Advanced Multiplex Loop Mediated Isothermal Amplification (mLAMP) Combined with Lateral Flow Detection (LFD) for Rapid Detection of Two Prevalent Malaria Species in India and Melting Curve Analysis
Abstract
:1. Introduction
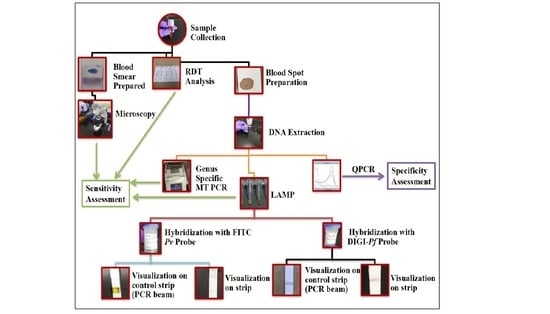
2. Materials and Methods
2.1. Study Samples
2.2. Microscopy and Rapid Diagnostic Test (RDT)
2.3. DNA Isolation for Polymerase Chain Reaction (PCR) and Loop-Mediated Isothermal Amplification (LAMP)
2.4. PCR
2.5. LAMP Primers
2.6. Reaction Mixture for LAMP
2.7. Melting Curve Analysis and Specificity Assessment
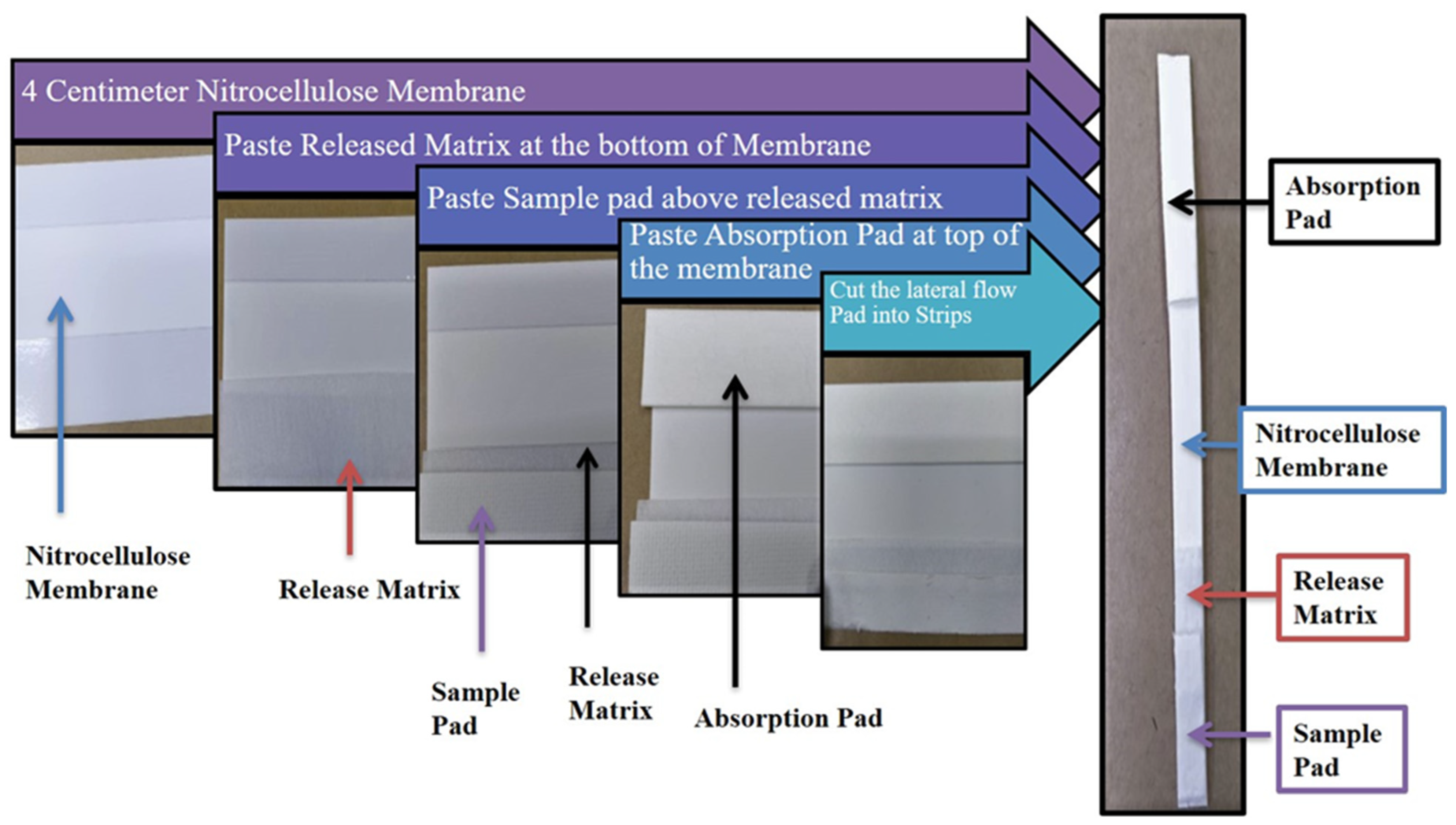
2.8. Construction of Lateral Flow Strip
2.8.1. Functionalization of Gold Nanoparticles
2.8.2. Hybridization Buffer Composition and Hybridization Protocol
2.8.3. Visualization on a Lateral Strip
2.9. Statistical Analyses
2.10. Ethics
3. Results
3.1. Microscopy, RDT, Mitochondrial PCR (Mt PCR) and LAMP Result
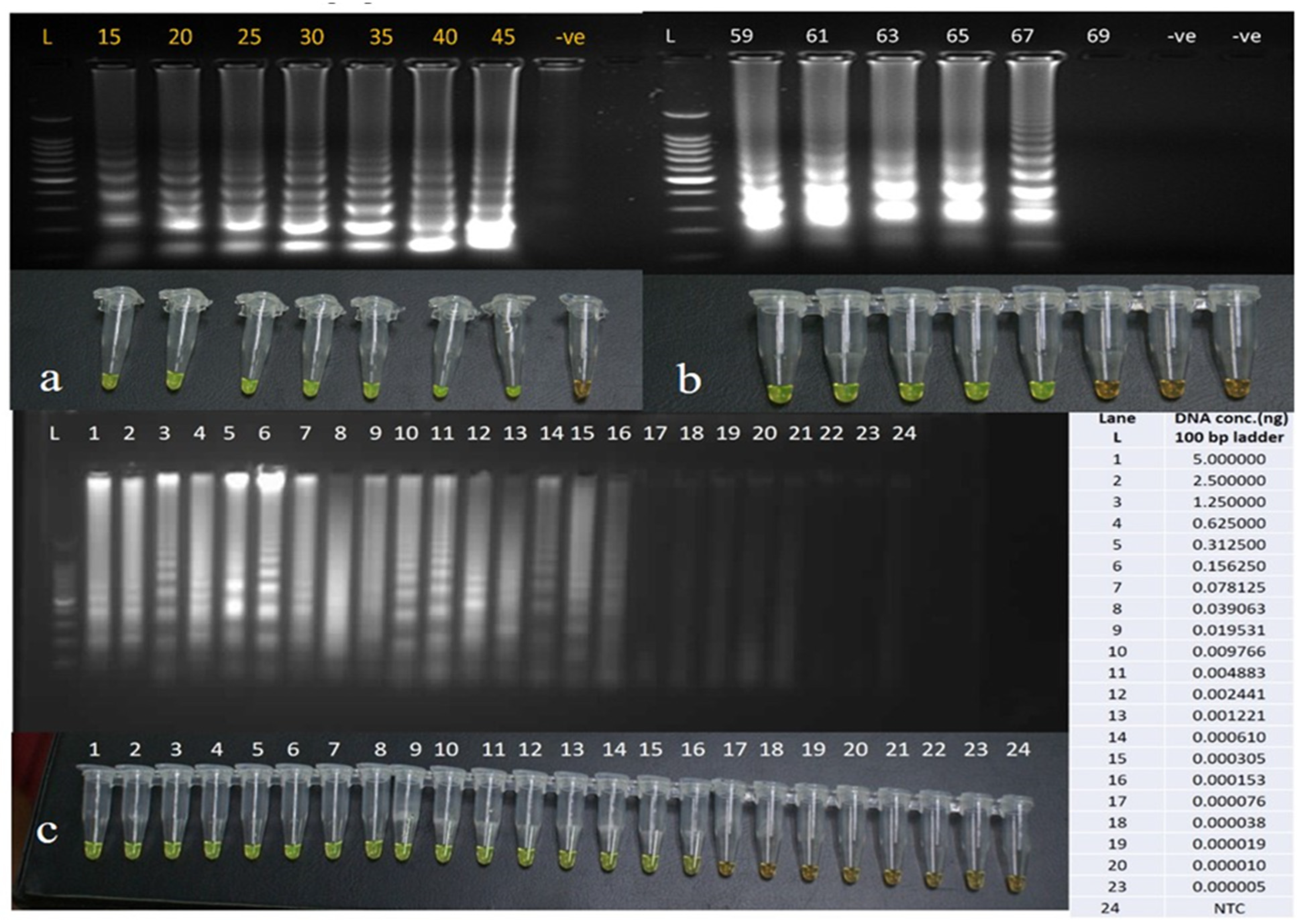
3.2. Optimizing the LAMP Assay Conditions and Its Sensitivity
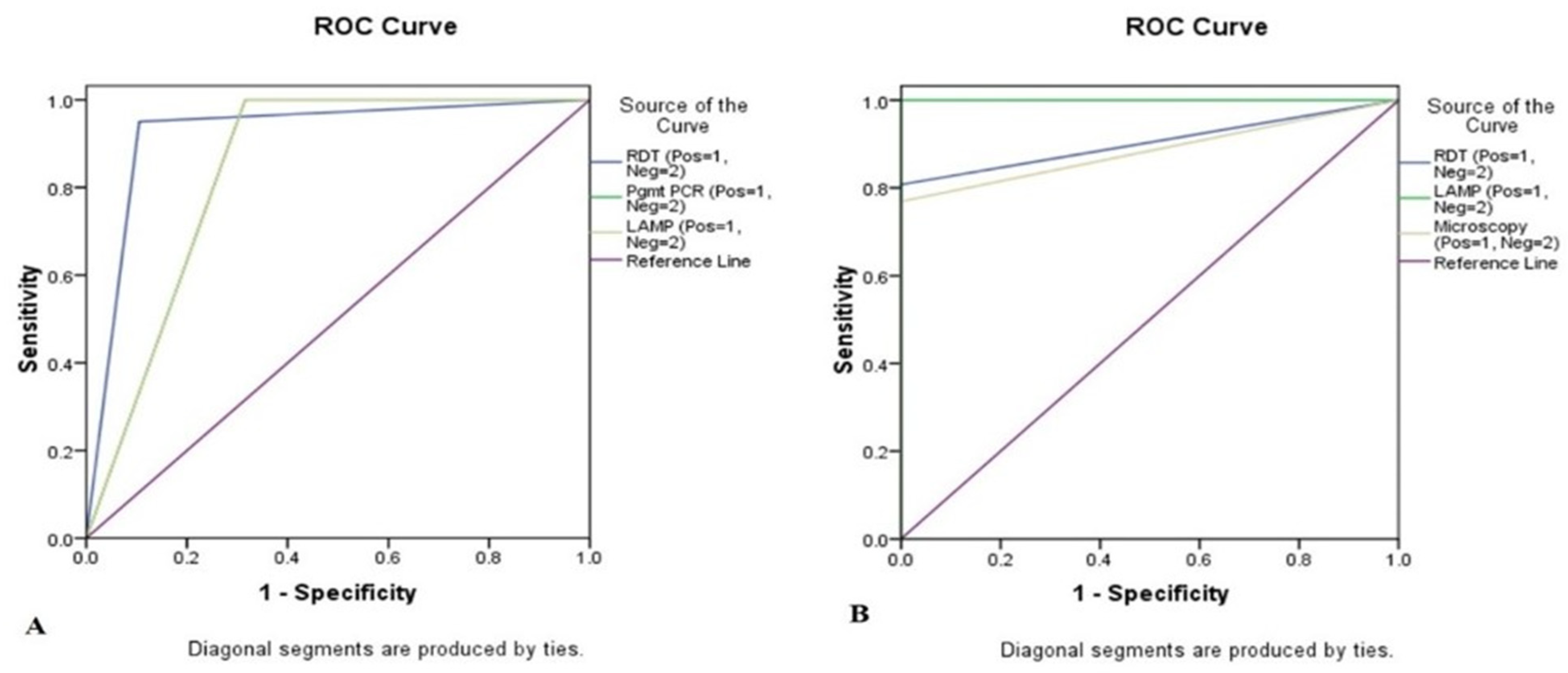
3.3. Sensitivity with Other Standard Methods
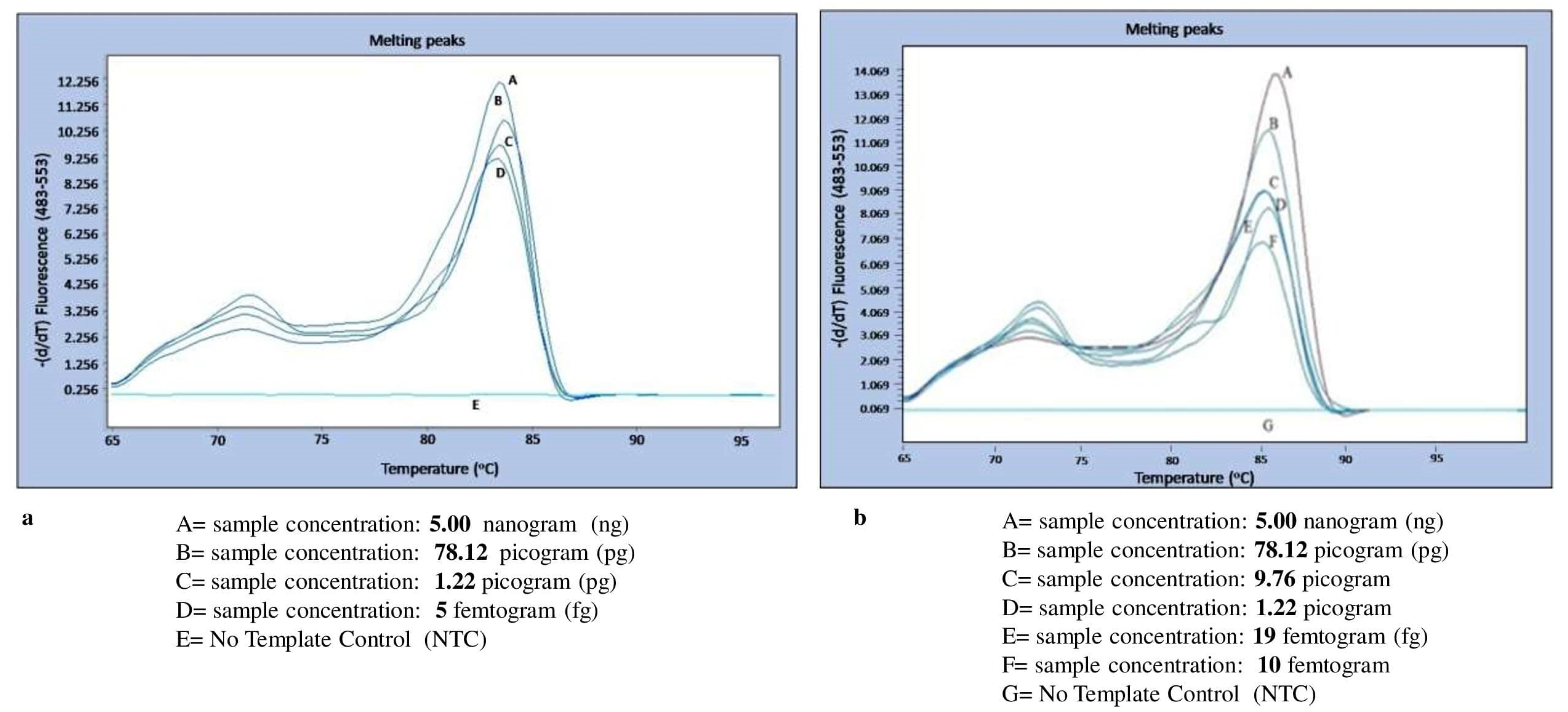
3.4. Melting Curve Analysis for Specificity Determination
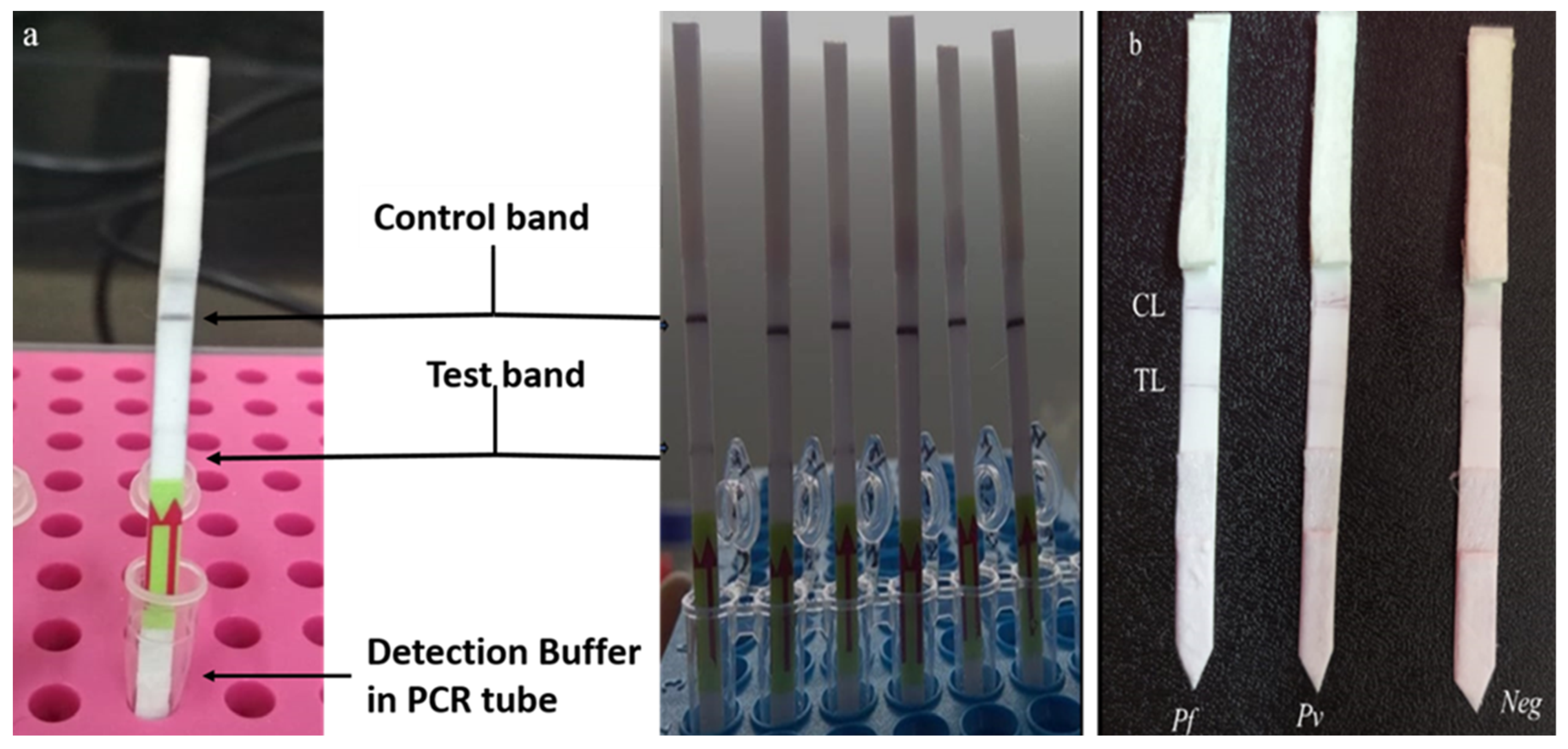
3.5. Principle and Result of mLAMP-LFD (Lateral Flow Detection)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- World Health Organization. World Malaria Report; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Berzosa, P.; de Lucio, A.; Romay-Barja, M.; Herrador, Z.; González, V.; García, L.; Fernández-Martínez, A.; Santana-Morales, M.; Ncogo, P.; Valladares, B.; et al. Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar. J. 2018, 17, 333. [Google Scholar] [CrossRef]
