The Detection of Colorectal Cancer through Machine Learning-Based Breath Sensor Analysis
Abstract
:1. Introduction
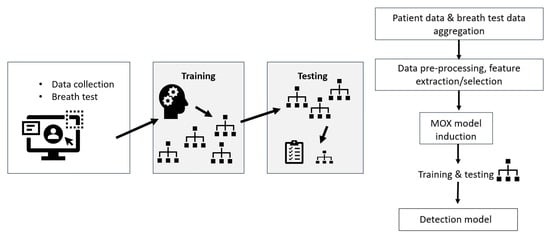
2. Materials and Methods
2.1. Study Recruitment Process
2.2. Study Group Description
2.3. Breath Analyzer and Sample Collection
2.4. Data Pre-Processing
- ▪ Minimum value of the curve.
- ▪ Average value of the curve.
- ▪ Maximum value of the curve.
- ▪ Mean value of the last 10 time points to characterize the sensor response after saturation.
- ▪ Area under the curve calculated using the trapezoidal rule.
2.5. Classification
2.6. Dimensionality Reduction
2.7. Experimental Setup
3. Results
3.1. Data Pre-Processing
3.2. Classification Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Sun, Q.; Xiong, K.; Su, Y.; Zhao, Q.; Zhang, C.; Zhang, L.; Li, X.; Fang, H. Optimization of screening strategies for colorectal cancer based on fecal DNA and occult blood testing. Eur. J. Public Health 2023, 33, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Issa, I.A.; Noureddine, M. Colorectal cancer screening: An updated review of the available options. World J. Gastroenterol. 2017, 23, 5086–5096. [Google Scholar] [CrossRef]
- Rees, C.J.; Bevan, R.; Zimmermann-Fraedrich, K.; Rutter, M.D.; Rex, D.; Dekker, E.; Ponchon, T.; Bretthauer, M.; Regula, J.; Saunders, B.; et al. Expert opinions and scientific evidence for colonoscopy key performance indicators. Gut 2016, 65, 2045–2060. [Google Scholar] [CrossRef]
- Senore, C.; Ederle, A.; Fantin, A.; Andreoni, B.; Bisanti, L.; Grazzini, G.; Zappa, M.; Ferrero, F.; Marutti, A.; Giuliani, O.; et al. Acceptability and side-effects of colonoscopy and sigmoidoscopy in a screening setting. J. Med. Screen. 2011, 18, 128–134. [Google Scholar] [CrossRef]
- Chini, A.; Manigrasso, M.; Cantore, G.; Maione, R.; Milone, M.; Maione, F.; De Palma, G.D. Can Computed Tomography Colonography Replace Optical Colonoscopy in Detecting Colorectal Lesions? State of the Art. Clin. Endosc. 2022, 55, 183–190. [Google Scholar] [CrossRef]
- Helsingen Lise, M.; Kalager, M. Colorectal Cancer Screening—Approach, Evidence, and Future Directions. NEJM Evid. 2022, 1, EVIDra2100035. [Google Scholar] [CrossRef]
- Shaukat, A.; Kahi, C.J.; Burke, C.A.; Rabeneck, L.; Sauer, B.G.; Rex, D.K. ACG Clinical Guidelines: Colorectal Cancer Screening 2021. Am. J. Gastroenterol. 2021, 116, 458–479. [Google Scholar] [CrossRef]
- Wen, Q.; Boshier, P.; Myridakis, A.; Belluomo, I.; Hanna, G.B. Urinary Volatile Organic Compound Analysis for the Diagnosis of Cancer: A Systematic Literature Review and Quality Assessment. Metabolites 2020, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Amann, A.; Costello, B.L.; Miekisch, W.; Schubert, J.; Buszewski, B.; Pleil, J.; Ratcliffe, N.; Risby, T. The human volatilome: Volatile organic compounds (VOCs) in exhaled breath, skin emanations, urine, feces and saliva. J. Breath Res. 2014, 8, 034001. [Google Scholar] [CrossRef] [PubMed]
- Dima, A.C.; Balaban, D.V.; Dima, A. Diagnostic Application of Volatile Organic Compounds as Potential Biomarkers for Detecting Digestive Neoplasia: A Systematic Review. Diagnostics 2021, 11, 2317. [Google Scholar] [CrossRef] [PubMed]
- Janfaza, S.; Khorsand, B.; Nikkhah, M.; Zahiri, J. Digging deeper into volatile organic compounds associated with cancer. Biol. Methods Protoc. 2019, 4, bpz014. [Google Scholar] [CrossRef]
- Sun, X.; Shao, K.; Wang, T. Detection of volatile organic compounds (VOCs) from exhaled breath as noninvasive methods for cancer diagnosis. Anal. Bioanal. Chem. 2015, 408, 2759–2780. [Google Scholar] [CrossRef]
