Machine Learning and Radiomics of Bone Scintigraphy: Their Role in Predicting Recurrence of Localized or Locally Advanced Prostate Cancer
Abstract
:1. Introduction
2. Methods
2.1. Study Population
2.2. Study Endpoints and Design
2.3. Clinical Features
2.4. Image Acquisition
2.4.1. Radiomic Analysis
2.4.2. Data Imbalance and Oversampling
2.4.3. K-Fold Cross-Validation
2.5. Machine Learning Model
- Logistic Regression: Logistic regression (LR) is a statistical model used for binary classification tasks. The LR model is an extension of linear regression, but instead of predicting continuous values, it predicts the probability of an event occurring. The predicted probability is then transformed using a logistic (sigmoid) function to ensure it lies between 0 and 1.
- Naive Bayes: Naive Bayes (NB) is a classification algorithm based on Bayes’ theorem with the assumption of independence among the predictor variables. It assumes that the probability of a certain class, given the occurrence of other features, can be calculated using prior probabilities and conditional probabilities. In the NB classification, we are focused on calculating the probability of a class.
- Random Forest: Random Forest (RF) is a supervised ML algorithm that is used widely in classification problems. It builds decision trees on different samples and takes their majority vote for classification or the average in the case of regression.
- Support Vector Classification: Support Vector Classification (SVC) is based on Support Vector Machine (SVM). The goal is to create the best line or decision boundary that can segregate n-dimensional space into classes. It aims to find a hyperplane in a high-dimensional feature space that can separate the feature of different classes with the largest margin.
- Xtreme Gradient Boosting: Xtreme Gradient Boosting (XGBoost) is based on Gradient Boosting, combined with the advantages of the bagging method of RF. Each decision tree is related to the other, and the previous errors are corrected by the decision tree generated later, with L1/L2 Regularization to avoid overfitting.
Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Patient-Based Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- NCCN Clinical Practice Guidelines in Oncology, Prostate Cancer, Version 3. 2023. Available online: https://www.nccn.org/login?ReturnURL=https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf (accessed on 24 August 2023).
- D’Amico, A.V.; Cote, K.; Loffredo, M.; Renshaw, A.A.; Schultz, D. Determinants of prostate cancer-specific survival after radiation therapy for patients with clinically localized prostate cancer. J. Clin. Oncol. 2002, 20, 4567–4573. [Google Scholar] [CrossRef]
- Bolton, D.; Cheng, Y.; Willems-Jones, A.J.; Li, J.; Niedermeyr, E.; Mitchell, G.; Clouston, D.; Lawrentschuk, N.; Sliwinski, A.; Fox, S.; et al. Altered significance of D’Amico risk classification in patients with prostate cancer linked to a familial breast cancer (kConFab) cohort. BJU Int. 2015, 116, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Remulla, D.; Nguyen, J.H.; Aastha, D.; Liu, Y.; Dasgupta, P.; Hung, A.J. Current status of artificial intelligence applications in urology and their potential to influence clinical practice. BJU Int. 2019, 124, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.J.; Chen, J.; Che, Z.; Nilanon, T.; Jarc, A.; Titus, M.; Oh, P.J.; Gill, I.S.; Liu, Y. Utilizing Machine Learning and Automated Performance Metrics to Evaluate Robot-Assisted Radical Prostatectomy Performance and Predict Outcomes. J. Endourol. 2018, 32, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, H.; Mofid, B.; Shiri, I.; Razzaghdoust, A.; Saadipoor, A.; Mahdavi, A.; Galandooz, H.M.; Mahdavi, S.R. Machine learning-based radiomic models to predict intensity-modulated radiation therapy response, Gleason score and stage in prostate cancer. Radiol. Med. 2019, 124, 555–567. [Google Scholar] [CrossRef]
- Wong, N.C.; Lam, C.; Patterson, L.; Shayegan, B. Use of machine learning to predict early biochemical recurrence after robot-assisted prostatectomy. BJU Int. 2019, 123, 51–57. [Google Scholar] [CrossRef]
- Acar, E.; Leblebici, A.; Ellidokuz, B.E.; Başbınar, Y.; Kaya, G. Machine learning for differentiating metastatic and completely responded sclerotic bone lesion in prostate cancer: A retrospective radiomics study. Br. J. Radiol. 2019, 92, 20190286. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, E.; Gillessen, J.S.; van der Kwast, T.H.; Lam, T.B.; Mason, M.D.; Oprea-Lager, D.E.; Ploussard, G.; van der Poel, H.G.; Schoots, I.G.; et al. Wiegel: EAU Guidelines on Prostate Cancer 2022; EAU Guidelines Office: Arnhem, The Netherlands, 2022; Available online: http://uroweb.org/guidelines/compilations-of-all-guidelines/ (accessed on 24 August 2023).