- Sharma, S.; Kaitholia, K.; Bharti, R.S.; Singh, M.P.; Mishra, N. Novel molecular diagnostic technique for detecting the different species of Plasmodium. Infect. Genet. Evol. 2020, 78, 104122. [Google Scholar] [CrossRef] [PubMed]
- Bharti, P.K.; Chandel, H.S.; Ahmad, A.; Krishna, S.; Udhayakumar, V.; Singh, N. Prevalence of pfhrp2 and/or pfhrp3 Gene Deletion in Plasmodium falciparum Population in Eight Highly Endemic States in India. PLoS ONE 2016, 11, e0157949. [Google Scholar] [CrossRef]
- Saito, T.; Kikuchi, A.; Kaneko, A.; Isozumi, R.; Teramoto, I.; Kimura, M.; Hirasawa, N.; Hiratsuka, M. Rapid and sensitive multiplex single-tube nested PCR for the identification of five human Plasmodium species. Parasitol. Int. 2018, 67, 277–283. [Google Scholar] [CrossRef]
- Lee, P.C.; Chong, E.T.J.; Anderios, F.; AL Lim, Y.; Chew, C.H.; Chua, K.H. Molecular detection of human Plasmodium species in Sabah using PlasmoNex™ multiplex PCR and hydrolysis probes real-time PCR. Malar. J. 2015, 14, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, E.-T. Loop-mediated isothermal amplification test for the molecular diagnosis of malaria. Expert Rev. Mol. Diagn. 2013, 13, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Di Gregorio, E.; Ferrauto, G.; Schwarzer, E.; Gianolio, E.; Valente, E.; Ulliers, D.; Aime, S.; Skorokhod, O. Relaxometric studies of erythrocyte suspensions infected by Plasmodium falciparum: A tool for staging infection and testing anti-malarial drugs. Magn. Reson. Med. 2020, 84, 3366–3378. [Google Scholar] [CrossRef]
