Downregulation of Glycine N-Acyltransferase in Kidney Renal Clear Cell Carcinoma: A Bioinformatic-Based Screening
Abstract
:1. Introduction
2. Methods
2.1. Expression Level Analysis
2.2. Mutation Character and Methylation Analysis
2.3. Immune Cell Infiltration Analysis
2.4. Survival Data Analysis
2.5. Statistical Analysis
3. Results
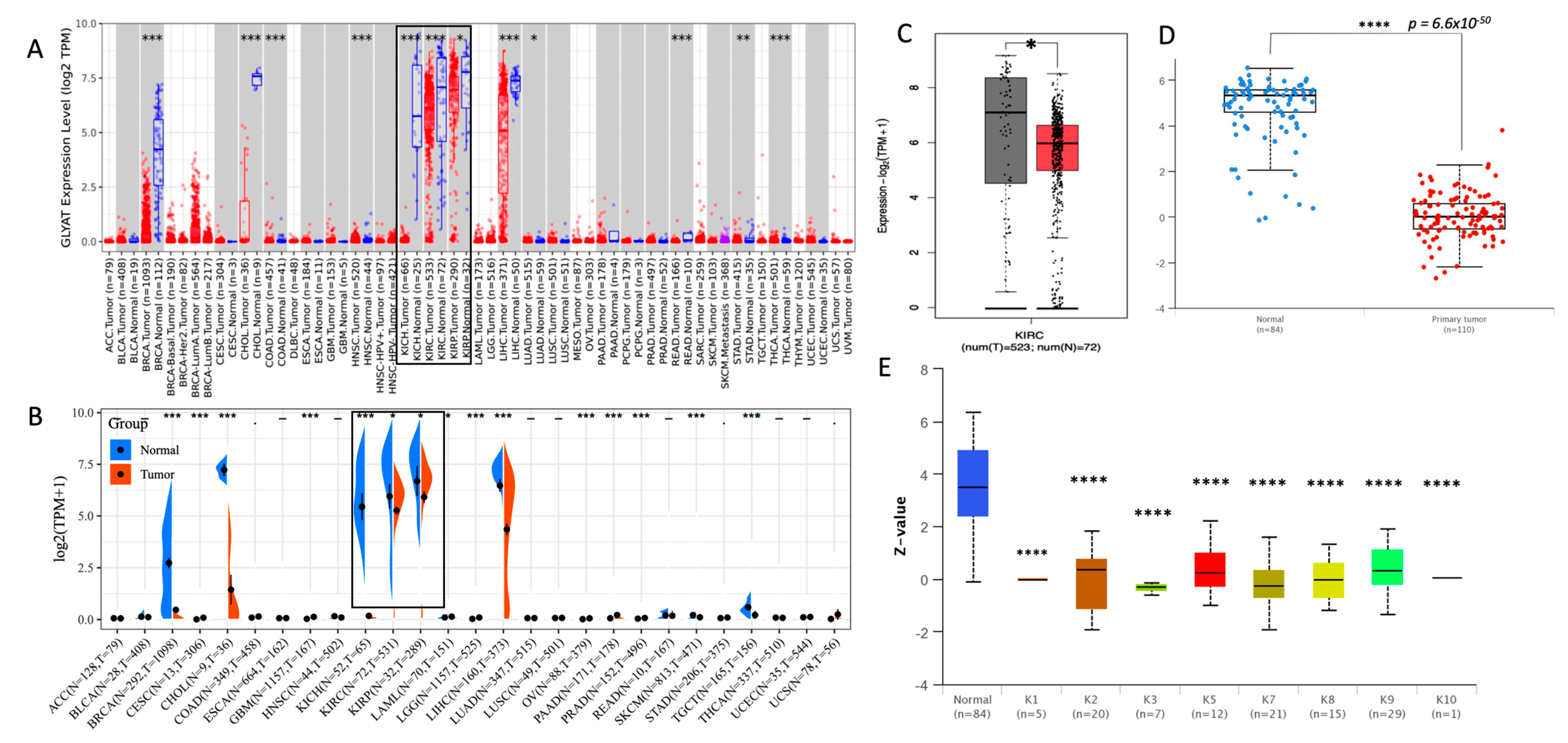
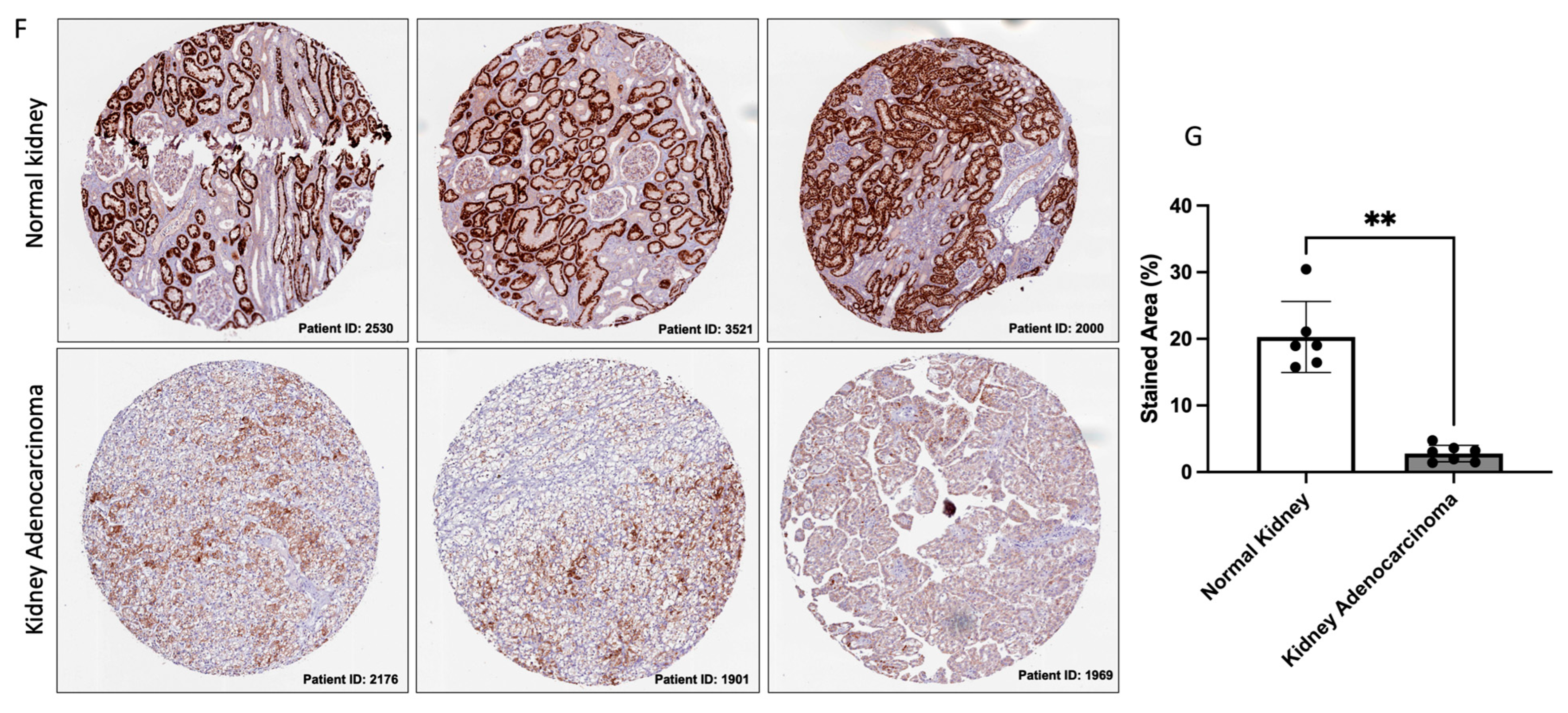
3.1. GLYAT Is Downregulated in KIRC
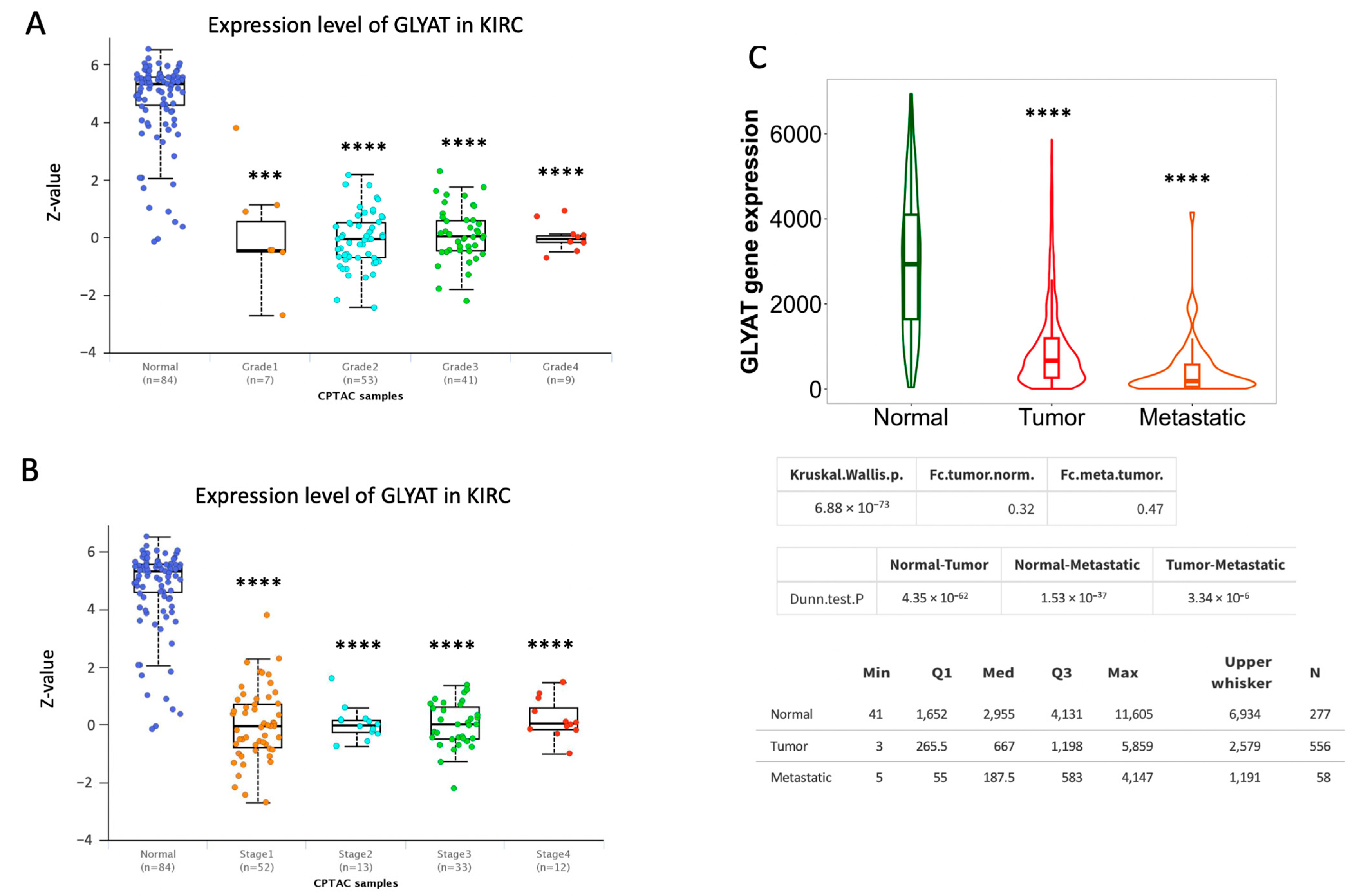
3.2. GLYAT Is Downregulated across Tumor Grade and Cancer Stages in KIRC Patients
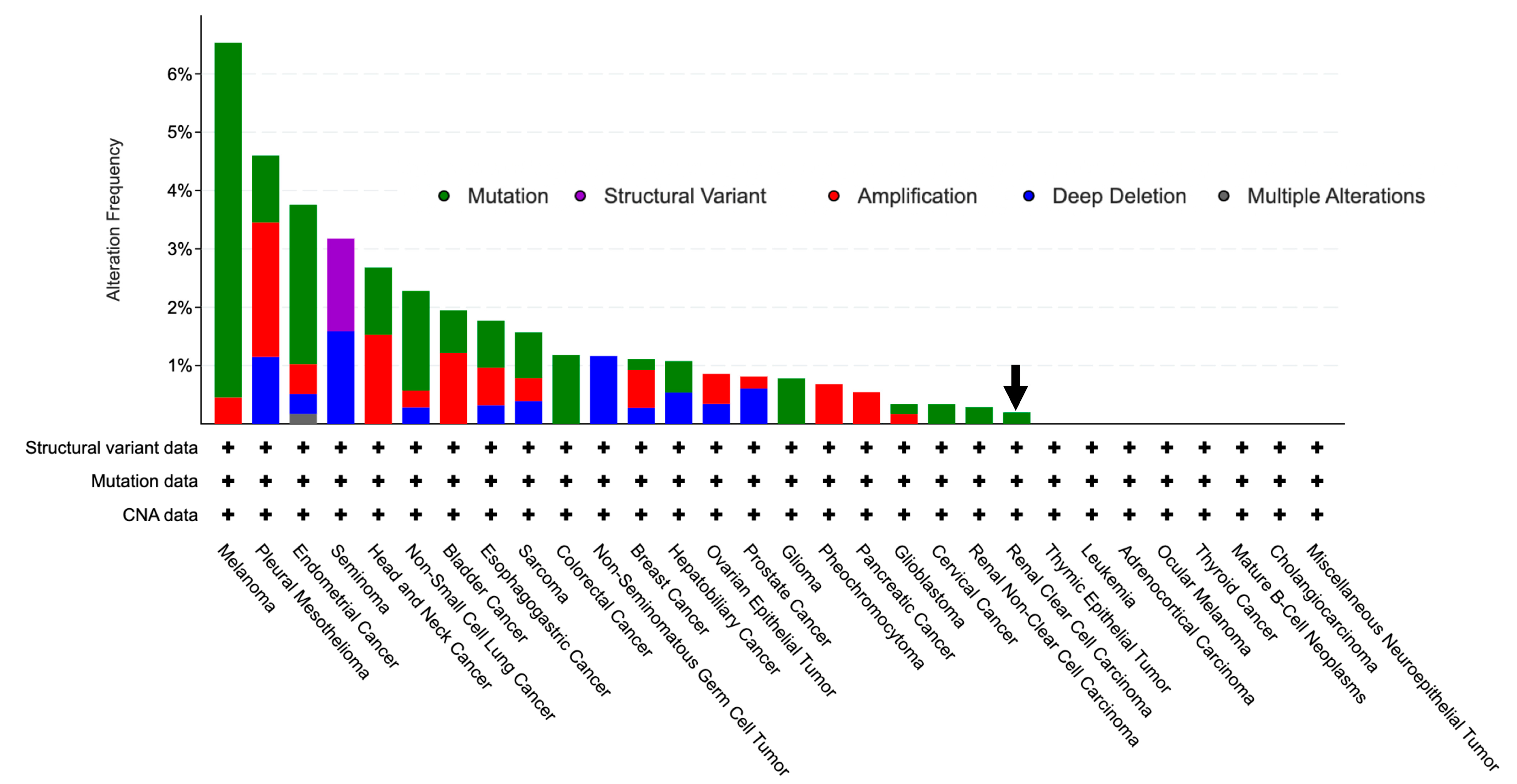
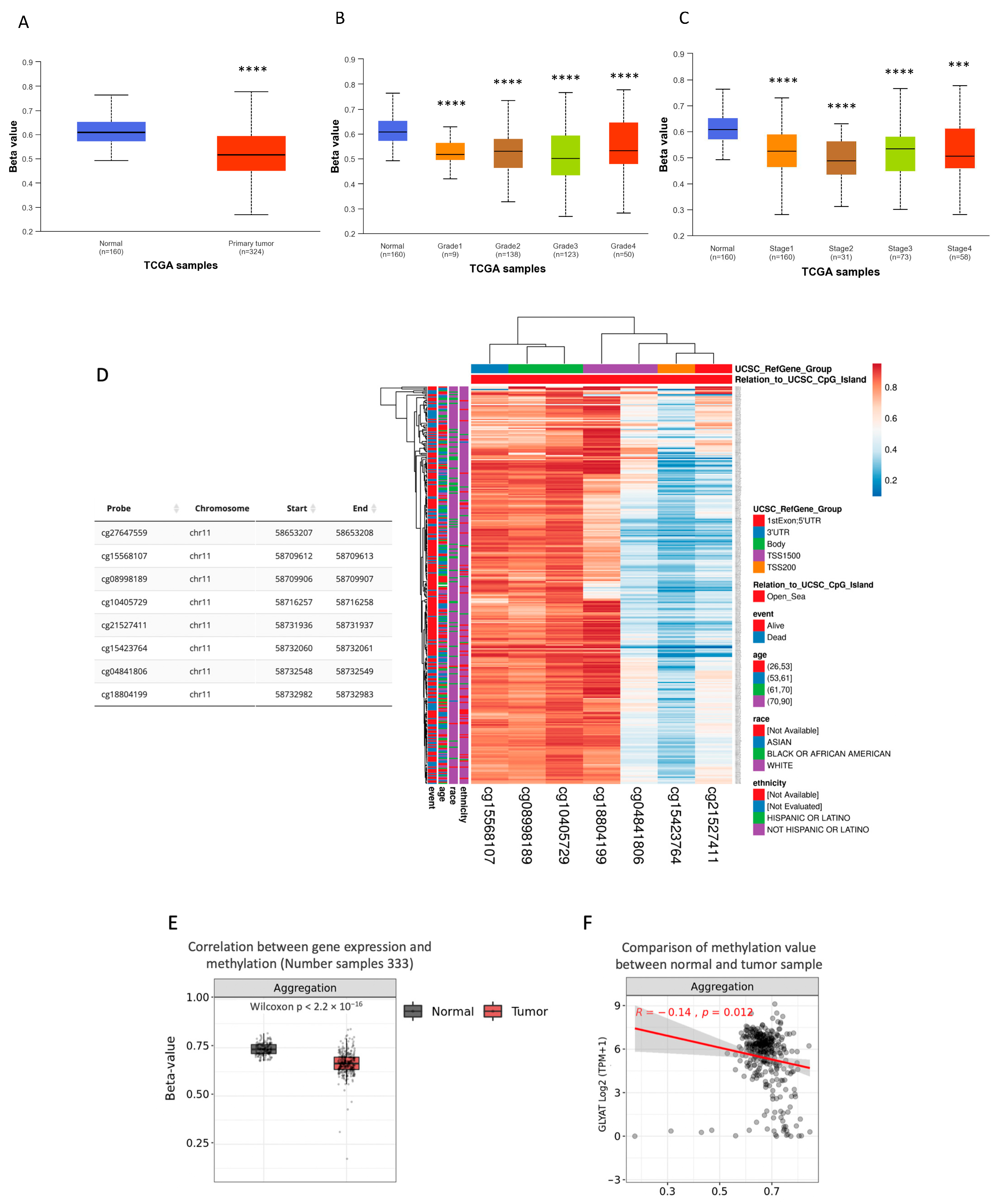
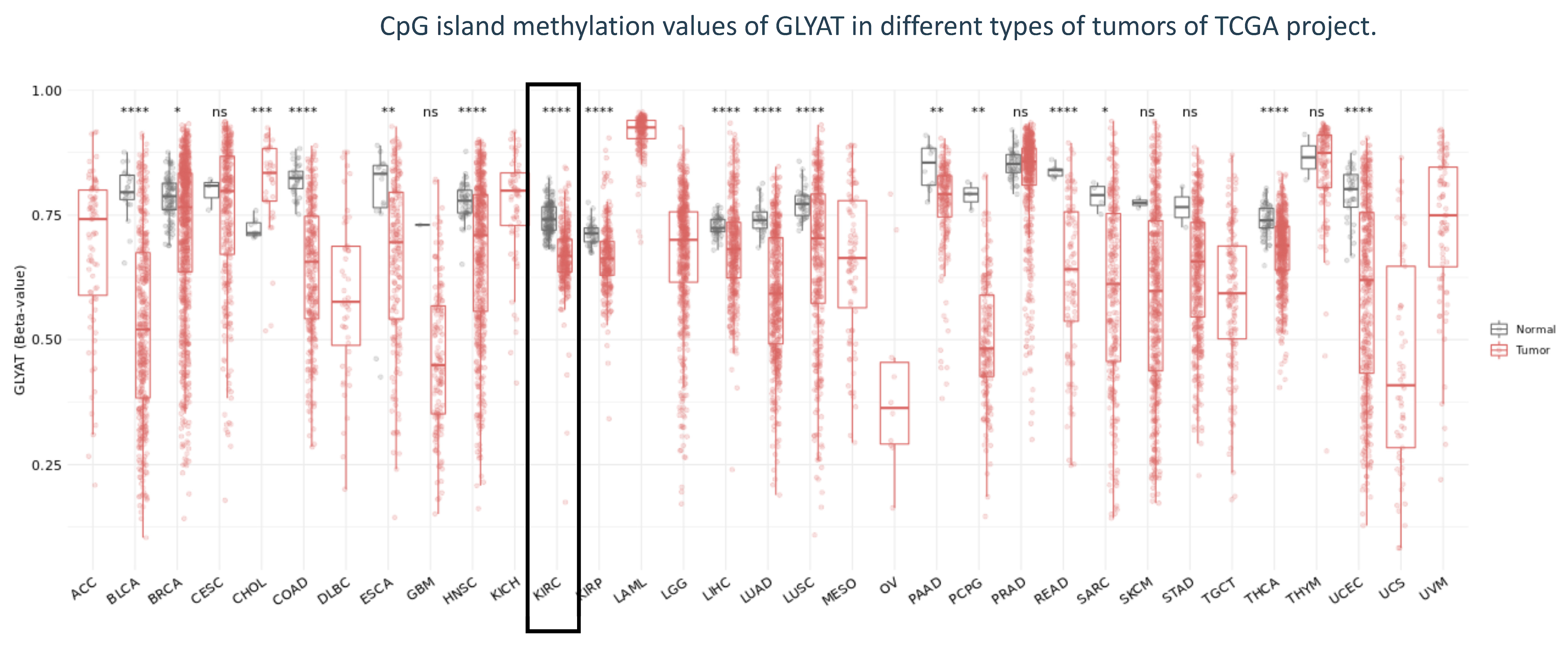
3.3. GLYAT Is Hypomethylated in KIRC Cases
3.4. miRNA Activity Associated to GLYAT Downregulation in KIRC
3.5. Association between GLYAT and Tumor-Infiltrating Immune Cells
