina Amplicon Sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2′s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F.; et al. The Biological Observation Matrix (BIOM) Format or: How I Learned to Stop Worrying and Love the Ome-Ome. Gigascience 2012, 464, 2047-217X-1-7. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Haendiges, J.; Gonzalez-Escalona, N.; Timme, R.; Balkey, M. Illumina DNA Prep (M) Tagmentation Library Preparation for Use on an Illumina MiSeq Sequencer.2020. Available online: https://www.researchgate.net/publication/354560175_Illumina_DNA_Prep_M_Tagmentation_Library_Preparation_for_use_on_an_Illumina_MiSeq_Sequencer_v2 (accessed on 14 April 2022).
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an Efficient Tool for Accurately Reconstructing Single Genomes from Complex Microbial Communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef]
- Chaumeil, P.; Mussing, A.; Hugenholtz, P.; Parks, D. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.-A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A Complete Domain-to-Species Taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and Sensitive Taxonomic Classification for Metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive Metagenomic Visualization in a Web Browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Garber, A.I.; Nealson, K.H.; Okamoto, A.; McAllister, S.M.; Chan, C.S.; Barco, R.A.; Merino, N. FeGenie: A Comprehensive Tool for the Identification of Iron Genes and Iron Gene Neighborhoods in Genome and Metagenome Assemblies. Front. Microbiol. 2020, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Garber, A.I.; Ramirez, G.; Merino, N.; Pavia, M.; McAllister, S. MagicLamp: Toolkit for Annotation of ’omics Datasets Using Curated HMM Setes. 2020. Available online: https://github.com/Arkadiy-Garber/MagicLamp (accessed on 14 April 2022).
- Wilke, C.O. Cowplot: Streamlined Plot Theme and Plot Annotations for “Ggplot2”; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Roden, E.E.; Zachara, J.M. Microbial Reduction of Crystalline Iron(III) Oxides: Influence of Oxide Surface Area and Potential for Cell Growth. Environ. Sci. Technol. 1996, 30, 1618–1628. [Google Scholar] [CrossRef]
- Roden, E.E.; Urrutia, M.M. Ferrous Iron Removal Promotes Microbial Reduction of Crystalline Iron(III) Oxides. Environ. Sci. Technol. 1999, 33, 1847–1853. [Google Scholar] [CrossRef]
- Vu, H.P.; Shaw, S.; Brinza, L.; Benning, L.G. Crystallization of Hematite (α-Fe2O3) under Alkaline Condition: The Effects of Pb. Cryst. Growth Des. 2010, 10, 1544–1551. [Google Scholar] [CrossRef]
- Fischer, M.A.; Güllert, S.; Neulinger, S.C.; Streit, W.R.; Schmitz, R.A. Evaluation of 16S RRNA Gene Primer Pairs for Monitoring Microbial Community Structures Showed High Reproducibility within and Low Comparability between Datasets Generated with Multiple Archaeal and Bacterial Primer Pairs. Front. Microbiol. 2016, 7, 211583. [Google Scholar] [CrossRef]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Mehta-Kolte, M.G.; Bond, D.R. Geothrix Fermentans Secretes Two Different Redox-Active Compounds to Utilize Electron Acceptors across a Wide Range of Redox Potentials. Appl. Environ. Microbiol. 2012, 78, 6987–6995. [Google Scholar] [CrossRef] [PubMed]
- Fuller, S.J.; McMillan, D.G.G.; Renz, M.B.; Schmidt, M.; Burke, I.T.; Stewart, D.I. Extracellular Electron Transport-Mediated Fe(III) Reduction by a Community of Alkaliphilic Bacteria That Use Flavins as Electron Shuttles. Appl. Environ. Microbiol. 2014, 80, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Mehta, T.; Coppi, M.V.; Childers, S.E.; Lovley, D.R. Outer Membrane C-Type Cytochromes Required for Fe(III) and Mn(IV) Oxide Reduction in Geobacter Sulfurreducens. Appl. Environ. Microbiol. 2005, 71, 8634–8641. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Lin, C.C.; Kukkadapu, R.; Engelhard, M.H.; Zhao, X.; Wang, Y.; Barkay, T.; Yee, N. Syntrophic Effects in a Subsurface Clostridial Consortium on Fe(III)-(Oxyhydr)Oxide Reduction and Secondary Mineralization. Geomicrobiol. J. 2014, 31, 101–115. [Google Scholar] [CrossRef]
