Stability of Multi-Parametric Prostate MRI Radiomic Features to Variations in Segmentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. Internal Dataset
2.1.2. External Dataset
2.2. In Silico Contour Generation
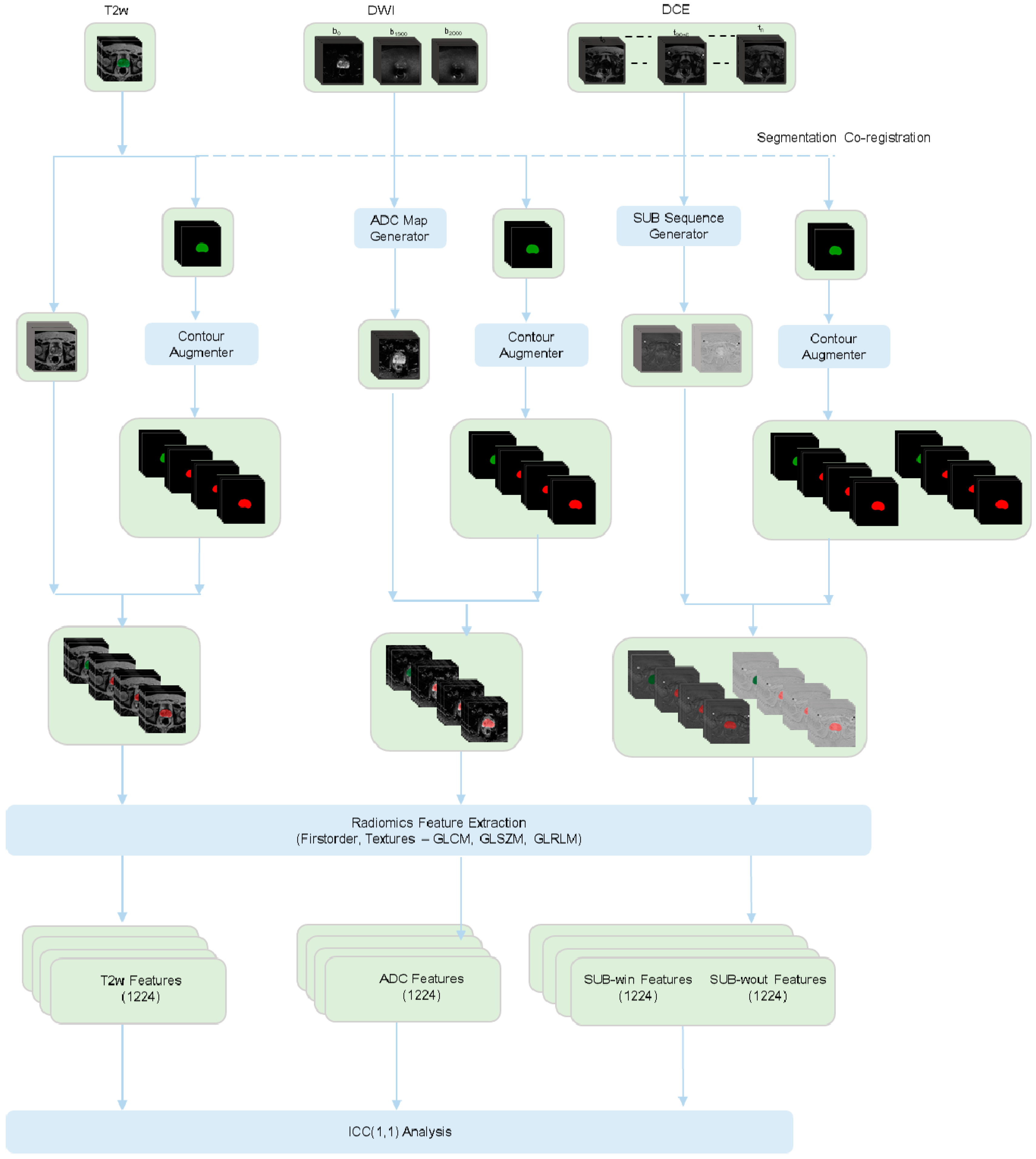
2.3. Image Processing Pipeline
2.4. Radiomics Feature Extraction Pipeline
- First-order statistics (FO, n = 18) providing information about the histogram of the grey values inside the prostate ROI; and
- Texture features, providing information about the spatial distribution of grey values. We used the following textural matrices to compute the textural features: Gray Level Co-occurrence Matrix (GLCM, n = 22 features); Gray Level Run Length Matrix (GLRLM, n = 16 features); Gray Level Size Zone Matrix (GLSZM, n = 16 features).