- Rud, E.; Noor, D.; Galtung, K.F.; Ottosson, F.; Jacewicz, M.; Baco, E.; Lauritzen, P.M. Validating the screening criteria for bone metastases in treatment-naive unfavorable intermediate and high-risk prostate cancer—The prevalence and location of bone- and lymph node metastases. Eur. Radiol. 2022, 32, 8266–8275. [Google Scholar] [CrossRef]
- Sevcenco, S.; Grubmuller, B.; Sonneck-Koenne, C.; Ahmadi, Y.; Knoll, P.; Floth, A.; Pokieser, W.; Zandieh, S.; Christoph Klingler, H.; Shariat, S.; et al. Bone Scintigraphy in Staging of Newly Diagnosed Prostate Cancer in Regard of Different Risk Groups. Asia Ocean. J. Nucl. Med. Biol. 2019, 7, 149–152. [Google Scholar]
- Pomykala, K.L.; Czernin, J.; Grogan, T.R.; Armstrong, W.R.; Williams, J.; Calais, J. Total-Body (68)Ga-PSMA-11 PET/CT for Bone Metastasis Detection in Prostate Cancer Patients: Potential Impact on Bone Scan Guidelines. J. Nucl. Med. 2020, 61, 405–411. [Google Scholar] [CrossRef]
- Pound, C.R.; Partin, A.W.; Eisenberger, M.A.; Chan, D.W.; Pearson, J.D.; Walsh, P.C. Natural history of progression after PSA elevation following radical prostatectomy. JAMA 1999, 281, 1591–1597. [Google Scholar] [CrossRef]
- Liberini, V.; Laudicella, R.; Balma, M.; Nicolotti, D.G.; Buschiazzo, A.; Grimaldi, S.; Lorenzon, L.; Bianchi, A.; Peano, S.; Bartolotta, T.V.; et al. Radiomics and artificial intelligence in prostate cancer: New tools for molecular hybrid imaging and theragnostics. Eur. Radiol. Exp. 2022, 6, 27. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; de Cobelli, O.; Musi, G.; Del Giudice, F.; Carrieri, G.; Busetto, G.M.; Falagario, U.G.; Sciarra, A.; Maggi, M.; Crocetto, F.; et al. Radiomics in prostate cancer: An up-to-date review. Ther. Adv. Urol. 2022, 14, 17562872221109020. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Schabath, M.B. Radiomics Improves Cancer Screening and Early Detection. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2556–2567. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Zelic, R.; Garmo, H.; Zugna, D.; Stattin, P.; Richiardi, L.; Akre, O.; Pettersson, A. Predicting Prostate Cancer Death with Different Pretreatment Risk Stratification Tools: A Head-to-head Comparison in a Nationwide Cohort Study. Eur. Urol. 2020, 77, 180–188. [Google Scholar] [CrossRef]