- Peng, W.K.; Kong, T.F.; Ng, C.S.; Chen, L.; Huang, Y.; Bhagat, A.A.; Nguyen, N.-T.; Preiser, P.R.; Han, J. Micromagnetic resonance relaxometry for rapid label-free malaria diagnosis. Nat. Med. 2014, 20, 1069–1073. [Google Scholar] [CrossRef] [PubMed]
- Fescenko, I.; Laraoui, A.; Smits, J.; Mosavian, N.; Kehayias, P.; Seto, J.; Bougas, L.; Jarmola, A.; Acosta, V.M. Diamond mag-netic microscopy of malarial hemozoin nanocrystals. Phys. Rev. Appl. 2019, 11, 034029. [Google Scholar] [CrossRef] [Green Version]
- Murzin, D.; Mapps, D.J.; Levada, K.; Belyaev, V.; Omelyanchik, A.; Panina, L.; Rodionova, V. Ultrasensitive Magnetic Field Sensors for Biomedical Applications. Sensors 2020, 20, 1569. [Google Scholar] [CrossRef] [Green Version]
- Kong, T.F.; Ye, W.; Peng, W.K.; Hou, H.W.; Preiser, P.R.; Nguyen, N.-T.; Han, J. Enhancing malaria diagnosis through microfluidic cell enrichment and magnetic resonance relaxometry detection. Sci. Rep. 2015, 5, 11425. [Google Scholar]
- Gupta, M.; Singh, K.; Lobiyal, D.K.; Safvan, C.P.; Sahu, B.K.; Yadav, P.; Singh, S. A sensitive on-chip probe–based portable nuclear magnetic resonance for detecting low parasitaemia plasmodium falciparum in human blood. Med. Devices Sens. 2020, 3, 10098. [Google Scholar] [CrossRef]
- Yamamoto, T.; Hashimoto, M.; Nagatomi, K.; Nogami, T.; Sofue, Y.; Hayashi, T.; Ido, Y.; Yatsushiro, S.; Abe, K.; Kajimoto, K. Field evaluation of a quantitative, and rapid malaria diagnostic system using a fluorescent Blue-ray optical device. bioRxiv 2019, 721076. [Google Scholar] [CrossRef]