- Tyagi, H.; Daulton, E.; Bannaga, A.S.; Arasaradnam, R.P.; Covington, J.A. Non-Invasive Detection and Staging of Colorectal Cancer Using a Portable Electronic Nose. Sensors 2021, 21, 5440. [Google Scholar] [CrossRef]
- Chapman, E.A.; Thomas, P.S.; Stone, E.; Lewis, C.; Yates, D.H. A breath test for malignant mesothelioma using an electronic nose. Eur. Respir. J. 2012, 40, 448–454. [Google Scholar] [CrossRef]
- Krilaviciute, A.; Heiss, J.A.; Leja, M.; Kupcinskas, J.; Haick, H.; Brenner, H. Detection of cancer through exhaled breath: A systematic review. Oncotarget 2015, 6, 38643–38657. [Google Scholar] [CrossRef]
- Ramgir, N. Electronic Nose Based on Nanomaterials: Issues, Challenges, and Prospects. ISRN Nanomater. 2013, 1, 21. [Google Scholar] [CrossRef]
- Hu, W.; Wan, L.; Jian, Y.; Ren, C.; Jin, K.; Su, X.; Bai, X.; Haick, H.; Yao, M.; Wu, W. Electronic Noses: From Advanced Materials to Sensors Aided with Data Processing. Adv. Mater. Technol. 2018, 4, 1800488. [Google Scholar] [CrossRef]
- Pelling, M.; Chandrapalan, S.; West, E.; Arasaradnam, R.P. A Systematic Review and Meta-Analysis: Volatile Organic Compound Analysis in the Detection of Hepatobiliary and Pancreatic Cancers. Cancers 2023, 15, 2308. [Google Scholar] [CrossRef] [PubMed]
- Salzberg, S.L. C4.5: Programs for Machine Learning by J. Ross Quinlan. Morgan Kaufmann Publishers, Inc. Mach Learn 1994, 16, 235–240. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- John, G.; Langley, P. Estimating Continuous Distributions in Bayesian Classifiers. In Proceedings of the 11th Conference on Uncertainty in Artificial Intelligence, Montreal, QC, Canada, 20 February 2013; Volume 1. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 20 September 2023).
- Awais, M. Data Mining: Practical machine learning tools and techniques. Data Min. 2016, 2, 4. [Google Scholar]
- Amal, H.; Leja, M.; Funka, K.; Lasina, I.; Skapars, R.; Sivins, A.; Ancans, G.; Kikuste, I.; Vanags, A.; Tolmanis, I.; et al. Breath testing as potential colorectal cancer screening tool. Int. J. Cancer 2016, 138, 229–236. [Google Scholar] [CrossRef]
- van Keulen, K.E.; Jansen, M.E.; Schrauwen, R.W.M.; Kolkman, J.J.; Siersema, P.D. Volatile organic compounds in breath can serve as a non-invasive diagnostic biomarker for the detection of advanced adenomas and colorectal cancer. Aliment. Pharmacol. Ther. 2020, 51, 334–346. [Google Scholar] [CrossRef]
- Steenhuis, E.G.M.; Schoenaker, I.J.H.; de Groot, J.W.B.; Fiebrich, H.B.; de Graaf, J.C.; Brohet, R.M.; van Dijk, J.D.; van Westreenen, H.L.; Siersema, P.D.; de Vos, T.N.; et al. Feasibility of volatile organic compound in breath analysis in the follow-up of colorectal cancer: A pilot study. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 2020, 46, 2068–2073. [Google Scholar] [CrossRef]
- Ergen, W.F.; Pasricha, T.; Hubbard, F.J.; Higginbotham, T.; Givens, T.; Slaughter, J.C.; Obstein, K.L. Providing Hospitalized Patients With an Educational Booklet Increases the Quality of Colonoscopy Bowel Preparation. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2016, 14, 858–864. [Google Scholar] [CrossRef]
- Ersöz, F.; Toros, A.B.; Aydoğan, G.; Bektaş, H.; Ozcan, O.; Arikan, S. Assessment of anxiety levels in patients during elective upper gastrointestinal endoscopy and colonoscopy. Turk. J. Gastroenterol. Off. J. Turk. Soc. Gastroenterol. 2010, 21, 29–33. [Google Scholar] [CrossRef]
- Chiu, H.M.; Lee, Y.C.; Tu, C.H.; Chen, C.C.; Tseng, P.H.; Liang, J.T.; Shun, C.T.; Lin, J.T.; Wu, M.S. Association between early stage colon neoplasms and false-negative results from the fecal immunochemical test. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2013, 11, 832–838.e2. [Google Scholar] [CrossRef]