2.5. Stability Analysis
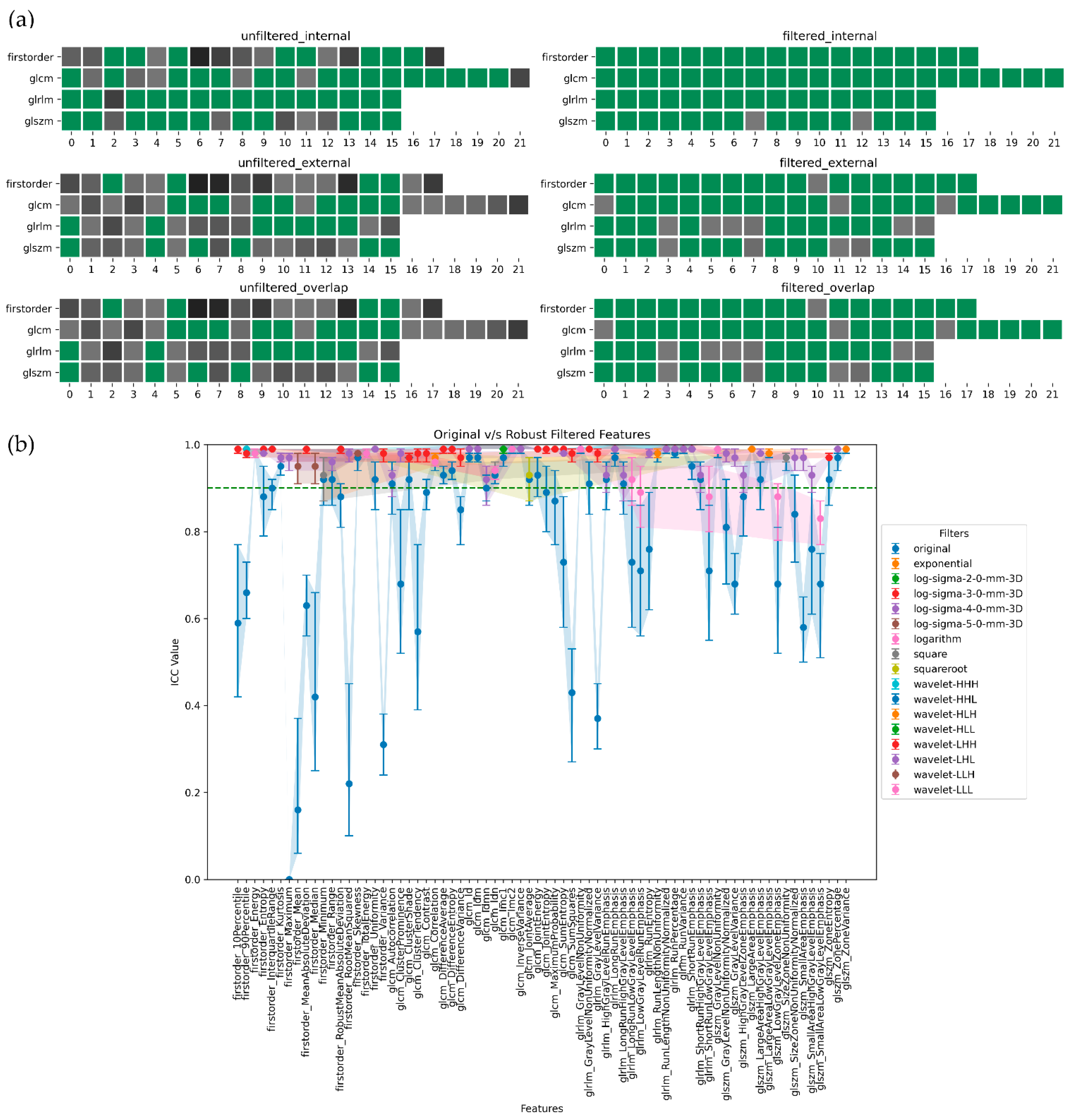
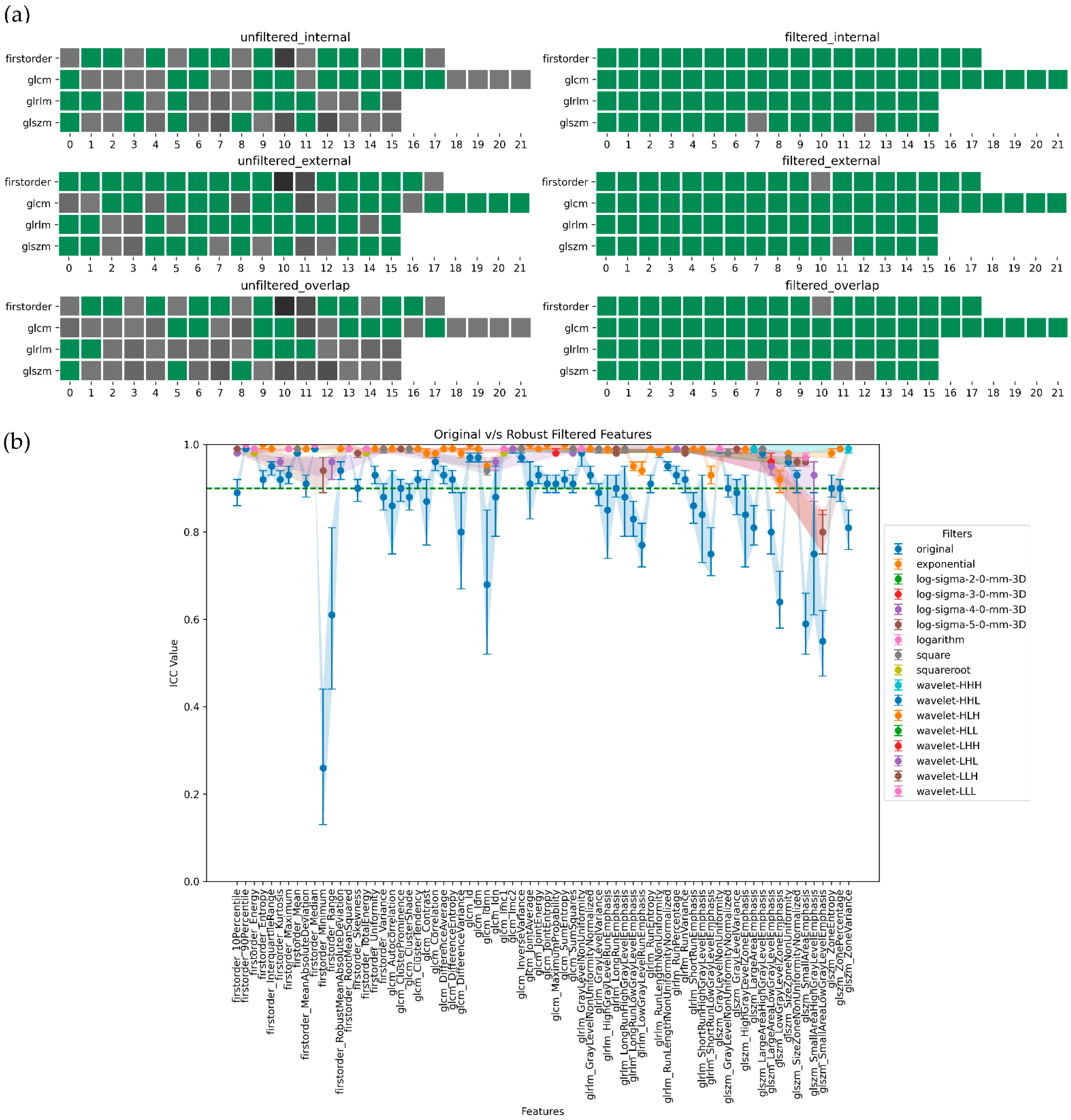
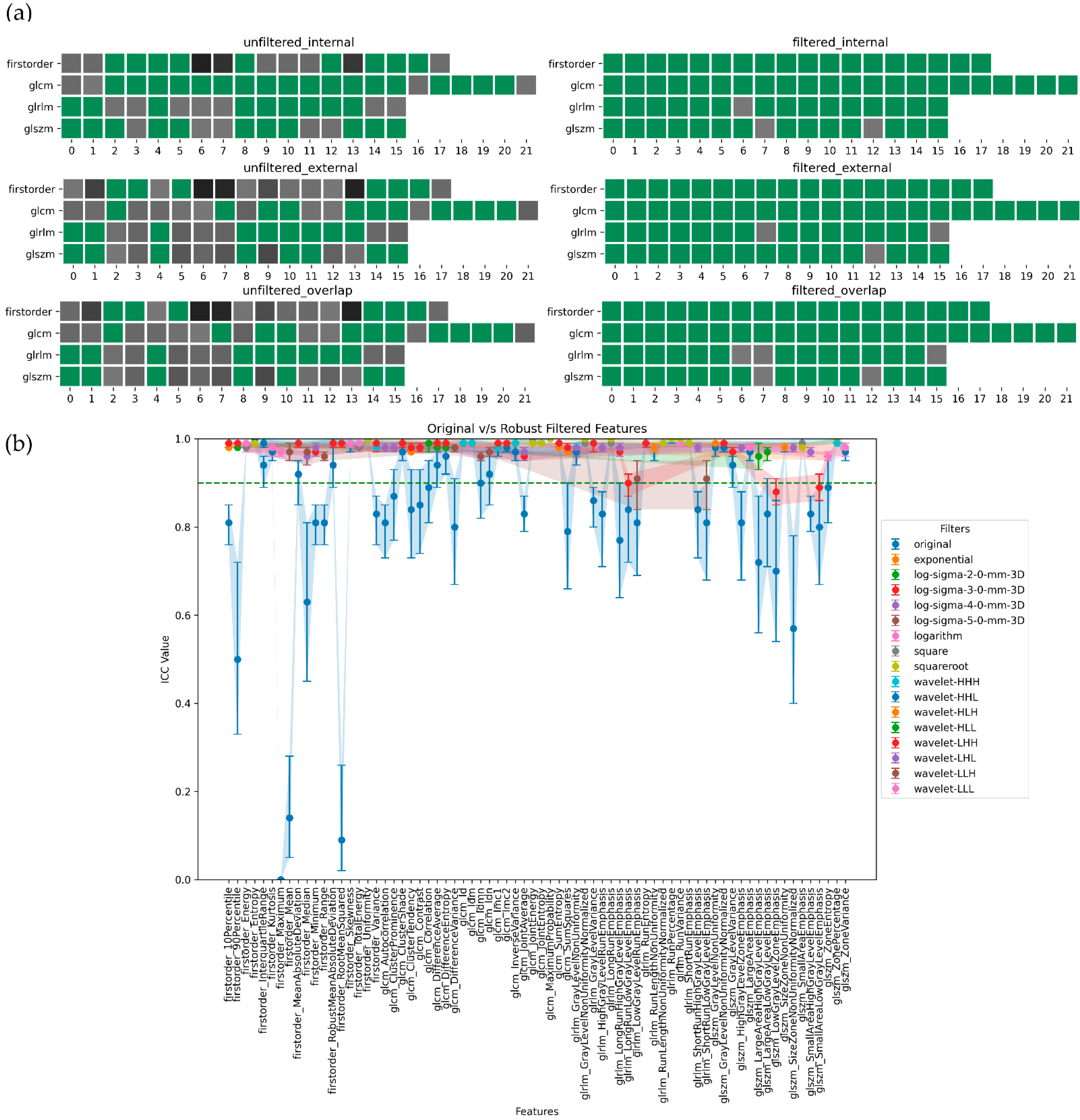
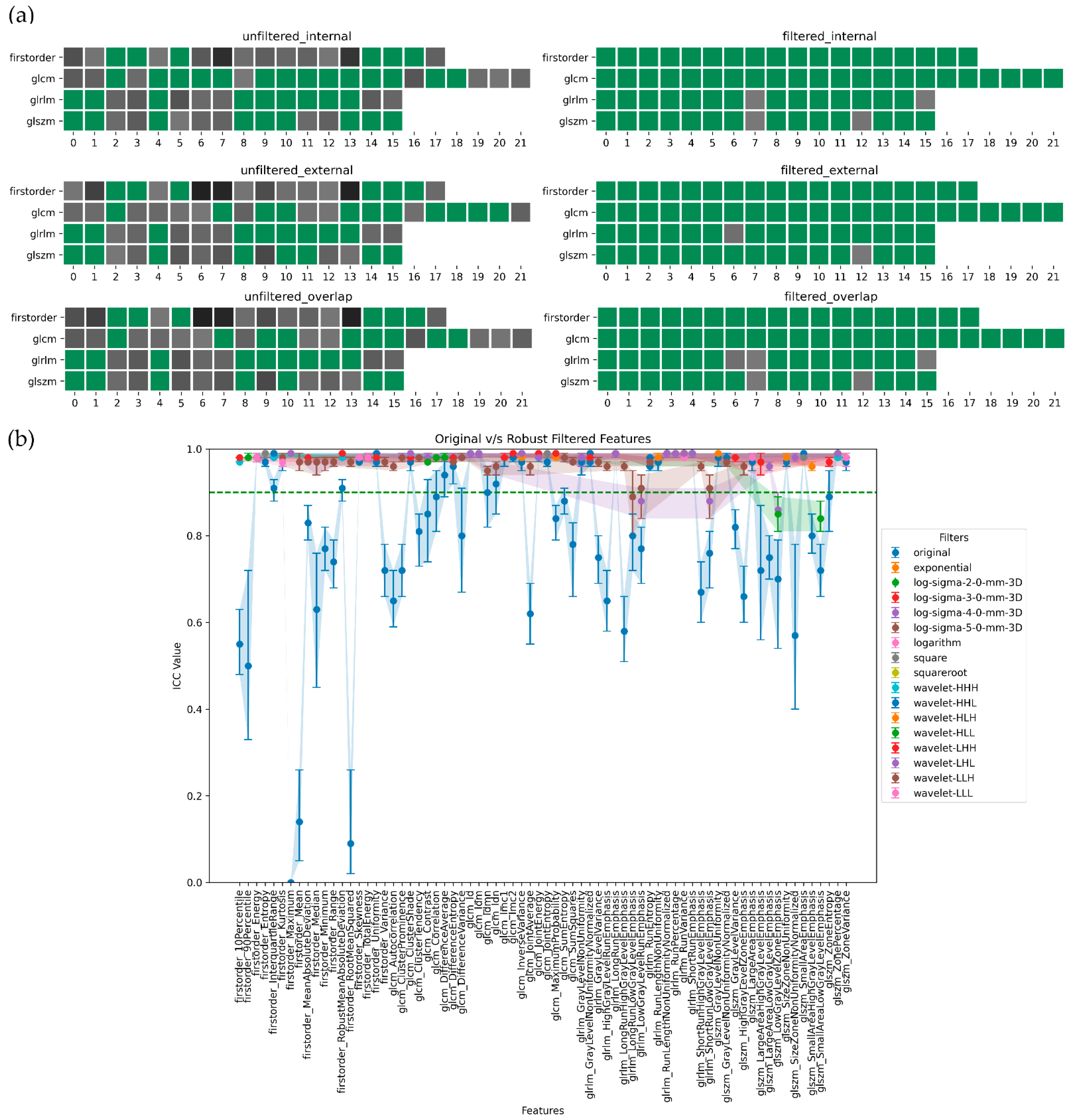
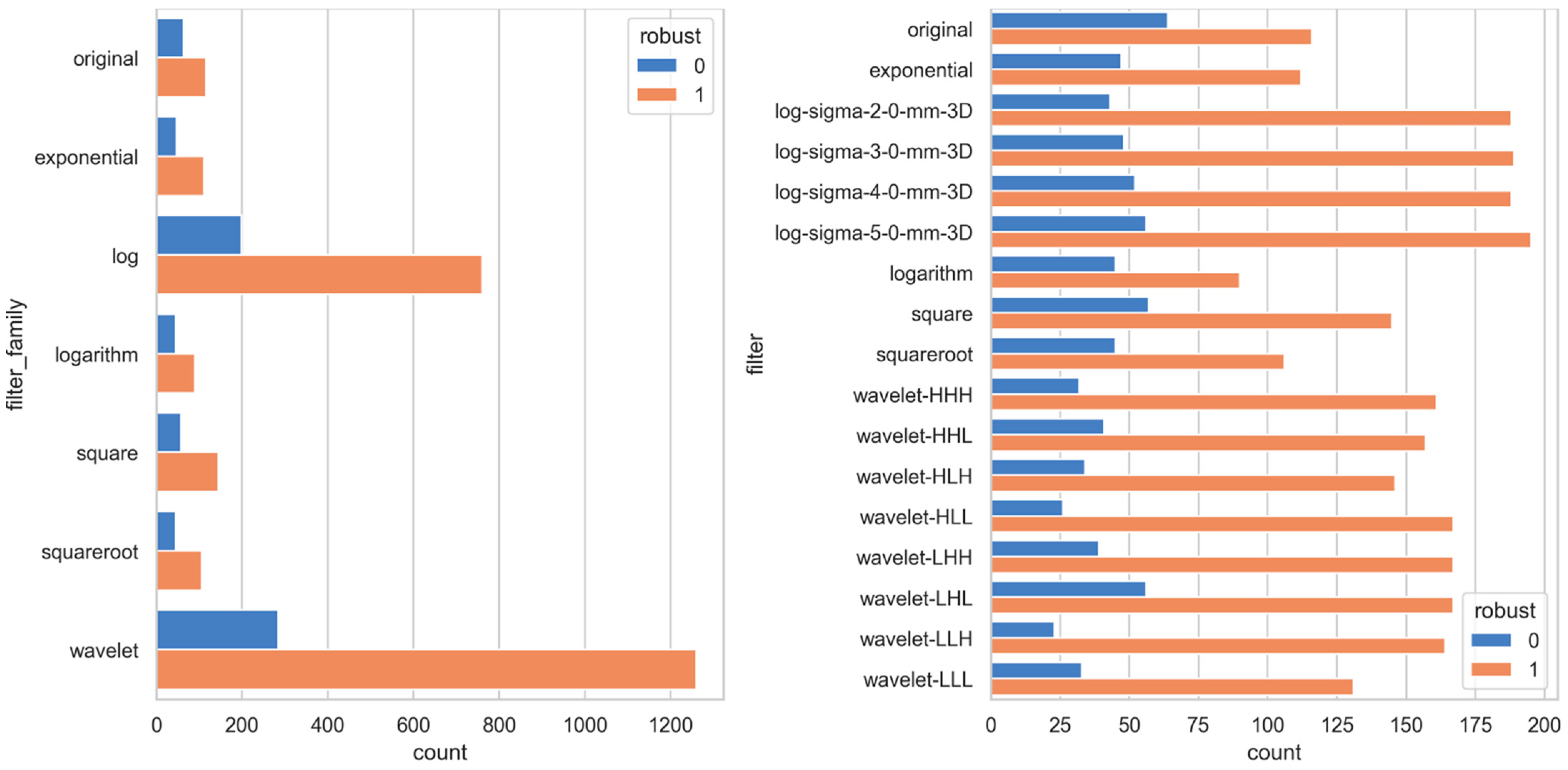
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, L.M.; Turkbey, B.; Figg, W.D.; Choyke, P.L. Multi-parametric MRI in Prostate Cancer Management. Nat. Rev. Clin. Oncol. 2014, 11, 346–353. [Google Scholar] [CrossRef]
- Barrett, T.; Haider, M.A. The Emerging Role of MRI in Prostate Cancer Active Surveillance and Ongoing Challenges. AJR Am. J. Roentgenol. 2017, 208, 131–139. [Google Scholar] [CrossRef]
- Thurtle, D.; Barrett, T.; Thankappan-Nair, V.; Koo, B.; Warren, A.; Kastner, C.; Saeb-Parsy, K.; Kimberley-Duffell, J.; Gnanapragasam, V.J. Progression and Treatment Rates Using an Active Surveillance Protocol Incorporating Image-Guided Baseline Biopsies and Multi-parametric Magnetic Resonance Imaging Monitoring for Men with Favourable-Risk Prostate Cancer. BJU Int. 2018, 122, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Schoots, I.G.; Petrides, N.; Giganti, F.; Bokhorst, L.P.; Rannikko, A.; Klotz, L.; Villers, A.; Hugosson, J.; Moore, C.M. Magnetic Resonance Imaging in Active Surveillance of Prostate Cancer: A Systematic Review. Eur. Urol. 2015, 67, 627–636. [Google Scholar] [CrossRef]
- Schoots, I.G.; Nieboer, D.; Giganti, F.; Moore, C.M.; Bangma, C.H.; Roobol, M.J. Is Magnetic Resonance Imaging-Targeted Biopsy a Useful Addition to Systematic Confirmatory Biopsy in Men on Active Surveillance for Low-Risk Prostate Cancer? A Systematic Review and Meta-Analysis. BJU Int. 2018, 122, 946–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghavimi, S.; Abdi, H.; Waterhouse, J.; Savdie, R.; Chang, S.; Harris, A.; Machan, L.; Gleave, M.; So, A.I.; Goldenberg, L.; et al. Natural History of Prostatic Lesions on Serial Multi-parametric Magnetic Resonance Imaging. Can. Urol. Assoc. J. 2018, 12, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Non-invasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sushentsev, N.; Rundo, L.; Blyuss, O.; Gnanapragasam, V.J.; Sala, E.; Barrett, T. MRI-Derived Radiomics Model for Baseline Prediction of Prostate Cancer Progression on Active Surveillance. Sci. Rep. 2021, 11, 12917. [Google Scholar] [CrossRef]
