Dynomics: A Novel and Promising Approach for Improved Breast Cancer Prognosis Prediction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset
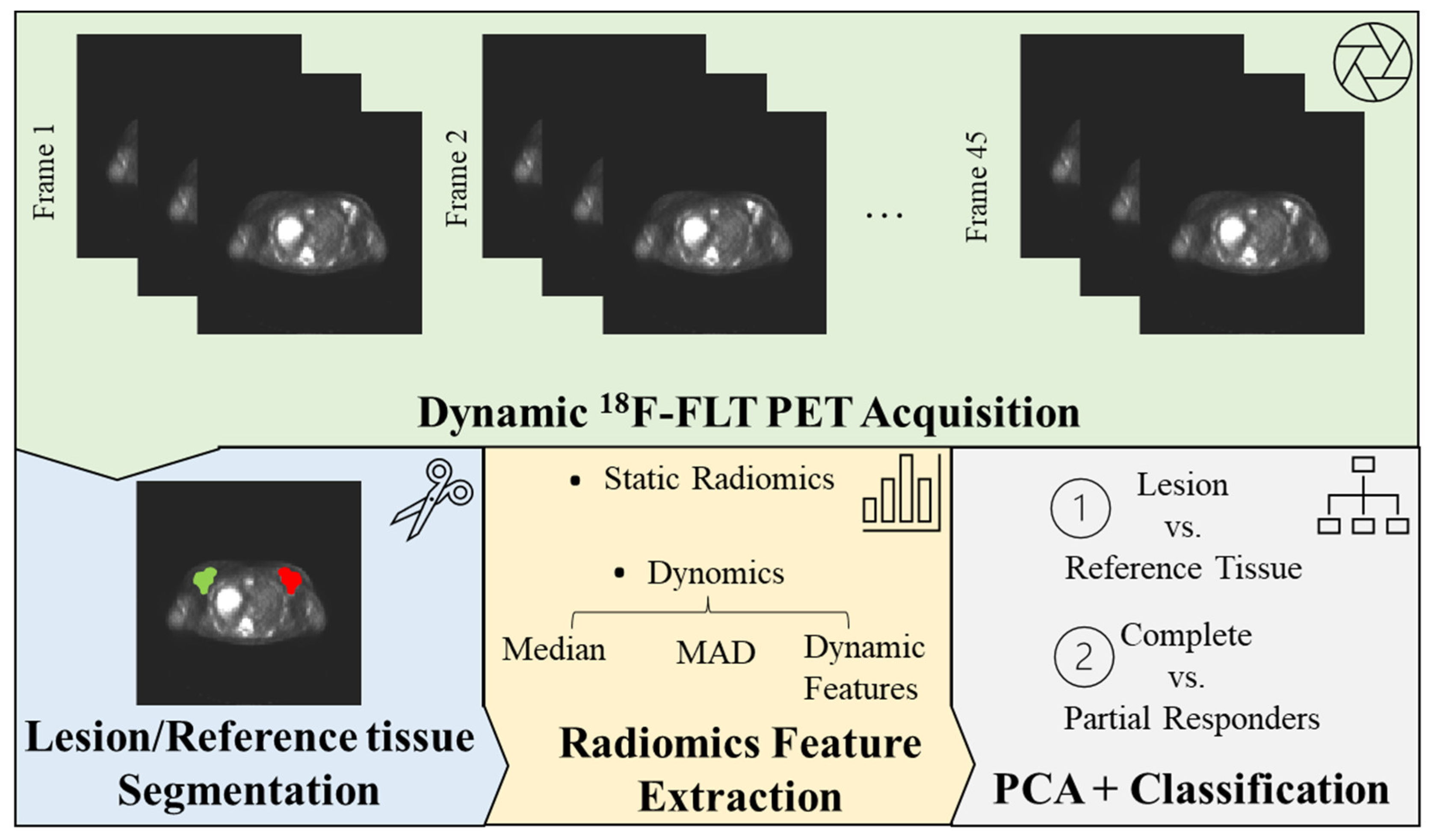
2.2. PET Data Pre-Processing
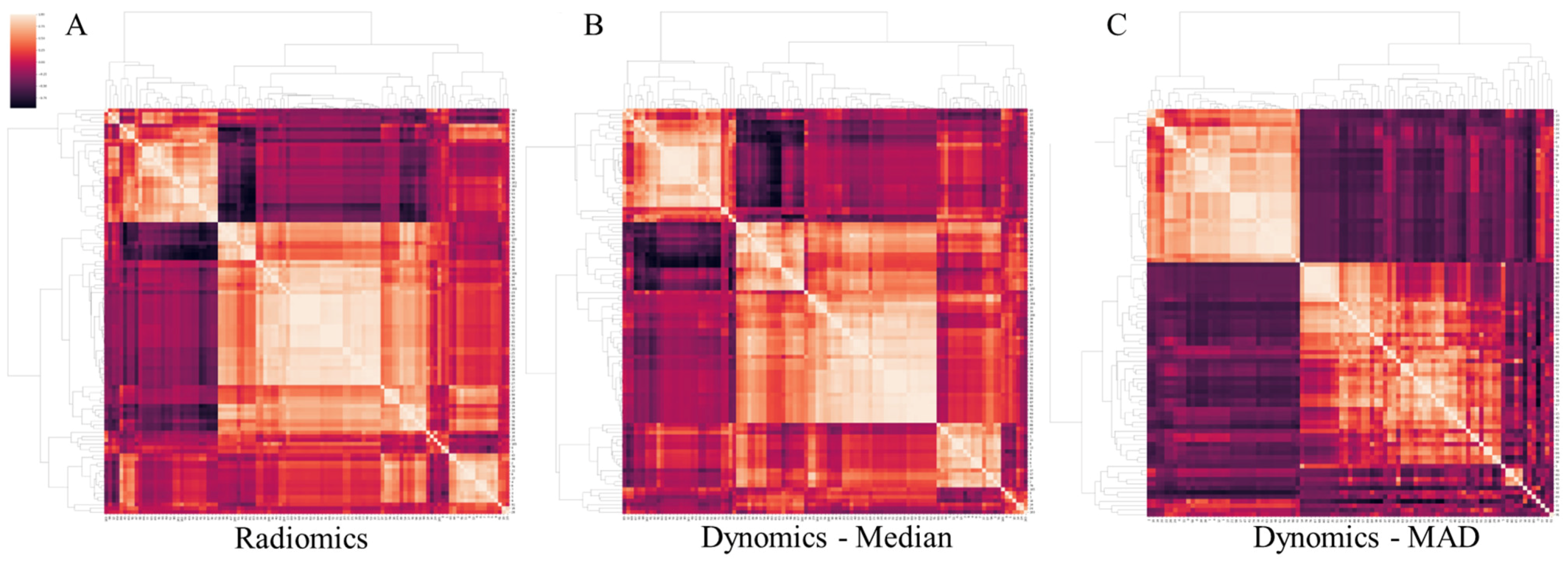
2.3. Radiomic Feature Extraction
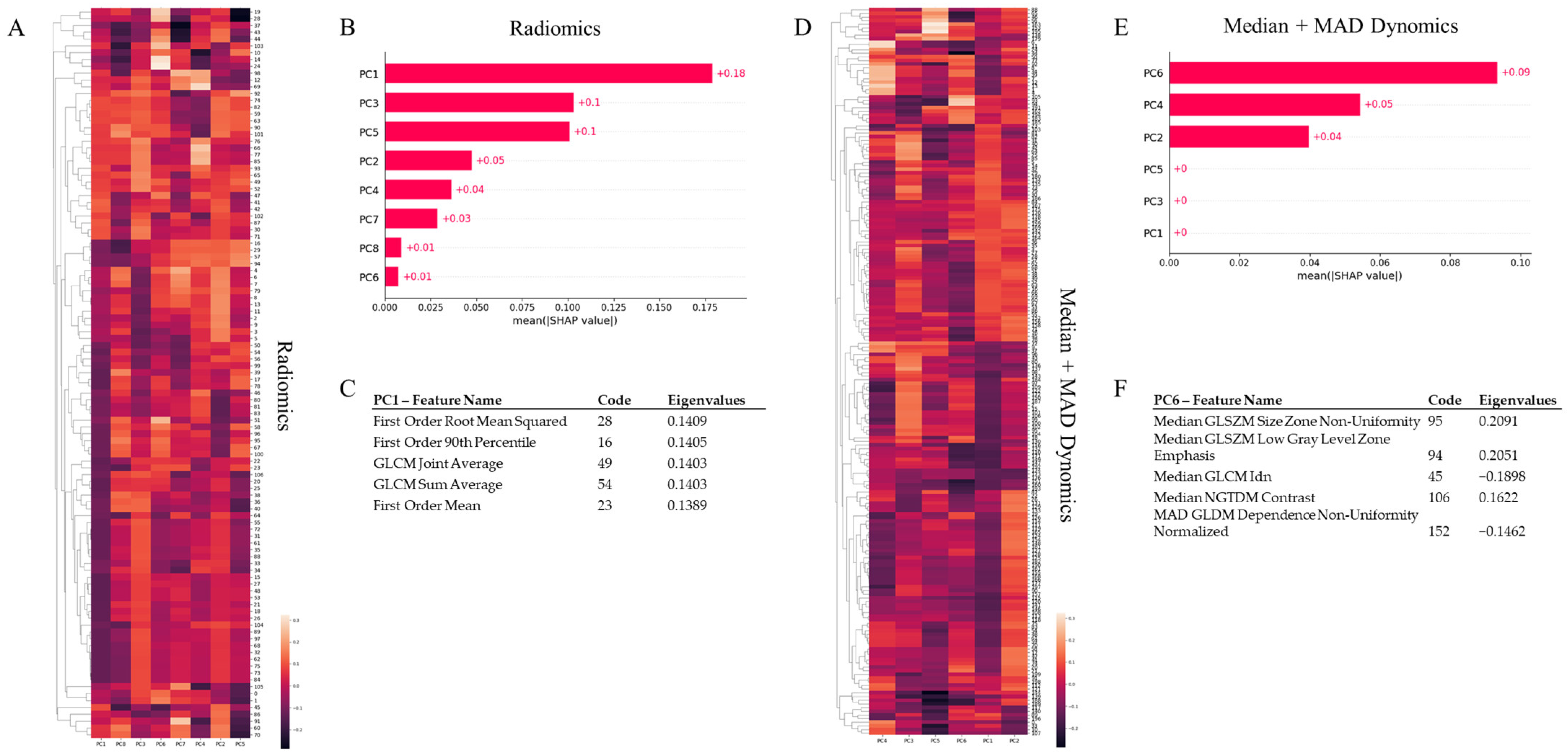
2.4. Machine Learning Models
3. Results
3.1. Tumour vs. Reference Tissue Classification
3.2. Complete vs. Partial Responders Classification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sollini, M.; Cozzi, L.; Ninatti, G.; Antunovic, L.; Cavinato, L.; Chiti, A.; Kirienko, M. PET/CT radiomics in breast cancer: Mind the step. Methods 2021, 188, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Conti, A.; Duggento, A.; Indovina, I.; Guerrisi, M.; Toschi, N. Radiomics in breast cancer classification and prediction. Semin. Cancer Biol. 2021, 72, 238–250. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Mariscotti, G.; Valdora, F.; Durando, M.; Nori, J.; La Forgia, D.; Rosenberg, I.; Caumo, F.; Gandolfo, N.; Sormani, M.P.; et al. A prospective comparative trial of adjunct screening with tomosynthesis or ultrasound in women with mammography-negative dense breasts (ASTOUND-2). Eur. J. Cancer 2018, 104, 39–46. [Google Scholar] [CrossRef]
- Yang, S.K.; Cho, N.; Moon, W.K. The Role of PET/CT for Evaluating Breast Cancer. Korean J. Radiol. 2007, 8, 429–437. [Google Scholar] [CrossRef] [Green Version]
- Kong, E.; Chun, K.A.; Cho, I.H. Quantitative assessment of simultaneous F-18 FDG PET/MRI in patients with various types of hepatic tumors: Correlation between glucose metabolism and apparent diffusion coefficient. PLoS ONE 2017, 12, e0180184. [Google Scholar] [CrossRef] [Green Version]
- Boers, J.; de Vries, E.F.J.; Glaudemans, A.W.J.M.; Hospers, G.A.P.; Schröder, C.P. Application of PET Tracers in Molecular Imaging for Breast Cancer. Curr. Oncol. Rep. 2020, 22, 85. [Google Scholar] [CrossRef]