- List, C.; Hosseini, Z.; Lederballe Meibom, K.; Hatzimanikatis, V.; Bernier-Latmani, R. Impact of Iron Reduction on the Metabolism of Clostridium acetobutylicum. Environ. Microbiol. 2019, 21, 3548–3563. [Google Scholar] [CrossRef] [PubMed]
- Morra, S. Fantastic [FeFe]-Hydrogenases and Where to Find Them. Front. Microbiol. 2022, 13, 853626. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, W.A.M.; Van Dongen, W.M.A.M.; Veeger, C. Reduction of the Amount of Periplasmic Hydrogenase in Desulfovibrio vulgaris (Hildenborough) with Antisense RNA: Direct Evidence for an Important Role of This Hydrogenase in Lactate Metabolism. J. Bacteriol. 1991, 173, 3688–3694. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhou, Y.; Rittmann, B.E. Reductive Precipitation of Sulfate and Soluble Fe(III) by Desulfovibrio vulgaris: Electron Donor Regulates Intracellular Electron Flow and Nano-FeS Crystallization. Water Res. 2017, 119, 91–101. [Google Scholar] [CrossRef]
- Lentini, C.J.; Wankel, S.D.; Hansel, C.M. Enriched Iron(III)-Reducing Bacterial Communities Are Shaped by Carbon Substrate and Iron Oxide Mineralogy. Front. Microbiol. 2012, 3, 36789. [Google Scholar] [CrossRef]
- Parker, C.W.; Senko, J.; Sasowsky, I.D.; Auler, A.; Barton, H.A. Can Fermenting Bacteria Drive Speleogenesis in Fe(III)-Rich Rocks? In Proceedings of the Union Internationale de Speleologie 17th Annuyal International Congress of Speleology, Sydney, Australia, 23–29 July 2017; pp. 399–400. [Google Scholar]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef]
- Coupland, K.; Johnson, D.B. Evidence That the Potential for Dissimilatory Ferric Iron Reduction Is Widespread among Acidophilic Heterotrophic Bacteria. FEMS Microbiol. Lett. 2008, 279, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Nevin, K.P.; Lovley, D.R. Mechanisms for Accessing Insoluble Fe(III) Oxide during Dissimilatory Fe(III) Reduction by Geothrix fermentans. Appl. Environ. Microbiol. 2002, 68, 2294–2299. [Google Scholar] [CrossRef] [PubMed]
- Coates, J.D.; Ellis, D.J.; Gaw, C.V.; Lovley, D.R. Geothrix fermentans gen. nov., sp. nov., a Novel Fe(III)-Reducing Bacterium from a Hydrocarbon-Contaminated Aquifer. Int. J. Syst. Bacteriol. 1999, 49, 1615–1622. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Vieira, S.; Bunk, B.; Riedel, T.; Spröer, C.; Overmann, J. First Complete Genome Sequence of a Subdivision 6 Acidobacterium Strain. Genome Announc. 2016, 4, 10-1128. [Google Scholar] [CrossRef]
- Wasmund, K.; Mußmann, M.; Loy, A. The Life Sulfuric: Microbial Ecology of Sulfur Cycling in Marine Sediments. Environ. Microbiol. Rep. 2017, 9, 323–344. [Google Scholar] [CrossRef]
- Anantharaman, K.; Hausmann, B.; Jungbluth, S.P.; Kantor, R.S.; Lavy, A.; Warren, L.A.; Rappé, M.S.; Pester, M.; Loy, A.; Thomas, B.C.; et al. Expanded Diversity of Microbial Groups That Shape the Dissimilatory Sulfur Cycle. ISME J. 2018, 12, 1715–1728. [Google Scholar] [CrossRef]
- Hausmann, B.; Pelikan, C.; Herbold, C.W.; Köstlbacher, S.; Albertsen, M.; Eichorst, S.A.; Glavina Del Rio, T.; Huemer, M.; Nielsen, P.H.; Rattei, T.; et al. Peatland Acidobacteria with a Dissimilatory Sulfur Metabolism. ISME J. 2018, 12, 1729–1742. [Google Scholar] [CrossRef]
- Kappler, A.; Bryce, C.; Mansor, M.; Lueder, U.; Byrne, J.M.; Swanner, E.D. An Evolving View on Biogeochemical Cycling of Iron. Nat. Rev. Microbiol. 2021, 19, 360–374. [Google Scholar] [CrossRef]
- Jackson, B.E.; McInerney, M.J. Anaerobic Microbial Metabolism Can Proceed Close to Thermodynamic Limits. Nature 2002, 415, 454–456. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 171794. [Google Scholar] [CrossRef] [PubMed]