- Arndt, L.; Koleala, T.; Orbán, Á.; Ibam, C.; Lufele, E.; Timinao, L.; Lorry, L.; Butykai, Á.; Kaman, P.; Molnár, A. Magne-to-optical diagnosis of symptomatic malaria in Papua New Guinea. Nat. Commun. 2021, 12, 969. [Google Scholar] [CrossRef]
- Wang, S.; Yang, C.; Preiser, P.; Zheng, Y. A Photoacoustic-Surface-Acoustic-Wave Sensor for Ring-Stage Malaria Parasite Detection. IEEE Trans. Circuits Syst. II Express Briefs 2020, 67, 881–885. [Google Scholar] [CrossRef]
- Polley, S.D.; González, I.J. Malaria Diagnostic Platform, LAMP. In Encyclopedia of Malaria; Hommel, M., Kremsner, P.G., Eds.; Springer: New York, NY, USA, 2014; pp. 1–13. [Google Scholar]
- Bell, D.; Visser, T. Malaria Diagnostic Platform: Lateral Flow Assays. In Encyclopedia of Malaria; Hommel, M., Kremsner, P.G., Eds.; Springer: New York, NY, USA, 2016; pp. 1–9. [Google Scholar]
- Yongkiettrakul, S.; Kolié, F.R.; Kongkasuriyachai, D.; Sattabongkot, J.; Nguitragool, W.; Nawattanapaibool, N.; Suansomjit, C.; Warit, S.; Kangwanrangsan, N.; Buates, S. Validation of PfSNP-LAMP-Lateral Flow Dipstick for Detection of Single Nu-cleotide Polymorphism Associated with Pyrimethamine Resistance in Plasmodium falciparum. Diagnostics 2020, 10, 948. [Google Scholar] [CrossRef]
- Morris, U.; Aydin-Schmidt, B. Performance and Application of Commercially Available Loop-Mediated Isothermal Amplifi-cation (LAMP) Kits in Malaria Endemic and Non-Endemic Settings. Diagnostics 2021, 11, 336. [Google Scholar] [CrossRef]