- Adejolu, M.; Huo, L.; Rohren, E.; Santiago, L.; Yang, W.T. False-Positive Lesions Mimicking Breast Cancer on FDG PET and PET/CT. Am. J. Roentgenol. 2012, 198, W304–W314. [Google Scholar] [CrossRef]
- Carlson, R.W.; Allred, D.C.; Anderson, B.O.; Burstein, H.J.; Edge, S.B.; Farrar, W.B.; Forero, A.; Giordano, S.H.; Goldstein, L.J.; Gradishar, W.J.; et al. Metastatic Breast Cancer, Version 1.2012. J. Natl. Compr. Cancer Netw. 2012, 10, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Venema, C.M.; Apollonio, G.; Hospers, G.A.P.; Schröder, C.P.; Dierckx, R.A.J.O.; de Vries, E.F.J.; Glaudemans, A.W.J.M. Recommendations and Technical Aspects of 16α-[18F]Fluoro-17β-Estradiol PET to Image the Estrogen Receptor In Vivo: The Groningen Experience. Clin. Nucl. Med. 2016, 41, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, L.; Guarneri, V.; Conte, P.F. 18F-Fluoroestradiol Positron Emission Tomography in Breast Cancer Patients: Systematic Review of the Literature & Meta-Analysis. Curr. Radiopharm. 2016, 9, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Dehdashti, F.; Wu, N.; Bose, R.; Naughton, M.J.; Ma, C.X.; Marquez-Nostra, B.V.; Diebolder, P.; Mpoy, C.; Rogers, B.E.; Lapi, S.E.; et al. Evaluation of [89Zr]Trastuzumab-PET/CT in Differentiating HER2-Positive from HER2-Negative Breast Cancer. Breast Cancer Res. Treat. 2018, 169, 523–530. [Google Scholar] [CrossRef]
- Romine, P.E.; Peterson, L.M.; Kurland, B.F.; Byrd, D.W.; Novakova-Jiresova, A.; Muzi, M.; Specht, J.M.; Doot, R.K.; Link, J.M.; Krohn, K.A.; et al. 18F-fluorodeoxyglucose (FDG) PET or 18F-fluorothymidine (FLT) PET to assess early response to aromatase inhibitors (AI) in women with ER+ operable breast cancer in a window-of-opportunity study. Breast Cancer Res. 2021, 23, 88. [Google Scholar] [CrossRef]
- Sanghera, B.; Wong, W.L.; Sonoda, L.I.; Beynon, G.; Makris, A.; Woolf, D.; Ardeshna, K. FLT PET-CT in evaluation of treatment response. Indian J. Nucl. Med. 2014, 29, 65–73. [Google Scholar] [CrossRef] [Green Version]