- Sushentsev, N.; Rundo, L.; Blyuss, O.; Nazarenko, T.; Suvorov, A.; Gnanapragasam, V.J.; Sala, E.; Barrett, T. Comparative Performance of MRI-Derived PRECISE Scores and Delta-Radiomics Models for the Prediction of Prostate Cancer Progression in Patients on Active Surveillance. Eur. Radiol. 2022, 32, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Fave, X.; Zhang, L.; Yang, J.; Mackin, D.; Balter, P.; Gomez, D.; Followill, D.; Jones, A.K.; Stingo, F.; Liao, Z.; et al. Delta-Radiomics Features for the Prediction of Patient Outcomes in Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, 588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, C.M.; Giganti, F.; Albertsen, P.; Allen, C.; Bangma, C.; Briganti, A.; Carroll, P.; Haider, M.; Kasivisvanathan, V.; Kirkham, A.; et al. Reporting Magnetic Resonance Imaging in Men on Active Surveillance for Prostate Cancer: The PRECISE Recommendations—A Report of a European School of Oncology Task Force. Eur. Urol. 2017, 71, 648–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Algohary, A.; Viswanath, S.; Shiradkar, R.; Ghose, S.; Pahwa, S.; Moses, D.; Jambor, I.; Shnier, R.; Böhm, M.; Haynes, A.-M.; et al. Radiomic Features on MRI Enable Risk Categorization of Prostate Cancer Patients on Active Surveillance: Preliminary Findings. J. Magn. Reson. Imaging 2018, 48, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Sushentsev, N.; Caglic, I.; Rundo, L.; Kozlov, V.; Sala, E.; Gnanapragasam, V.J.; Barrett, T. Serial Changes in Tumour Measurements and Apparent Diffusion Coefficients in Prostate Cancer Patients on Active Surveillance with and without Histopathological Progression. Br. J. Radiol. 2022, 95, 20210842. [Google Scholar] [CrossRef] [PubMed]