3.6. GLYAT Expression on Disease Prognosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Khaleel, S.; Katims, A.; Cumarasamy, S.; Rosenzweig, S.; Attalla, K.; Hakimi, A.A.; Mehrazin, R. Radiogenomics in Clear Cell Renal Cell Carcinoma: A Review of the Current Status and Future Directions. Cancers 2022, 14, 2085. [Google Scholar] [CrossRef]
- Linehan, W.M.; Schmidt, L.S.; Crooks, D.R.; Wei, D.; Srinivasan, R.; Lang, M.; Ricketts, C.J. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019, 9, 1006–1021. [Google Scholar] [CrossRef]
- Finelli, A.; Ismaila, N.; Bro, B.; Durack, J.; Eggener, S.; Evans, A.; Gill, I.; Graham, D.; Huang, W.; Jewett, M.A.; et al. Management of Small Renal Masses: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2017, 35, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.I.; Lam, J.S.; Figlin, R.A.; Belldegrun, A.S. Surveillance strategies for renal cell carcinoma patients following nephrectomy. Rev. Urol. 2006, 8, 1–7. [Google Scholar]
- Sanchez, D.J.; Simon, M.C. Genetic and metabolic hallmarks of clear cell renal cell carcinoma. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Chandra, A.; Chatterjee, I.B. Pathobiology of cigarette smoke-induced invasive cancer of the renal pelvis and its prevention by vitamin C. Toxicol. Rep. 2018, 5, 1002–1010. [Google Scholar] [CrossRef]
- Wettersten, H.I.; Hakimi, A.A.; Morin, D.; Bianchi, C.; Johnstone, M.E.; Donohoe, D.R.; Trott, J.F.; Aboud, O.A.; Stirdivant, S.; Neri, B.; et al. Grade-Dependent Metabolic Reprogramming in Kidney Cancer Revealed by Combined Proteomics and Metabolomics Analysis. Cancer Res. 2015, 75, 2541–2552. [Google Scholar] [CrossRef]
- Perroud, B.; Ishimaru, T.; Borowsky, A.D.; Weiss, R.H. Grade-dependent proteomics characterization of kidney cancer. Mol. Cell Proteom. 2009, 8, 971–985. [Google Scholar] [CrossRef]
- Unwin, R.D.; Craven, R.A.; Harnden, P.; Hanrahan, S.; Totty, N.; Knowles, M.; Eardley, I.; Selby, P.J.; Banks, R.E. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 2003, 3, 1620–1632. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326.e5. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Reznik, E.; Lee, C.