- Fantini, L.; Belli, M.L.; Azzali, I.; Loi, E.; Bettinelli, A.; Feliciani, G.; Mezzenga, E.; Fedeli, A.; Asioli, S.; Paganelli, G.; et al. Exploratory Analysis of 18F-3’-deoxy-3’-fluorothymidine (18F-FLT) PET/CT-Based Radiomics for the Early Evaluation of Response to Neoadjuvant Chemotherapy in Patients with Locally Advanced Breast Cancer. Front. Oncol. 2021, 11, 601053. [Google Scholar] [CrossRef]
- Crippa, F.; Agresti, R.; Sandri, M.; Mariani, G.; Padovano, B.; Alessi, A.; Bianchi, G.; Bombardieri, E.; Maugeri, I.; Rampa, M.; et al. 18F-FLT PET/CT as an imaging tool for early prediction of pathological response in patients with locally advanced breast cancer treated with neoadjuvant chemotherapy: A pilot study. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 818–830. [Google Scholar] [CrossRef]
- Bailly, C.; Bodet-Milin, C.; Bourgeois, M.; Gouard, S.; Ansquer, C.; Barbaud, M.; Sébille, J.-C.; Chérel, M.; Kraeber-Bodéré, F.; Carlier, T. Exploring Tumor Heterogeneity Using PET Imaging: The Big Picture. Cancers 2019, 11, 1282. [Google Scholar] [CrossRef] [Green Version]
- Jamal-Hanjani, M.; Quezada, S.A.; Larkin, J.; Swanton, C. Translational implications of tumor heterogeneity. Clin. Cancer Res. 2015, 21, 1258–1266. [Google Scholar] [CrossRef] [Green Version]
- Dimitrakopoulou-Strauss, A.; Pan, L.; Sachpekidis, C. Kinetic modeling and parametric imaging with dynamic PET for oncological applications: General considerations, current clinical applications, and future perspectives. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Antunovic, L.; De Sanctis, R.; Cozzi, L.; Kirienko, M.; Sagona, A.; Torrisi, R.; Tinterri, C.; Santoro, A.; Chiti, A.; Zelic, R.; et al. PET/CT radiomics in breast cancer: Promising tool for prediction of pathological response to neoadjuvant chemotherapy. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Do Clinical, Histological or Immunohistochemical Primary Tumour Characteristics Translate into Different 18F-FDG PET/CT Volumetric and Heterogeneity Features in Stage II/III Breast Cancer?|SpringerLink. Available online: https://link.springer.com/article/10.1007/s00259-015-3110-x (accessed on 12 October 2022).