- Nayan, M.; Salari, K.; Bozzo, A.; Ganglberger, W.; Lu, G.; Carvalho, F.; Gusev, A.; Schneider, A.; Westover, B.M.; Feldman, A.S. A Machine Learning Approach to Predict Progression on Active Surveillance for Prostate Cancer. Urol. Oncol. 2022, 40, 161.e1–161.e7. [Google Scholar] [CrossRef]
- Radtke, J.P.; Kuru, T.H.; Boxler, S.; Alt, C.D.; Popeneciu, I.V.; Huettenbrink, C.; Klein, T.; Steinemann, S.; Bergstraesser, C.; Roethke, M.; et al. Comparative Analysis of Transperineal Template Saturation Prostate Biopsy versus Magnetic Resonance Imaging Targeted Biopsy with Magnetic Resonance Imaging-Ultrasound Fusion Guidance. J. Urol. 2015, 193, 87–94. [Google Scholar] [CrossRef]
- Filson, C.P.; Natarajan, S.; Margolis, D.J.A.; Huang, J.; Lieu, P.; Dorey, F.J.; Reiter, R.E.; Marks, L.S. Prostate Cancer Detection with Magnetic Resonance-Ultrasound Fusion Biopsy: The Role of Systematic and Targeted Biopsies. Cancer 2016, 122, 884–892. [Google Scholar] [CrossRef]
- Johnson, D.C.; Raman, S.S.; Mirak, S.A.; Kwan, L.; Bajgiran, A.M.; Hsu, W.; Maehara, C.K.; Ahuja, P.; Faiena, I.; Pooli, A.; et al. Detection of Individual Prostate Cancer Foci via Multi-parametric Magnetic Resonance Imaging. Eur. Urol. 2019, 75, 712–720. [Google Scholar] [CrossRef] [Green Version]
- de Rooij, M.; Israël, B.; Bomers, J.G.R.; Schoots, I.G.; Barentsz, J.O. Can Biparametric Prostate Magnetic Resonance Imaging Fulfill Its PROMIS? Eur. Urol. 2020, 78, 512–514. [Google Scholar] [CrossRef]
- Peerlings, J.; Woodruff, H.C.; Winfield, J.M.; Ibrahim, A.; Van Beers, B.E.; Heerschap, A.; Jackson, A.; Wildberger, J.E.; Mottaghy, F.M.; DeSouza, N.M.; et al. Stability of Radiomics Features in Apparent Diffusion Coefficient Maps from a Multi-Centre Test-Retest Trial. Sci. Rep. 2019, 9, 4800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, R.; Holloway, L.C.; Brink, C.; Field, M.; Christiansen, R.L.; Sun, Y.; Barton, M.B.; Liney, G.P. Multicenter Evaluation of MRI-Based Radiomic Features: A Phantom Study. Med. Phys. 2020, 47, 3054–3063. [Google Scholar] [CrossRef] [PubMed]