-H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Linehan, W.M.; Srinivasan, R.; Schmidt, L.S. The genetic basis of kidney cancer: A metabolic disease. Nat. Rev. Urol. 2010, 7, 277–285. [Google Scholar] [CrossRef]
- Matsuo, M.; Terai, K.; Kameda, N.; Matsumoto, A.; Kurokawa, Y.; Funase, Y.; Nishikawa, K.; Sugaya, N.; Hiruta, N.; Kishimoto, T. Designation of enzyme activity of glycine-N-acyltransferase family genes and depression of glycine-N-acyltransferase in human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2012, 420, 901–906. [Google Scholar] [CrossRef] [PubMed]
- van der Sluis, R.; Badenhorst, C.P.S.; van der Westhuizen, F.H.; van Dijk, A.A. Characterisation of the influence of genetic variations on the enzyme activity of a recombinant human glycine N-acyltransferase. Gene 2013, 515, 447–453. [Google Scholar] [CrossRef]
- Schulke, D.; Sass, J.O. Frequent sequence variants of human glycine N-acyltransferase (GLYAT) and inborn errors of metabolism. Biochimie 2021, 183, 30–34. [Google Scholar] [CrossRef]
- Mitchell, G.A.; Gauthier, N.; Lesimple, A.; Wang, S.P.; Mamer, O.; Qureshi, I. Hereditary and acquired diseases of acyl-coenzyme A metabolism. Mol. Genet. Metab. 2008, 94, 4–15. [Google Scholar] [CrossRef]
- Yang, H.; Zhao, C.; Tang, M.-C.; Wang, Y.; Wang, S.P.; Allard, P.; Furtos, A.; Mitchell, G.A. Inborn errors of mitochondrial acyl-coenzyme a metabolism: Acyl-CoA biology meets the clinic. Mol. Genet. Metab. 2019, 128, 30–44. [Google Scholar] [CrossRef]
- Mawal, Y.; Paradis, K.; Qureshi, I.A. Developmental profile of mitochondrial glycine N-acyltransferase in human liver. J. Pediatr. 1997, 130, 1003–1007. [Google Scholar] [CrossRef]
- Xu, M.; Ren, P.; Tian, J.; Xiao, L.; Hu, P.; Chen, P.; Li, W.; Xue, L. dGLYAT modulates Gadd45-mediated JNK activation and cell invasion. Cell Div. 2022, 17, 4. [Google Scholar] [CrossRef]
- Tian, X.; Wu, L.; Jiang, M.; Zhang, Z.; Wu, R.; Miao, J.; Liu, C.; Gao, S. Downregulation of GLYAT Facilitates Tumor Growth and Metastasis and Poor Clinical Outcomes Through the PI3K/AKT/Snail Pathway in Human Breast Cancer. Front. Oncol. 