- Lee, H.; Lee, D.; Park, S.; Kim, T.S.; Jung, S.-Y.; Lee, S.; Kang, H.S.; Lee, E.S.; Sim, S.H.; Park, I.H.; et al. Predicting Response to Neoadjuvant Chemotherapy in Patients with Breast Cancer: Combined Statistical Modeling Using Clinicopathological Factors and FDG PET/CT Texture Parameters. Clin. Nucl. Med. 2019, 44, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Kinahan, P.E.; Fletcher, J.W. PET/CT Standardized Uptake Values (SUVs) in Clinical Practice and Assessing Response to Therapy. Semin. Ultrasound CT MR 2010, 31, 496–505. [Google Scholar] [CrossRef] [Green Version]
- Kostakoglu, L.; Duan, F.; Idowu, M.O.; Jolles, P.R.; Bear, H.D.; Muzi, M.; Cormack, J.; Muzi, J.P.; Pryma, D.A.; Specht, J.M.; et al. A Phase II Study of 3’-Deoxy-3’-18F-Fluorothymidine PET in the Assessment of Early Response of Breast Cancer to Neoadjuvant Chemotherapy: Results from ACRIN 6688. J. Nucl. Med. 2015, 56, 1681–1689. [Google Scholar] [CrossRef] [Green Version]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westerterp, M.; Pruim, J.; Oyen, W.; Hoekstra, O.; Paans, A.; Visser, E.; van Lanschot, J.; Sloof, G.; Boellaard, R. Quantification of FDG PET studies using standardised uptake values in multi-centre trials: Effects of image reconstruction, resolution and ROI definition parameters. Eur. J. Nucl. Med. Mol. Imaging 2007, 34, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Inglese, M.; Ferrante, M.; Duggento, A.; Boccato, T.; Toschi, N. Spatiotemporal learning of dynamic positron emission tomography data improves diagnostic accuracy in breast cancer. IEEE Trans. Radiat. Plasma Med. Sci. 2023, 1. [Google Scholar] [CrossRef]
- Šimundić, A.-M. Measures of Diagnostic Accuracy: Basic Definitions. EJIFCC 2009, 19, 203–211. [Google Scholar]
- Yuan, H.; Yu, H.; Wang, J.; Li, K.; Ji, S. On Explainability of Graph Neural Networks via Subgraph Explorations. arXiv 2021. [Google Scholar] [CrossRef]