- Bologna, M.; Corino, V.; Mainardi, L. Technical Note: Virtual Phantom Analyses for Preprocessing Evaluation and Detection of a Robust Feature Set for MRI-Radiomics of the Brain. Med. Phys. 2019, 46, 5116–5123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Park, C.M.; Lee, M.; Park, S.J.; Song, Y.S.; Lee, J.H.; Hwang, E.J.; Goo, J.M. Impact of Reconstruction Algorithms on CT Radiomic Features of Pulmonary Tumors: Analysis of Intra- and Inter-Reader Variability and Inter-Reconstruction Algorithm Variability. PLoS ONE 2016, 11, e0164924. [Google Scholar] [CrossRef] [Green Version]
- Choe, J.; Lee, S.M.; Do, K.-H.; Lee, G.; Lee, J.-G.; Lee, S.M.; Seo, J.B. Deep Learning-Based Image Conversion of CT Reconstruction Kernels Improves Radiomics Reproducibility for Pulmonary Nodules or Masses. Radiology 2019, 292, 365–373. [Google Scholar] [CrossRef]
- Schwier, M.; van Griethuysen, J.; Vangel, M.G.; Pieper, S.; Peled, S.; Tempany, C.; Aerts, H.J.W.; Kikinis, R.; Fennessy, F.M.; Fedorov, A. Repeatability of Multi-parametric Prostate MRI Radiomics Features. Sci. Rep. 2019, 9, 9441. [Google Scholar] [CrossRef] [Green Version]
- Scalco, E.; Belfatto, A.; Mastropietro, A.; Rancati, T.; Avuzzi, B.; Messina, A.; Valdagni, R.; Rizzo, G. T2w-MRI Signal Normalization Affects Radiomics Features Reproducibility. Med. Phys. 2020, 47, 1680–1691. [Google Scholar] [CrossRef]
- Saha, A.; Harowicz, M.R.; Mazurowski, M.A. Breast Cancer MRI Radiomics: An Overview of Algorithmic Features and Impact of Inter-reader Variability in Annotating Tumors. Med. Phys. 2018, 45, 3076–3085. [Google Scholar] [CrossRef]
- Haarburger, C.; Müller-Franzes, G.; Weninger, L.; Kuhl, C.; Truhn, D.; Merhof, D. Radiomics Feature Reproducibility under Inter-Rater Variability in Segmentations of CT Images. Sci. Rep. 2020, 10, 12688. [Google Scholar] [CrossRef]
- Baeßler, B.; Weiss, K.; Pinto Dos Santos, D. Robustness and Reproducibility of Radiomics in Magnetic Resonance Imaging: A Phantom Study. Investig. Radiol. 2019, 54, 221–228. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in Medical Imaging-“how-to” Guide and Critical Reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Galavis, P.E. Reproducibility and Standardization in Radiomics: Are We There Yet? In Proceedings of the XVI Mexican Symposium On Medical Physics, Mexico City, Mexico, 26–30 October 2020; AIP Publishing: Melville, NY, USA, 2021.