- Churpek, M.M.; Yuen, T.C.; Winslow, C.; Meltzer, D.O.; Kattan, M.W.; Edelson, D.P. Multicenter Comparison of Machine Learning Methods and Conventional Regression for Predicting Clinical Deterioration on the Wards. Crit. Care Med. 2016, 44, 368–374. [Google Scholar] [CrossRef]
- Deist, T.M.; Dankers, F.J.W.M.; Valdes, G.; Wijsman, R.; Hsu, I.C.; Oberije, C.; Lustberg, T.; van Soest, J.; Hoebers, F.; Jochems, A.; et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers. Med. Phys. 2018, 45, 3449–3459. [Google Scholar] [CrossRef]
- Schonlau, M.; Zou, R.Y. The random forest algorithm for statistical learning. Stata J. 2020, 20, 3–29. [Google Scholar] [CrossRef]
- Esmaily, H.; Tayefi, M.; Doosti, H.; Ghayour-Mobarhan, M.; Nezami, H.; Amirabadizadeh, A. A Comparison between Decision Tree and Random Forest in Determining the Risk Factors Associated with Type 2 Diabetes. J. Res. Health Sci. 2018, 18, e00412. [Google Scholar] [PubMed]
- Daskivich, T.J.; Fan, K.H.; Koyama, T.; Albertsen, P.C.; Goodman, M.; Hamilton, A.S.; Hoffman, R.M.; Stanford, J.L.; Stroup, A.M.; Litwin, M.S.; et al. Effect of age, tumor risk, and comorbidity on competing risks for survival in a U.S. population-based cohort of men with prostate cancer. Ann. Intern. Med. 2013, 158, 709–717. [Google Scholar] [CrossRef]
- Kastner, C.; Armitage, J.; Kimble, A.; Rawal, J.; Carter, P.G.; Venn, S. The Charlson comorbidity score: A superior comorbidity assessment tool for the prostate cancer multidisciplinary meeting. Prostate Cancer Prostatic Dis. 2006, 9, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.M.; Rais-Bahrami, S.; Turkbey, B.; George, A.K.; Rothwax, J.; Shakir, N.; Okoro, C.; Raskolnikov, D.; Parnes, H.L.; Linehan, W.M.; et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015, 313, 390–397. [Google Scholar] [CrossRef]
- Constantinou, J.; Feneley, M.R. PSA testing: An evolving relationship with prostate cancer screening. Prostate Cancer Prostatic Dis. 2006, 9, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Saberi-Karimian, M.; Khorasanchi, Z.; Ghazizadeh, H.; Tayefi, M.; Saffar, S.; Ferns, G.A.; Ghayour-Mobarhan, M. Potential value and impact of data mining and machine learning in clinical diagnostics. Crit. Rev. Clin. Lab. Sci. 2021, 58, 275–296. [Google Scholar] [CrossRef]