- Seal, A.; Bhattacharjee, D.; Nasipuri, M. Predictive and probabilistic model for cancer detection using computer tomography images. Multimed Tools Appl. 2018, 77, 3991–4010. [Google Scholar] [CrossRef]
- Bonadonna, G.; Valagussa, P.; Brambilla, C.; Ferrari, L.; Moliterni, A.; Terenziani, M.; Zambetti, M. Primary chemotherapy in operable breast cancer: Eight-year experience at the Milan Cancer Institute. J. Clin. Oncol. 1998, 16, 93–100. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Sharma, R.; Valls, P.O.; Inglese, M.; Dubash, S.; Chen, M.; Gabra, H.; Montes, A.; Challapalli, A.; Arshad, M.; Tharakan, G.; et al. [18F]Fluciclatide PET as a biomarker of response to combination therapy of pazopanib and paclitaxel in platinum-resistant/refractory ovarian cancer. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1239–1251. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Inglese, M.; Dubash, S.; Lu, H.; Pinato, D.J.; Sanghera, C.; Patel, N.; Chung, A.; Tait, P.D.; Mauri, F.; et al. Monitoring response to transarterial chemoembolization in hepatocellular carcinoma using 18F-fluorothymidine PET. J. Nucl. Med. 2020, 61, 1743–1748. [Google Scholar] [CrossRef]
- Dubash, S.; Inglese, M.; Mauri, F.; Kozlowski, K.; Trivedi, P.; Arshad, M.; Challapalli, A.; Barwick, T.; Al-Nahhas, A.; Stanbridge, R.; et al. Spatial heterogeneity of radiolabeled choline positron emission tomography in tumors of patients with non-small cell lung cancer: First-in-patient evaluation of [18F]fluoromethyl-(1,2-2H4)-choline. Theranostics 2020, 10, 8677–8690. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Inglese, M.; Dubash, S.; Barnes, C.; Brickute, D.; Braga, M.C.; Wang, N.; Beckley, A.; Heinzmann, K.; Allott, L.; et al. Consideration of metabolite efflux in radiolabelled choline kinetics. Pharmaceutics 2021, 13, 1246. [Google Scholar] [CrossRef] [PubMed]
- Urso, L.; Manco, L.; Castello, A.; Evangelista, L.; Guidi, G.; Castellani, M.; Florimonte, L.; Cittanti, C.; Turra, A.; Panareo, S. PET-Derived Radiomics and Artificial Intelligence in Breast Cancer: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 13409. [Google Scholar] [CrossRef] [PubMed]