- Xu, L.; Zhang, G.; Zhao, L.; Mao, L.; Li, X.; Yan, W.; Xiao, Y.; Lei, J.; Sun, H.; Jin, Z. Radiomics Based on Multi-parametric Magnetic Resonance Imaging to Predict Extraprostatic Extension of Prostate Cancer. Front. Oncol. 2020, 10, 940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-M.-Y.; Han, Y.-Q.; Wei, J.-W.; Qi, Y.-F.; Gu, D.-S.; Lei, J.; Yan, W.-G.; Xiao, Y.; Xue, H.-D.; Feng, F.; et al. Radiomics Based on MRI as a Biomarker to Guide Therapy by Predicting Upgrading of Prostate Cancer from Biopsy to Radical Prostatectomy. J. Magn. Reson. Imaging 2020, 52, 1239–1248. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Li, M.; Gu, Y.; Zhang, Y.; Yang, S.; Wei, C.; Wu, J.; Li, X.; Zhao, W.; Shen, J. Prostate Cancer Differentiation and Aggressiveness: Assessment with a Radiomic-Based Model vs. PI-RADS V2. J. Magn. Reson. Imaging 2019, 49, 875–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fehr, D.; Veeraraghavan, H.; Wibmer, A.; Gondo, T.; Matsumoto, K.; Vargas, H.A.; Sala, E.; Hricak, H.; Deasy, J.O. Automatic Classification of Prostate Cancer Gleason Scores from Multi-parametric Magnetic Resonance Images. Proc. Natl. Acad. Sci. USA 2015, 112, E6265–E6273. [Google Scholar] [CrossRef]
- Khoo, E.L.H.; Schick, K.; Plank, A.W.; Poulsen, M.; Wong, W.W.G.; Middleton, M.; Martin, J.M. Prostate Contouring Variation: Can It Be Fixed? Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 1923–1929. [Google Scholar] [CrossRef]
- Balagurunathan, Y.; Gu, Y.; Wang, H.; Kumar, V.; Grove, O.; Hawkins, S.; Kim, J.; Goldgof, D.B.; Hall, L.O.; Gatenby, R.A.; et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl. Oncol. 2014, 7, 72–87. [Google Scholar] [CrossRef] [Green Version]