- Gower, H.; Danielson, K.; Dennett, A.P.E.; Deere, J. Potential role of volatile organic compound breath testing in the Australasian colorectal cancer pathway. ANZ J. Surg. 2023, 93, 1159–1161. [Google Scholar] [CrossRef] [PubMed]
- van de Goor, R.M.; Leunis, N.; van Hooren, M.R.; Francisca, E.; Masclee, A.; Kremer, B.; Kross, K.W. Feasibility of electronic nose technology for discriminating between head and neck, bladder, and colon carcinomas. Eur. Arch. Oto-Rhino-Laryngol. 2016, 274, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Glöckler, J.; Mizaikoff, B.; Díaz de León-Martínez, L. SARS CoV-2 infection screening via the exhaled breath fingerprint obtained by FTIR spectroscopic gas-phase analysis. A proof of concept. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 302, 123066. [Google Scholar] [CrossRef] [PubMed]
| Gender | Median Age | Colorectal Cancer Group | Non-Cancer Group | ||||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Males | 113 | 39 | 64 | 57 | 54 | 56 | 30 |
| Females | 178 | 61 | 63 | 48 | 46 | 130 | 70 |
| Total | 291 | 100 | 63 | 105 | 100 | 186 | 100 |
| Classification Method | Overall Accuracy | Sensitivity | Specificity | AUC ROC |
|---|---|---|---|---|
| C4.5 | 60.9% | 46.7% | 68.4% | 0.567 |
| Naïve Bayes (NB) | 47.1% | 86.7% | 26.3% | 0.593 |
| Artificial Neural Networks (ANNs) | 64.4% | 46.7% | 73.7% | 0.584 |
| Random Forest (RF) | 75.9% | 43.3% | 93.0% | 0.684 |
| Classification Method | Overall Accuracy | Sensitivity | Specificity | AUC ROC |
|---|---|---|---|---|
| C4.5 | 65.1% | 65.7% | 64.3% | 0.657 |
| Naïve Bayes (NB) | 60.3% | 94.3% | 17.9% | 0.627 |
| Artificial Neural Networks (ANNs) | 66.7% | 57.1% | 78.6% | 0.713 |
| Random Forest (RF) | 60.3% | 48.6% | 75.0% | 0.658 |
| Classification Method | Number of Features | Overall Accuracy | Sensitivity | Specificity | AUC ROC | Feature Selection |
|---|---|---|---|---|---|---|
| C4.5 | 9 | 77.0% | 63.3% | 84.2% | 0.759 | MOX, Greedy sel. |
| Naïve Bayes (NB) | 4 | 71.59% | 34.3% | 96.2% | 0.671 | GNP, Greedy sel. |
| Artificial Neural Networks (ANN) | 52 | 72.4% | 46.7% | 86.0% | 0.705 | MOX, Evolutionary |
| Random Forest (RF) | 8 | 72.4% | 56.7% | 80.7% | 0.685 | MOX, Greedy sel. |
| Classification Method | Number of Features | Overall Accuracy | Sensitivity | Specificity | AUC ROC | Feature Selection |
|---|---|---|---|---|---|---|
| C4.5 | 9 | 77.0% | 63.3% | 84.2% | 0.759 | MOX, Greedy sel. |
| Naïve Bayes (NB) | 1 | 72.4% | 40.0% | 89.5% | 0.711 | All, Greedy sel. |
| Neural Networks (NNs) | 5 | 78.2% | 43.3% | 96.5% | 0.735 | All, Greedy sel. |
| Random Forest (RF) | 75 | 79.3% | 53.3% | 93.0% | 0.734 | All, Evolutionary |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poļaka, I.; Mežmale, L.; Anarkulova, L.; Kononova, E.; Vilkoite, I.; Veliks, V.; Ļeščinska, A.M.; Stonāns, I.; Pčolkins, A.; Tolmanis, I.; et al. The Detection of Colorectal Cancer through Machine Learning-Based Breath Sensor Analysis. Diagnostics 2023, 13, 3355. https://doi.org/10.3390/diagnostics13213355
Poļaka I, Mežmale L, Anarkulova L, Kononova E, Vilkoite I, Veliks V, Ļeščinska AM, Stonāns I, Pčolkins A, Tolmanis I, et al. The Detection of Colorectal Cancer through Machine Learning-Based Breath Sensor Analysis. Diagnostics. 2023; 13(21):3355. https://doi.org/10.3390/diagnostics13213355
Chicago/Turabian StylePoļaka, Inese, Linda Mežmale, Linda Anarkulova, Elīna Kononova, Ilona Vilkoite, Viktors Veliks, Anna Marija Ļeščinska, Ilmārs Stonāns, Andrejs Pčolkins, Ivars Tolmanis, and et al. 2023. "The Detection of Colorectal Cancer through Machine Learning-Based Breath Sensor Analysis" Diagnostics 13, no. 21: 3355. https://doi.org/10.3390/diagnostics13213355