- Baliu-Piqué, M.; Pandiella, A.; Ocana, A. Breast Cancer Heterogeneity and Response to Novel Therapeutics. Cancers 2020, 12, 3271. [Google Scholar] [CrossRef]
- Muzi, M.; Vesselle, H.; Grierson, J.R.; Mankoff, D.A.; Schmidt, R.A.; Peterson, L.; Wells, J.M.; Krohn, K.A. Kinetic Analysis of 3′-Deoxy-3′-Fluorothymidine PET Studies: Validation Studies in Patients with Lung Cancer. J. Nucl. Med. 2005, 46, 274–282. [Google Scholar]
- Muzi, M.; Mankoff, D.A.; Grierson, J.R.; Wells, J.M.; Vesselle, H.; Krohn, K.A. Kinetic Modeling of 3′-Deoxy-3′-Fluorothymidine in Somatic Tumors: Mathematical Studies. J. Nucl. Med. 2005, 46, 371–380. [Google Scholar]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef]
- Mascalchi, M.; Ginestroni, A.; Toschi, N.; Poggesi, A.; Cecchi, P.; Salvadori, E.; Tessa, C.; Cosottini, M.; De Stefano, N.; Pracucci, G.; et al. The burden of microstructural damage modulates cortical activation in elderly subjects with MCI and leuko-araiosis. A DTI and fMRI study. Hum. Brain Mapp. 2014, 35, 819–830. [Google Scholar] [CrossRef]
- Nigro, S.; Passamonti, L.; Riccelli, R.; Toschi, N.; Rocca, F.; Valentino, P.; Nisticò, R.; Fera, F.; Quattrone, A. Structural “connectomic” alterations in the limbic system of multiple sclerosis patients with major depression. Mult. Scler. J. 2015, 21, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Vergallo, A.; Bun, R.-S.; Toschi, N.; Baldacci, F.; Zetterberg, H.; Blennow, K.; Cavedo, E.; Lamari, F.; Habert, M.-O.; Dubois, B.; et al. Association of cerebrospinal fluid α-synuclein with total and phospho-tau181 protein concentrations and brain amyloid load in cognitively normal subjective memory complainers stratified by Alzheimer’s disease biomarkers. Alzheimer’s Dement. 2018, 14, 1623–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morland, D.; Triumbari, E.K.A.; Boldrini, L.; Gatta, R.; Pizzuto, D.; Annunziata, S. Radiomics in Oncological PET Imaging: A Systematic Review-Part 1, Supradiaphragmatic Cancers. Diagnostics 2022, 12, 1329. [Google Scholar] [CrossRef]