- Lee, C.; Light, A.; Alaa, A.; Thurtle, D.; van der Schaar, M.; Gnanapragasam, V.J. Application of a novel machine learning framework for predicting non-metastatic prostate cancer-specific mortality in men using the Surveillance, Epidemiology, and End Results (SEER) database. Lancet Digit. Health 2021, 3, e158–e165. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Cheng, D.C.; Hsieh, T.C.; Yen, K.Y.; Kao, C.H. Lesion-Based Bone Metastasis Detection in Chest Bone Scintigraphy Images of Prostate Cancer Patients Using Pre-Train, Negative Mining, and Deep Learning. Diagnostics 2021, 11, 518. [Google Scholar] [CrossRef]
- Ferro, M.; de Cobelli, O.; Vartolomei, M.D.; Lucarelli, G.; Crocetto, F.; Barone, B.; Sciarra, A.; Del Giudice, F.; Muto, M.; Maggi, M.; et al. Prostate Cancer Radiogenomics-From Imaging to Molecular Characterization. Int. J. Mol. Sci. 2021, 22, 9971. [Google Scholar] [CrossRef]
- Shen, C.; Liu, Z.; Guan, M.; Song, J.; Lian, Y.; Wang, S.; Tang, Z.; Dong, D.; Kong, L.; Wang, M.; et al. 2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer. Transl. Oncol. 2017, 10, 886–894. [Google Scholar] [CrossRef]
- Spadarella, G.; Stanzione, A.; Akinci D’Antonoli, T.; Andreychenko, A.; Fanni, S.C.; Ugga, L.; Kotter, E.; Cuocolo, R. Systematic review of the radiomics quality score applications: An EuSoMII Radiomics Auditing Group Initiative. Eur. Radiol. 2023, 33, 1884–1894. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, J.; Francis, R.; Hassan, G.M.; Rowshanfarzad, P.; Jeraj, R.; Kasisi, C.; Rusanov, B.; Ebert, M. Radiomics for Identification and Prediction in Metastatic Prostate Cancer: A Review of Studies. Front. Oncol. 2021, 11, 771787. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, T.; Chen, S.; Zhang, X.; Serrano Sosa, M.; Liu, J.; Mo, X.; Chen, X.; Huang, M.; Li, S.; et al. Fully automated radiomic screening pipeline for osteoporosis and abnormal bone density with a deep learning-based segmentation using a short lumbar mDixon sequence. Quant. Imaging Med. Surg. 2022, 12, 1198–1213. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, Y.; Qin, S.; Liu, X.; Liu, K.; Xin, P.; Zhao, W.; Yuan, H.; Lang, N. Prognostic Value and Quantitative CT Analysis in RANKL Expression of Spinal GCTB in the Denosumab Era: A Machine Learning Approach. Cancers 2022, 14, 5201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Bado, I.L.; Hu, J.; Wan, Y.W.; Wu, L.; Wang, H.; Gao, Y.; Jeong, H.H.; Xu, Z.; Hao, X.; et al. The bone microenvironment invigorates metastatic seeds for further dissemination. Cell 2021, 184, 2471–2486.e20. [Google Scholar] [CrossRef]
- Spohn, S.K.B.; Bettermann, A.S.; Bamberg, F.; Benndorf, M.; Mix, M.; Nicolay, N.H.; Fechter, T.; Hölscher, T.; Grosu, R.; Chiti, A.; et al. Radiomics in prostate cancer imaging for a personalized treatment approach—Current aspects of methodology and a systematic review on validated studies. Theranostics 2021, 11, 8027–8042. [Google Scholar] [CrossRef]
- Azad, G.K.; Cousin, F.; Siddique, M.; Taylor, B.; Goh, V.; Cook, G.J.R. Does Measurement of First-Order and Heterogeneity Parameters Improve Response Assessment of Bone Metastases in Breast Cancer Compared to SUV(max) in [(18)F]fluoride and [(18)F]FDG PET? Mol. Imaging Biol. 2019, 21, 781–789. [Google Scholar] [CrossRef]
- Rebbeck, T.R. Prostate Cancer Genetics: Variation by Race, Ethnicity, and Geography. Semin. Radiat. Oncol. 2017, 27, 3–10. [Google Scholar] [CrossRef]
- Abramowitz, M.C.; Li, T.; Buyyounouski, M.K.; Ross, E.; Uzzo, R.G.; Pollack, A.; Horwitz, E.M. The Phoenix definition of biochemical failure predicts for overall survival in patients with prostate cancer. Cancer 2008, 112, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Nematzadeh, Z.; Ibrahim, R.; Selamat, A. Comparative studies on breast cancer classifications with k-fold cross validations using machine learning techniques. In Proceedings of the 2015 10th Asian Control Conference (ASCC), Kota Kinabalu, Malaysia, 31 May–3 June 2015; pp. 1–6. [Google Scholar]

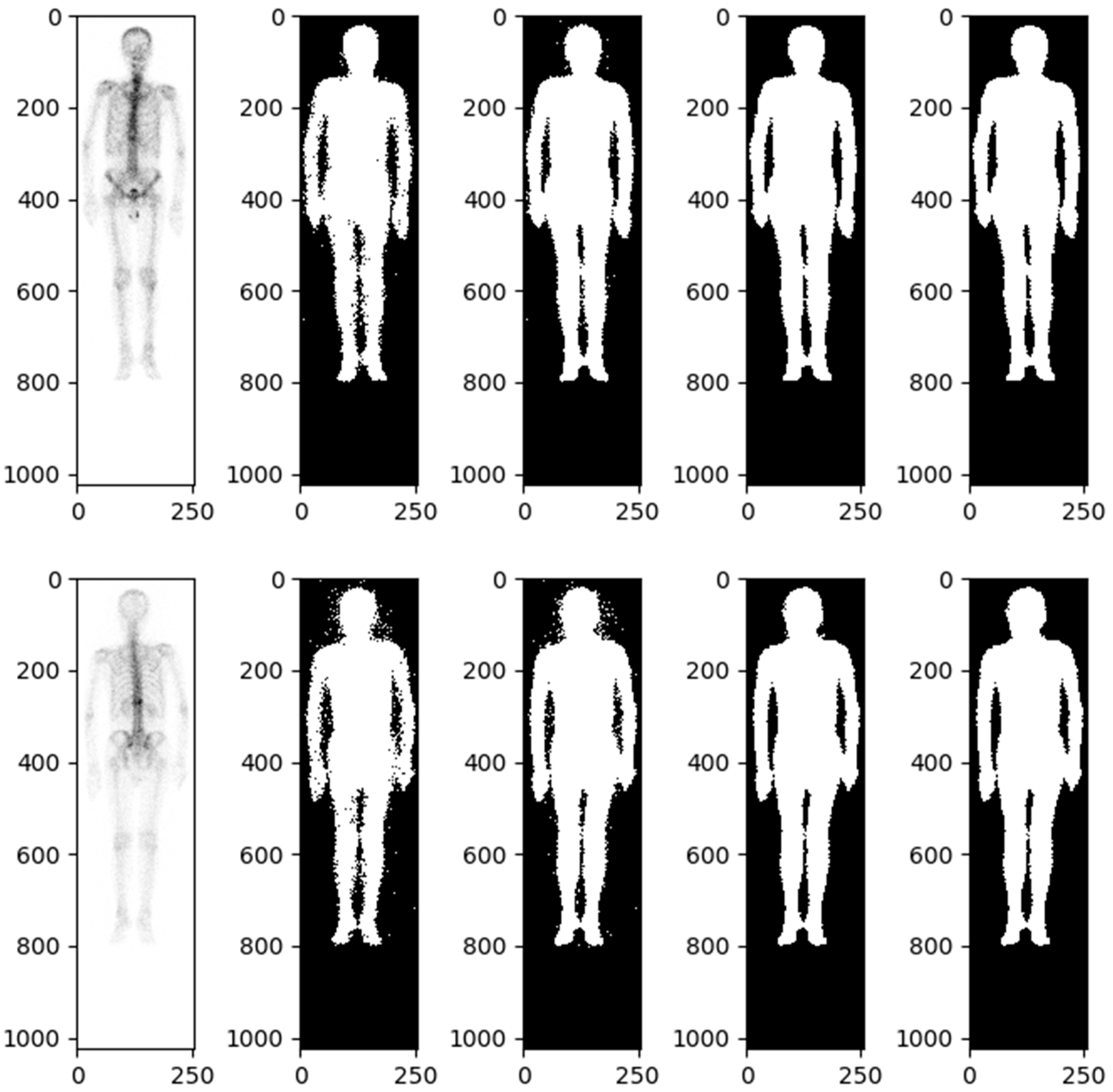

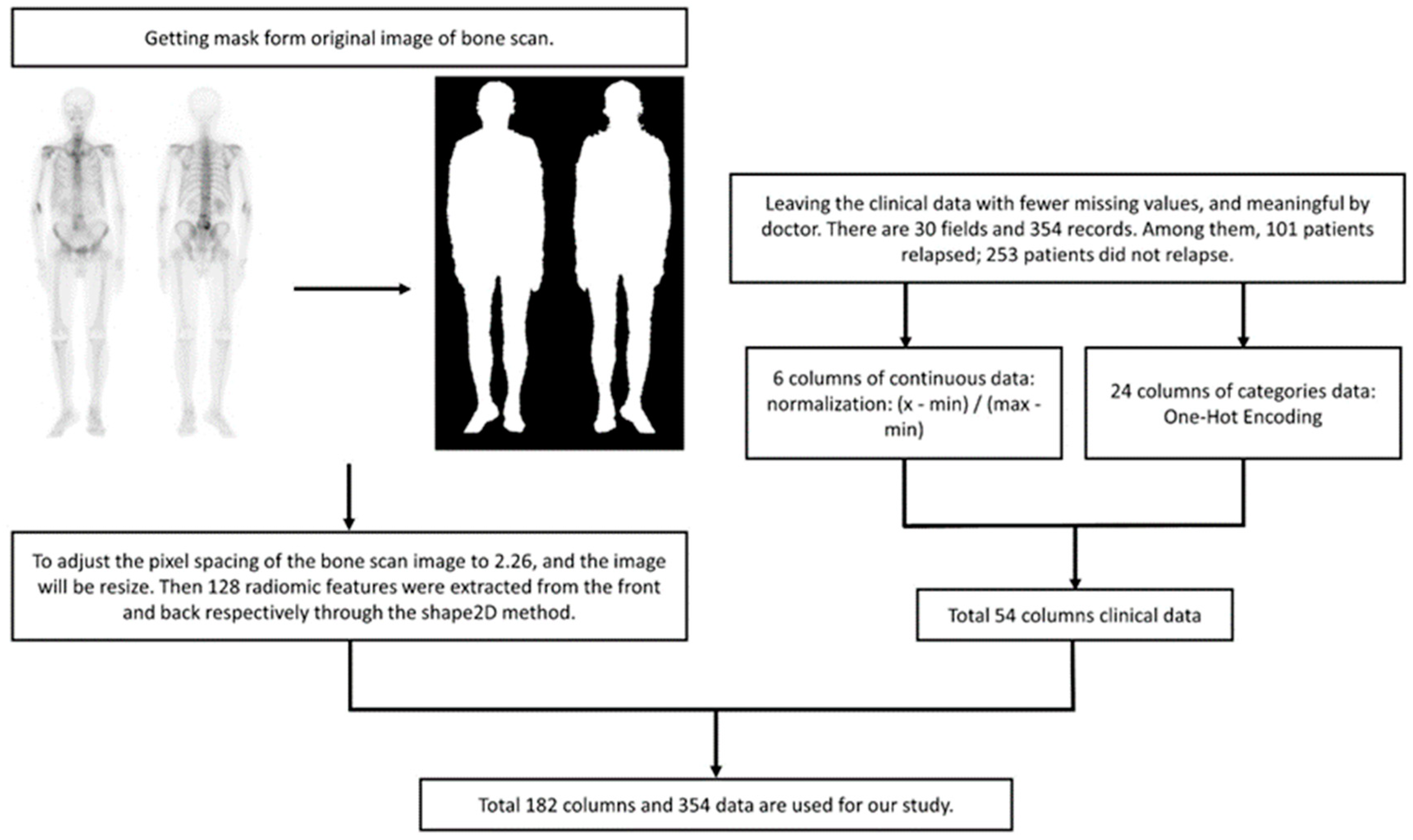
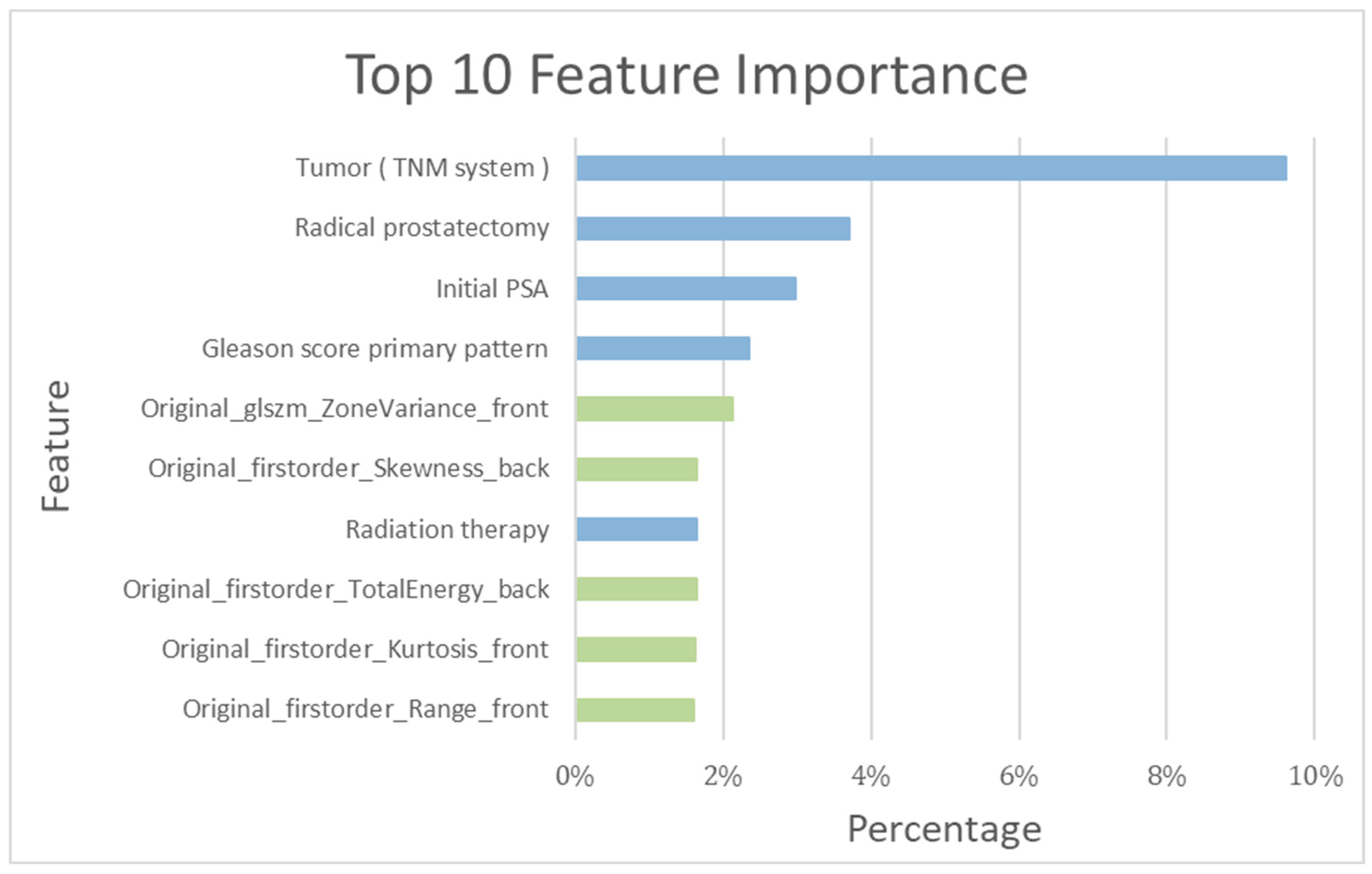
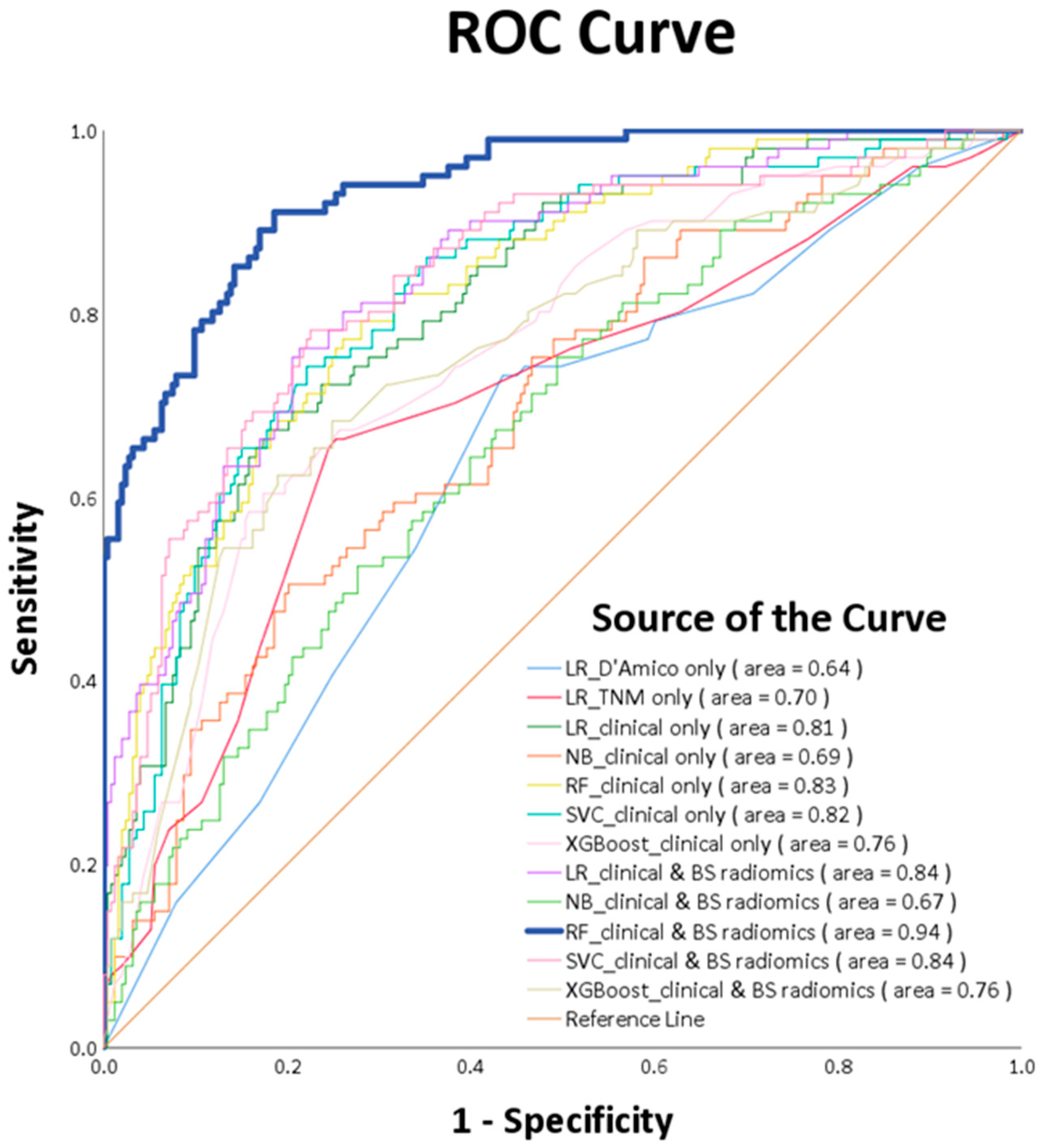
| PCa Population | |||
|---|---|---|---|
| No Recurrence | Recurrence | ||
| Population | N = 253 (71.47%) | N = 101 (28.53%) | p Value |