- Haanshuus, C.G.; Mohn, S.C.; Mørch, K.; Langeland, N.; Blomberg, B.; Hanevik, K. A novel, single-amplification PCR targeting mitochondrial genome highly sensitive and specific in diagnosing malaria among returned travellers in Bergen, Norway. Malar. J. 2013, 12, 26. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.Z.; Bhardwaj, N.; Sharma, S.; Pande, V.; Anvikar, A.R. Transcriptional Modulation of the Host Immunity Mediated by Cytokines and Transcriptional Factors in Plasmodium falciparum-Infected Patients of North-East India. Biomolecules 2019, 9, 600. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C T method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Ang, G.Y.; Yu, C.Y.; Yean, C.Y. Ambient temperature detection of PCR amplicons with a novel sequence-specific nucleic acid lateral flow biosensor. Biosens. Bioelectron. 2012, 38, 151–156. [Google Scholar] [CrossRef]
- Singh, U.S.; Siwal, N.; Pande, V.; Das, A. Can mixed parasite infections thwart targeted malaria elimination program in India? BioMed Res. Int. 2017, 2017, 2847548. [Google Scholar] [CrossRef] [Green Version]
- Lau, Y.L.; Lai, M.-Y.; Fong, M.-Y.; Jelip, J.; Mahmud, R. Loop-Mediated Isothermal Amplification Assay for Identification of Five Human Plasmodium Species in Malaysia. Am. J. Trop. Med. Hyg. 2016, 94, 336–339. [Google Scholar] [CrossRef] [Green Version]
- Yamamura, M.; Makimura, K.; Ota, Y. Evaluation of a new rapid molecular diagnostic system for Plasmodium falciparum combined with DNA filter paper, loop-mediated isothermal amplification, and melting curve analysis. Jpn. J. Infect. Dis. 2009, 62, 20–25. [Google Scholar]