- Brescia, E.; Costantino, D.; Marzo, F.; Massenio, P.R.; Cascella, G.L.; Naso, D. Automated Multistep Parameter Identification of SPMSMs in Large-Scale Applications Using Cloud Computing Resources. Sensors 2021, 21, 4699. [Google Scholar] [CrossRef] [PubMed]
- Araz, M.; Soydal, Ç.; Gündüz, P.; Kırmızı, A.; Bakırarar, B.; Dizbay Sak, S.; Özkan, E. Can Radiomics Analyses in 18F-FDG PET/CT Images of Primary Breast Carcinoma Predict Hormone Receptor Status? Mol. Imaging Radionucl. Ther. 2022, 31, 49–56. [Google Scholar] [CrossRef] [PubMed]
| Demographic | Variable | Patients (n = 44) |
|---|---|---|
| Mean age ± SD (y) | 52.5 ± 10.6 | |
| Mean tumor size ± SD (cm) | 4.6 ± 2.5 | |
| Menopausal status | Premenopausal | 16 (36.4%) |
| Postmenopausal | 28 (63.6%) | |
| Initial diagnosis | Invasive breast cancer NOS | 1 (2.27%) |
| Invasive ductal | 39 (88.63%) | |
| Invasive lobular + mixed invasive and lobular | 4 (9.09%) | |
| Estrogen receptor status | Positive | 24 (54.54%) |
| Negative | 20 (45.45%) | |
| Progesterone receptor status | Positive | 25 (56.81%) |
| Negative | 29 (65.91%) | |
| HER2 status | Positive | 12 (27.27%) |
| Negative | 22 (50.00%) | |
| Receptor status | Triple negative | 9 (20.45%) |
| Other | 35 (79.54%) | |
| Grade at diagnosis | 1 | 1 (2.27%) |
| 2 | 9 (20.45%) | |
| 3 | 27 (61.36%) |
| AUC | Accuracy | Precision | Recall | |||||
|---|---|---|---|---|---|---|---|---|
| PET Acquisition | Input Data | Model | Lesion | Reference | Lesion | Reference | ||
| Static | PET image | CONV3D | 0.59 (±0.09) | 0.61 (±0.99) | 0.56 (±0.13) | 0.61 (±0.21) | 0.67 (±0.17) | 0.48 (±0.15) |
| Radiomics | XGBoost | 0.94 | 0.94 | 1.00 | 0.90 | 0.89 | 1.00 | |
| Dynamic | PET image | CONV3D + LSTM | 0.81 (±0.08) | 0.75 (±0.09) | 0.69 (±0.09) | 0.91 (±0.09) | 0.96 (±0.03) | 0.55 (±0.17) |
| Dynomics—Median | XGBoost | 0.94 | 0.94 | 0.90 | 1.00 | 1.00 | 0.89 | |