| a Diagnosis Age * | 70 (64–75) | 69 (64–74) | 0.38 |
| b Comorbidity | |||
| Cancer (exclude Leukemia/Lymphoma) | 45 (17.79%) | 15 (14.85%) | 0.51 |
| Lymphoma | 1 (0.40%) | 0 (0.00%) | 0.53 |
| Metastatic solid tumor | 4 (1.58%) | 1 (0.99%) | 0.67 |
| Chronic pulmonary disease | 41 (16.21%) | 10 (9.90%) | 0.13 |
| Peripheral vascular disease | 3 (1.19%) | 0 (0.00%) | 0.27 |
| Prior myocardial infarction | 5 (1.98%) | 3 (2.97%) | 0.57 |
| Congestive heart failure | 7 (2.77%) | 2 (1.98%) | 0.67 |
| Cerebrovascular disease | 33 (13.04%) | 13 (12.87%) | 0.97 |
| Dementia | 16 (6.32%) | 6 (5.94%) | 0.89 |
| Peptic ulcer disease | 57 (22.53%) | 16 (15.84%) | 0.16 |
| Diabetes | 39 (15.42%) | 19 (18.81%) | 0.44 |
| Diabetes with chronic complications | 19 (7.51%) | 7 (6.93%) | 0.85 |
| Mild liver disease | 15 (5.93%) | 6 (5.94%) | 1.00 |
| Moderate or severe liver disease | 0 (0.00%) | 1 (0.99%) | 0.11 |
| Moderate-to-severe renal disease | 44 (17.39%) | 14 (13.86%) | 0.42 |
| Rheumatologic disease | 2 (0.79%) | 0 (0.00%) | 0.37 |
| a Initial PSA * | 10.56 (7.02–19.77) | 10.23 (9.34–29.42) | 0.05 |
| b Value ≤ 10 ng/mL | 116 (45.85%) | 30 (29.70%) | <0.01 |
| b Value > 10 ng/mL and Value ≤ 20 ng/mL | 76 (30.04%) | 32 (31.68%) | 0.76 |
| b Value > 20 ng/mL | 61 (24.11%) | 39 (38.61%) | <0.01 |
| b Gleason score primary pattern * | 4 (3–4) | 4 (4–4) | <0.01 |
| 1 | 0 (0.00%) | 0 (0.00%) | |
| 2 | 6 (2.37%) | 0 (0.00%) | |
| 3 | 90 (35.57%) | 19 (18.63%) | |
| 4 | 146 (57.71%) | 75 (74.26%) | |
| 5 | 11 (4.35%) | 7 (6.93%) | |
| b Gleason score secondary pattern * | 4 (3–4) | 4 (3–5) | <0.01 |
| 1 | 0 (0.00%) | 0 (0.00%) | |
| 2 | 6 (2.37%) | 1 (0.99%) | |
| 3 | 90 (35.57%) | 43 (17.00%) | |
| 4 | 146 (57.71%) | 28 (27.72%) | |
| 5 | 11 (4.35%) | 29 (28.71%) | |
| b Gleason Score_Grade Group | <0.01 | ||
| I | 27 (10.67%) | 4 (3.96%) | |
| II | 68 (26.88%) | 14 (13.86%) | |
| III | 96 (37.94%) | 40 (39.60%) | |
| IV | 19 (7.51%) | 9 (8.91%) | |
| V | 43 (17.00%) | 34 (33.66%) | |
| b T | <0.01 | ||
| T1 | 30 (11.86%) | 4 (3.96%) | |
| T2 | 161 (63.64%) | 31 (30.69%) | |
| T3 | 60 (23.72%) | 58 (57.43%) | |
| T4 | 2 (0.79%) | 8 (7.92%) | |
| b N | <0.01 | ||