- Mangold, K.; Manson, R.U.; Koay, E.S.C.; Stephens, L.; Regner, M.; Thomson, R.B.; Peterson, L.R.; Kaul, K.L. Real-Time PCR for Detection and Identification of Plasmodium spp. J. Clin. Microbiol. 2005, 43, 2435–2440. [Google Scholar] [CrossRef] [Green Version]
- Uemura, N.; Makimura, K.; Onozaki, M.; Otsuka, Y.; Shibuya, Y.; Yazaki, H.; Kikuchi, Y.; Abe, S.; Kudoh, S. Development of a loop-mediated isothermal amplification method for diagnosing Pneumocystis pneumonia. J. Med Microbiol. 2008, 57, 50–57. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.Y.; Ang, G.Y.; Chua, A.L.; Tan, E.H.; Lee, S.Y.; Falero-Diaz, G.; Otero, O.; Rodríguez, I.; Reyes, F.; Acosta, A.; et al. Dry-reagent gold nanoparticle-based lateral flow biosensor for the simultaneous detection of Vibrio cholerae serogroups O1 and O139. J. Microbiol. Methods 2011, 86, 277–282. [Google Scholar] [CrossRef]
- Mallepaddi, P.C.; Lai, M.-Y.; Lau, Y.-L.; Liew, J.W.-K.; Polavarapu, R.; Podha, S.; Ooi, C.-H. Development of Loop-Mediated Isothermal Amplification–Based Lateral Flow Device Method for the Detection of Malaria. Am. J. Trop. Med. Hyg. 2018, 99, 704–708. [Google Scholar] [CrossRef] [Green Version]
- Yongkiettrakul, S.; Jaroenram, W.; Arunrut, N.; Chareanchim, W.; Pannengpetch, S.; Suebsing, R.; Kiatpathomchai, W.; Pornthanakasem, W.; Yuthavong, Y.; Kongkasuriyachai, D. Application of loop-mediated isothermal amplification assay combined with lateral flow dipstick for detection of Plasmodium falciparum and Plasmodium vivax. Parasitol. Int. 2014, 63, 777–784. [Google Scholar] [CrossRef]