| Dynomics—MAD | XGBoost | 0.89 | 0.89 | 1 | 0.82 | 0.78 | 1.00 | |
| Dynomics—Median + MAD | XGBoost | 0.94 | 0.94 | 1 | 0.90 | 0.89 | 1 | |
| Dynomics | CONV1D | 0.49 (±0.05) | 0.49 (±0.05) | 0.49 (±0.03) | 0.48 (±0.02) | 0.84 (±0.11) | 0.15 (±0.10) | |
| AUC | Accuracy | Precision | Recall | |||||
|---|---|---|---|---|---|---|---|---|
| PET Acquisition | Input Data | Model | CR | PR | CR | PR | ||
| Static | PET image | CONV3D | 0.50 (±0.00) | 0.59 (±0.18) | 0.30 (±0.37) | 0.20 (±0.26) | 0.40 (±0.48) | 0.60 (±0.48) |
| Radiomics | XGBoost | 0.67 | 0.71 | 1.00 | 0.67 | 0.33 | 1.00 | |
| Dynamic | PET image | CONV3D + LSTM | 0.50 (±0.00) | 0.59 (±0.18) | 0.30 (±0.37) | 0.29 (±0.27) | 0.40 (±0.49) | 0.60 (±0.49) |
| Dynomics—Median | XGBoost | 0.67 | 0.71 | 1.00 | 0.67 | 0.33 | 1.00 | |
| Dynomics—MAD | XGBoost | 0.58 | 0.57 | 0.50 | 0.67 | 0.67 | 0.50 | |
| Dynomics—Median + MAD | XGBoost | 0.83 | 0.86 | 1.00 | 0.80 | 0.67 | 1.00 | |
| Dynomics | CONV1D | 0.50 (±0.00) | 0.52 (±0.11) | 0.37 (±0.30) | 0.15 (±0.19) | 0.60 (±0.49) | 0.40 (±0.49) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inglese, M.; Ferrante, M.; Boccato, T.; Conti, A.; Pistolese, C.A.; Buonomo, O.C.; D’Angelillo, R.M.; Toschi, N. Dynomics: A Novel and Promising Approach for Improved Breast Cancer Prognosis Prediction. J. Pers. Med. 2023, 13, 1004. https://doi.org/10.3390/jpm13061004
Inglese M, Ferrante M, Boccato T, Conti A, Pistolese CA, Buonomo OC, D’Angelillo RM, Toschi N. Dynomics: A Novel and Promising Approach for Improved Breast Cancer Prognosis Prediction. Journal of Personalized Medicine. 2023; 13(6):1004. https://doi.org/10.3390/jpm13061004
Chicago/Turabian StyleInglese, Marianna, Matteo Ferrante, Tommaso Boccato, Allegra Conti, Chiara A. Pistolese, Oreste C. Buonomo, Rolando M. D’Angelillo, and Nicola Toschi. 2023. "Dynomics: A Novel and Promising Approach for Improved Breast Cancer Prognosis Prediction" Journal of Personalized Medicine 13, no. 6: 1004. https://doi.org/10.3390/jpm13061004