| N0 | 232 (91.70%) | 81 (80.20%) | |
| N1 | 21 (8.30%) | 20 (19.80%) | |
| b Risk classification | <0.01 | ||
| Low risk | 18 (7.11%) | 3 (2.97%) | |
| Intermediate risk | 125 (49.41%) | 24 (23.76%) | |
| High risk | 110 (43.48%) | 74 (73.27%) | |
| b Bone scintigraphy Result | 0.41 | ||
| No definite bone metastasis | 241 (95.26%) | 94 (93.07%) | |
| Equivocal findings | 12 (4.74%) | 7 (6.93%) | |
| b Cancer Treatment | |||
| RP | 141 (55.73%) | 81 (80.20%) | <0.01 |
| RP and RT | 10 (3.95%) | 0 (0.00%) | 0.04 |
| RP and ADT | 5 (1.98%) | 3 (2.97%) | 0.57 |
| RP and ADT and RT | 9 (3.56%) | 3 (2.97%) | 0.78 |
| RT | 56 (22.13%) | 7 (6.93%) | <0.01 |
| RT and ADT | 19 (7.51%) | 4 (3.96%) | 0.22 |
| HIFU | 1 (0.40%) | 0 (0.00%) | 0.53 |
| Cryotherapy | 3 (1.19%) | 1 (0.99%) | 0.88 |
| First-generation antiandrogen | 2 (0.79%) | 0 (0.00%) | 0.37 |
| AS | 7 (2.77%) | 2 (1.98%) | 0.67 |
| a Cancer Follow-up period (Years) * | 6.4 (5.51–7.54) | 2.08 (1.24–3.43) | <0.01 |
| k-Fold Test Result (K = 5) | ||||||
|---|---|---|---|---|---|---|
| Machine Learning Model | Accuracy | Sensitivity | Specificity | Precision | F1-Score | AUC |
| LR_D’Amico only | 0.61 (0.61–0.61) | 0.73 (0.67–0.79) | 0.56 (0.52–0.59) | 0.40 (0.32–0.44) | 0.52 (0.45–0.57) | 0.64 (0.62–0.68) |
| LR_TNM only | 0.73 (0.72–0.73) | 0.65 (0.53–0.81) | 0.75 (0.68–0.81) | 0.52 (0.50–0.57) | 0.58 (0.51–0.62) | 0.70 (0.69–0.74) |
| LR_clinical only | 0.75 (0.70–0.81) | 0.71 (0.57–0.79) | 0.76 (0.74–0.82) | 0.55 (0.48–0.63) | 0.62 (0.52–0.70) | 0.81 (0.74–0.85) |
| NB_clinical only | 0.52 (0.49–0.54) | 0.87 (0.75–1.00) | 0.38 (0.29–0.41) | 0.36 (0.33–0.39) | 0.51 (0.46–0.56) | 0.69 (0.61–0.81) |
| RF_clinical only | 0.76 (0.70–0.87) | 0.71 (0.58–0.90) | 0.78 (0.71–0.86) | 0.56 (0.48–0.71) | 0.63 (0.56–0.79) | 0.83 (0.76–0.94) |
| SVC_clinical only | 0.77 (0.74–0.81) | 0.71 (0.63–0.82) | 0.80 (0.74–0.88) | 0.59 (0.54–0.67) | 0.64 (0.60–0.70) | 0.82 (0.80–0.86) |
| XGBoost_clinical only | 0.73 (0.70–0.77) | 0.64 (0.53–0.73) | 0.77 (0.71–0.83) | 0.53 (0.46–0.59) | 0.58 (0.53–0.62) | 0.76 (0.74–0.79) |
| LR_clinical and BS radiomics | 0.78 (0.73–0.81) | 0.76 (0.57–0.90) | 0.79 (0.75–0.82) | 0.59 (0.52–0.64) | 0.66 (0.55–0.71) | 0.84 (0.76–0.89) |