- Piera, K.A.; Aziz, A.; William, T.; Bell, D.; González, I.J.; Barber, B.E.; Anstey, N.M.; Grigg, M.J. Detection of Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax using loop-mediated isothermal amplification (LAMP) in a co-endemic area in Malaysia. Malar. J. 2017, 16, 29. [Google Scholar] [CrossRef] [Green Version]

| Primer | Sequence |
|---|---|
| F3 | TCCATTAATCAAGAACGAAAGT |
| B3 | CCCAGAACCCAAAGACTT |
| FIP | TCCAAAGCCTAGTCGGCATAGAGGGAGTGAAGACGATCAGATACCGTCGTAAT |
| BIP-biotin | GAGTTTTTCTTTTCTCTCCGGAGA-TTCTCATAAGGCACTGAAGG |
| Pf Digoxigenin probe | AAGTCATCTTTCTAGGTGACT |
| Pv FITC probe | GAATTTTCTCTTCGGAGTTTATT |
| Method (n = 39) | RDT | Microscopy | Mt PCR | LAMP |
|---|---|---|---|---|
| Positive | 21 | 20 | 26 | 26 |
| Negative | 18 | 19 | 13 | 13 |
| A | Microscopy Positive (n = 20) | Microscopy Negative (n = 19) | Sensitivity% (95% C.I) | Specificity% (95% C.I) | PPV% (95% C.I) | NPV% (95% C.I) |
|---|---|---|---|---|---|---|
| RDT Positive | 19 (95.0%) | 2 (10.5%) | 95 (75.13–99.87) | 89.47 (66.86–98.70) | 90.48 (71.84–97.25) | 94.44 (71.44–99.14) |
| RDT Negative | 1 (5%) | 17 (89.5%) | ||||
| Mt PCR Positive | 20 (100%) | 6 (31.6%) | 100 (83.16–100.00) | 68.42 (43.45–87.42) | 76.92 (63.23–86.60) | 100 |
| Mt PCR Negative | 0 (0%) | 13(68.4%) | ||||
| LAMP Positive | 20 (100%) | 6 (31.6%) | 100 (83.16–100.00) | 68.42 (43.45–87.42) | 76.92 (63.23–86.60) | 100 |
| LAMP Negative | 0 (0%) | 13(68.4%) | ||||
| B | Mt PCR Positive (n = 26) | Mt PCR Negative (n = 13) | Sensitivity% (95% C.I) | Specificity% (95% C.I) | PPV% (95% C.I) | NPV% (95% C.I) |
| RDT Positive | 21 (80.8%) | 0 (0%) | 80.77 (60.65- 93.45) | 100 (75.29–100.00) | 100 | 72.22 (54.18–85.11) |