- Sait, M.; Davis, K.E.R.; Janssen, P.H. Effect of PH on Isolation and Distribution of Members of Subdivision 1 of the Phylum Acidobacteria Occurring in Soil. Appl. Environ. Microbiol. 2006, 72, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Dolfing, J.; Guo, Z.; Chen, R.; Wu, M.; Li, Z.; Lin, X.; Feng, Y. Important Ecophysiological Roles of Non-Dominant Actinobacteria in Plant Residue Decomposition, Especially in Less Fertile Soils. Microbiome 2021, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, P.; Sankar, P.M. Culture-Independent and Culture-Dependent Approaches in Symbiont Analysis. In Microbial Symbionts; Elsevier: Amsterdam, The Netherlands, 2023; pp. 743–763. [Google Scholar]
- Pallardy, S.G. Nitrogen Metabolism. In Physiology of Woody Plants; Elsevier: Amsterdam, The Netherlands, 2008; pp. 233–254. [Google Scholar]
- Steenhoudt, O.; Vanderleyden, J. Azospirillum, a Free-Living Nitrogen-Fixing Bacterium Closely Associated with Grasses: Genetic, Biochemical and Ecological Aspects. FEMS Microbiol. Rev. 2000, 24, 487–506. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, T.A.; Bophela, K.N. Tree Leaves as a Habitat for phyllobacteria. In Forest Microbiology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 133–144. [Google Scholar]
- Compant, S.; Nowak, J.; Coenye, T.; Clément, C.; Ait Barka, E. Diversity and Occurrence of Burkholderia spp. in the Natural Environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef]
- Pidot, S.J.; Coyne, S.; Kloss, F.; Hertweck, C. Antibiotics from Neglected Bacterial Sources. Int. J. Med. Microbiol. 2014, 304, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Bayer-Santos, E.; DeMoraes Ceseti, L.; Farah, C.S.; Alvarez-Martinez, C.E. Distribution, Function and Regulation of Type 6 Secretion Systems of Xanthomonadales. Front. Microbiol. 2019, 10, 458221. [Google Scholar] [CrossRef]
- Borriss, R. Bacillus. In Beneficial Microbes in Agro-Ecology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 107–132. [Google Scholar]
- Pan, Y.; Yang, X.; Xu, M.; Sun, G. The Role of Enriched Microbial Consortium on Iron-Reducing Bioaugmentation in Sediments. Front. Microbiol. 2017, 8, 462. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R.; Holmes, D.E.; Nevin, K.P. Dissimilatory Fe(III) and Mn(IV) Reduction. Adv. Microb. Physiol. 1991, 49, 219–286. [Google Scholar] [CrossRef]
- Gapes, J.R. Economics of AB Fermentation 27 The Economics of Acetone-Butanol Fermentation: Theoretical and Market Considerations Fermentation Symposium JMMB Minireview. J. Mol. Microbiol. Biotechnol. 2000, 2, 27–32. [Google Scholar]
- Parkes, R.J.; Sass, H. Deep Sub-Surface. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 64–79. [Google Scholar]
- Adriaens, P.; Gruden, C.; McCormick, M.L. Biogeochemistry of Halogenated Hydrocarbons. Treatise Geochem. 2003, 9, 612. [Google Scholar] [CrossRef]
- Hansel, C.M.; Lentini, C.J.; Tang, Y.; Johnston, D.T.; Wankel, S.D.; Jardine, P.M. Dominance of Sulfur-Fueled Iron Oxide Reduction in Low-Sulfate Freshwater Sediments. ISME J. 2015, 9, 2400–2412. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulford, M.K.; Mukherjee, A.; Auler, A.S.; Barton, H.A.; Senko, J.M. The Influences of O2 Availability on the Microbial Activities and Fe Transformations in the Iron Formation Caves of the Southern Espinhaço Range, Brazil. Minerals 2024, 14, 425. https://doi.org/10.3390/min14040425
Mulford MK, Mukherjee A, Auler AS, Barton HA, Senko JM. The Influences of O2 Availability on the Microbial Activities and Fe Transformations in the Iron Formation Caves of the Southern Espinhaço Range, Brazil. Minerals. 2024; 14(4):425. https://doi.org/10.3390/min14040425
Chicago/Turabian StyleMulford, Melissa K., Anela Mukherjee, Augusto S. Auler, Hazel A. Barton, and John M. Senko. 2024. "The Influences of O2 Availability on the Microbial Activities and Fe Transformations in the Iron Formation Caves of the Southern Espinhaço Range, Brazil" Minerals 14, no. 4: 425. https://doi.org/10.3390/min14040425