| NB_clinical and BS radiomics | 0.51 (0.44–0.54) | 0.82 (0.70–1.00) | 0.38 (0.29–0.47) | 0.35 (0.30–0.39) | 0.49 (0.42–0.56) | 0.67 (0.60–0.81) |
| RF_clinical and BS radiomics | 0.85 (0.76–0.99) | 0.81 (0.60–1.00) | 0.87 (0.73–0.98) | 0.71 (0.54–0.96) | 0.76 (0.60–0.98) | 0.94 (0.83–1.00) |
| SVC_clinical and BS radiomics | 0.77 (0.74–0.80) | 0.71 (0.62–0.79) | 0.80 (0.77–0.82) | 0.59 (0.54–0.61) | 0.64 (0.58–0.68) | 0.84 (0.75–0.90) |
| XGBoost_clinical and BS radiomics | 0.73 (0.70–0.76) | 0.65 (0.53–0.73) | 0.76 (0.71–0.84) | 0.52 (0.46–0.56) | 0.58 (0.54–0.62) | 0.76 (0.75–0.80) |
| Predicted | |||
|---|---|---|---|
| No | Yes | ||
| True | No | 219 | 34 |
| Yes | 19 | 82 | |
| The Best Model | Other Model | p-Value |
|---|---|---|
| RF_clinical and BS radiomics | LR_D’Amico only | <0.0001 |
| LR_TNM only | <0.0001 | |
| LR_clinical only | <0.0001 | |
| NB_clinica only | <0.0001 | |
| RF_clinical only | <0.0001 | |
| SVC_clinical only | <0.0001 | |
| XGBoost_clinical only | <0.0001 | |
| LR_clinical and BS radiomics | 0.0003 | |
| NB_clinical and BS radiomics | <0.0001 | |
| SVC_clinical and BS radiomics | 0.0003 | |
| XGBoost_clinical and BS radiomics | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-D.; Huang, C.-P.; Yang, Y.-R.; Wu, H.-C.; Hsu, Y.-J.; Yeh, Y.-C.; Yeh, P.-C.; Wu, K.-C.; Kao, C.-H. Machine Learning and Radiomics of Bone Scintigraphy: Their Role in Predicting Recurrence of Localized or Locally Advanced Prostate Cancer. Diagnostics 2023, 13, 3380. https://doi.org/10.3390/diagnostics13213380
Wang Y-D, Huang C-P, Yang Y-R, Wu H-C, Hsu Y-J, Yeh Y-C, Yeh P-C, Wu K-C, Kao C-H. Machine Learning and Radiomics of Bone Scintigraphy: Their Role in Predicting Recurrence of Localized or Locally Advanced Prostate Cancer. Diagnostics. 2023; 13(21):3380. https://doi.org/10.3390/diagnostics13213380
Chicago/Turabian StyleWang, Yu-De, Chi-Ping Huang, You-Rong Yang, Hsi-Chin Wu, Yu-Ju Hsu, Yi-Chun Yeh, Pei-Chun Yeh, Kuo-Chen Wu, and Chia-Hung Kao. 2023. "Machine Learning and Radiomics of Bone Scintigraphy: Their Role in Predicting Recurrence of Localized or Locally Advanced Prostate Cancer" Diagnostics 13, no. 21: 3380. https://doi.org/10.3390/diagnostics13213380