| RDT Negative | 5 (19.2%) | 13 (100%) | ||||
| LAMP Positive | 26 (100%) | 0 (0%) | 100 (86.77–100.00) | 100 (75.29–100.00) | 100 | 100 |
| LAMP Negative | 0 (0%) | 13 (100%) | ||||
| Microscopy Positive | 20 (76.9) | 0 (0%) | 76.92 (56.35–91.03) | 100 (75.29–100.00) | 100 | 68.42 (51.78–81.38) |
| Microscopy Negative | 6 (23.1%) | 13 (100%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, S.; Kumar, S.; Ahmed, M.Z.; Bhardwaj, N.; Singh, J.; Kumari, S.; Savargaonkar, D.; Anvikar, A.R.; Das, J. Advanced Multiplex Loop Mediated Isothermal Amplification (mLAMP) Combined with Lateral Flow Detection (LFD) for Rapid Detection of Two Prevalent Malaria Species in India and Melting Curve Analysis. Diagnostics 2022, 12, 32. https://doi.org/10.3390/diagnostics12010032
Sharma S, Kumar S, Ahmed MZ, Bhardwaj N, Singh J, Kumari S, Savargaonkar D, Anvikar AR, Das J. Advanced Multiplex Loop Mediated Isothermal Amplification (mLAMP) Combined with Lateral Flow Detection (LFD) for Rapid Detection of Two Prevalent Malaria Species in India and Melting Curve Analysis. Diagnostics. 2022; 12(1):32. https://doi.org/10.3390/diagnostics12010032
Chicago/Turabian StyleSharma, Supriya, Sandeep Kumar, Md Zohaib Ahmed, Nitin Bhardwaj, Jaskirat Singh, Sarita Kumari, Deepali Savargaonkar, Anupkumar R. Anvikar, and Jyoti Das. 2022. "Advanced Multiplex Loop Mediated Isothermal Amplification (mLAMP) Combined with Lateral Flow Detection (LFD) for Rapid Detection of Two Prevalent Malaria Species in India and Melting Curve Analysis" Diagnostics 12, no. 1: 32. https://doi.org/10.3390/diagnostics12010032