2021, 11, 641399. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.H.; Li, C.H.; Yeh, C.H.; Huang, P.C.; Fang, C.C.; Chen, Y.F.; Lee, K.J.; Chou, C.H.; Cheng, H.Y.; Huang, H.D.; et al. MicroRNA-224 down-regulates Glycine N-methyltransferase gene expression in Hepatocellular Carcinoma. Sci. Rep. 2018, 8, 12284. [Google Scholar] [CrossRef]
- Siddig, A.; Wan Abdul Rahman, W.F.; Mohd Nafi, S.N.; Sulong, S.; Yahya, M.M.; Al-Astani Tengku Din, T.A.D.; Razali, R.; Musa, K.I. Comparing the Biology of Young versus Old Age Estrogen-Receptor-Positive Breast Cancer through Gene and Protein Expression Analyses. Biomedicines 2023, 11, 200. [Google Scholar] [CrossRef]
- Guan, R.; Hong, W.; Huang, J.; Peng, T.; Zhao, Z.; Lin, Y.; Yu, M.; Jian, Z. The expression and prognostic value of GLYATL1 and its potential role in hepatocellular carcinoma. J. Gastrointest. Oncol. 2020, 11, 1305–1321. [Google Scholar] [CrossRef]
- Eich, M.L.; Chandrashekar, D.S.; Rodriguez Pen, A.M.; Robinson, A.D.; Siddiqui, J.; Daignault-Newton, S.; Chakravarthi, B.; Kunju, L.P.; Netto, G.J.; Varambally, S. Characterization of glycine-N-acyltransferase like 1 (GLYATL1) in prostate cancer. Prostate 2019, 79, 1629–1639. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023. [Google Scholar] [CrossRef]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife 2017, 6, e26476. [Google Scholar] [CrossRef]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Sun, D.; Wang, J.; Han, Y.; Dong, X.; Ge, J.; Zheng, R.; Shi, X.; Wang, B.; Li, Z.; Ren, P.; et al. TISCH: A comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2020, 49, D1420–D1430. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–w560. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef]
- Modhukur, V.; Iljasenko, T.; Metsalu, T.; Lokk, K.; Laisk-Podar, T.; Vilo, J. MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018, 10, 277–288. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Lino Cardenas, C.L.; Bourgine, J.; Cauffiez, C.; Allorge, D.; Lo-Guidice, J.M.; Broly, F.; Chevalier, D. Genetic polymorphisms of glycine N-acyltransferase (GLYAT) in a French Caucasian population. Xenobiotica 2010, 40, 853–861. [Google Scholar] [CrossRef]
- Ren, P.; Li, W.; Xue, L. GLYAT regulates JNK-mediated cell death in Drosophila. Sci. Rep. 2017, 7, 5183. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz, J.P.; Calaf, G.M. Downregulation of Glycine N-Acyltransferase in Kidney Renal Clear Cell Carcinoma: A Bioinformatic-Based Screening. Diagnostics 2023, 13, 3505. https://doi.org/10.3390/diagnostics13233505
Muñoz JP, Calaf GM. Downregulation of Glycine N-Acyltransferase in Kidney Renal Clear Cell Carcinoma: A Bioinformatic-Based Screening. Diagnostics. 2023; 13(23):3505. https://doi.org/10.3390/diagnostics13233505
Chicago/Turabian StyleMuñoz, Juan P., and Gloria M. Calaf. 2023. "Downregulation of Glycine N-Acyltransferase in Kidney Renal Clear Cell Carcinoma: A Bioinformatic-Based Screening" Diagnostics 13, no. 23: 3505. https://doi.org/10.3390/diagnostics13233505