- Kalpathy-Cramer, J.; Mamomov, A.; Zhao, B.; Lu, L.; Cherezov, D.; Napel, S.; Echegaray, S.; Rubin, D.; McNitt-Gray, M.; Lo, P.; et al. Radiomics of Lung Nodules: A Multi-Institutional Study of Robustness and Agreement of Quantitative Imaging Features. Tomography 2016, 2, 430–437. [Google Scholar] [CrossRef]
- Haarburger, C.; Schock, J.; Truhn, D.; Weitz, P.; Mueller-Franzes, G.; Weninger, L.; Merhof, D. Radiomic Feature Stability Analysis Based on Probabilistic Segmentations. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020. [Google Scholar]
- Berman, R.M.; Brown, A.M.; Chang, S.D.; Sankineni, S.; Kadakia, M.; Wood, B.J.; Pinto, P.A.; Choyke, P.L.; Turkbey, B. DCE MRI of prostate cancer. Abdom. Radiol. 2016, 41, 844–853. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [Green Version]
- Fedorov, A.; Vangel, M.G.; Tempany, C.M.; Fennessy, F.M. Multi-parametric Magnetic Resonance Imaging of the Prostate: Repeatability of Volume and Apparent Diffusion Coefficient Quantification. Investig. Radiol. 2017, 52, 538–546. [Google Scholar] [CrossRef] [Green Version]
- Fedorov, A.; Schwier, M.; Clunie, D.; Herz, C.; Pieper, S.; Kikinis, R.; Tempany, C.; Fennessy, F. An Annotated Test-Retest Collection of Prostate Multi-parametric MRI. Sci. Data 2018, 5, 180281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, L.; Wang, J. The Effectiveness of Data Augmentation in Image Classification Using Deep Learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef] [Green Version]
- Pérez-García, F.; Sparks, R.; Ourselin, S. TorchIO: A Python Library for Efficient Loading, Preprocessing, Augmentation and Patch-Based Sampling of Medical Images in Deep Learning. Comput. Methods Programs Biomed. 2021, 208, 106236. [Google Scholar] [CrossRef]
- Rasch, C.; Barillot, I.; Remeijer, P.; Touw, A.; van Herk, M.; Lebesque, J.V. Definition of the Prostate in CT and MRI: A Multi-Observer Study. Int. J. Radiat. Oncol. Biol. Phys. 1999, 43, 57–66. [Google Scholar] [CrossRef]
- Smith, W.L.; Lewis, C.; Bauman, G.; Rodrigues, G.; D’Souza, D.; Ash, R.; Ho, D.; Venkatesan, V.; Downey, D.; Fenster, A. Prostate Volume Contouring: A 3D Analysis of Segmentation Using 3DTRUS, CT, and MR. Int. J. Radiat. Oncol.*Biol.*Phys. 2007, 67, 1238–1247. [Google Scholar] [CrossRef] [PubMed]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Chen, Y.; Fedorov, A.; Li, X.; Jajamovich, G.H.; Malyarenko, D.I.; Aryal, M.P.; LaViolette, P.S.; Oborski, M.J.; O’Sullivan, F.; et al. The Impact of Arterial Input Function Determination Variations on Prostate Dynamic Contrast-Enhanced Magnetic Resonance Imaging Pharmacokinetic Modeling: A Multicenter Data Analysis Challenge. Tomography 2016, 2, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, L.; Botta, F.; Origgi, D.; Rizzo, S.; Mariani, M.; Summers, P.; García-Polo, P.; Cremonesi, M.; Lascialfari, A. PETER PHAN: An MRI Phantom for the Optimisation of Radiomic Studies of the Female Pelvis. Phys. Med. 2020, 71, 71–81. [Google Scholar] [CrossRef]
- Tixier, F.; Rest, L.; Hatt, C.C.; Albarghach, M. Intratumor Heterogeneity Characterized by Textural Features on Baseline 18F-FDG PET Images Predicts Response to Concomitant Radiochemotherapy in Esophageal. J. Nucl. 2011, 52, 369–378. [Google Scholar] [CrossRef] [Green Version]
- Vallat, R. Pingouin: Statistics in Python. J. Open Source Softw. 2018, 3, 1026. [Google Scholar] [CrossRef]
- Velazquez, R. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017, 77, 3922–3930. [Google Scholar] [CrossRef] [Green Version]
- Eklund, M.; Jäderling, F.; Discacciati, A.; Bergman, M.; Annerstedt, M.; Aly, M.; Glaessgen, A.; Carlsson, S.; Grönberg, H.; Nordström, T.; et al. MRI-Targeted or Standard Biopsy in Prostate Cancer Screening. N. Engl. J. Med. 2021, 385, 908–920. [Google Scholar] [CrossRef]
- Zhou, X.; Gao, F.; Duan, S.; Zhang, L.; Liu, Y.; Zhou, J.; Bai, G.; Tao, W. Radiomic Features of Pk-DCE MRI Parameters Based on the Extensive Tofts Model in Application of Breast Cancer. Phys. Eng. Sci. Med. 2020, 43, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Mithun, S.; Jaiswar, V.; Sherkhane, U.B.; Purandare, N.C.; Prabhash, K.; Rangarajan, V.; Dekker, A.; Wee, L.; Traverso, A. Repeatability and Reproducibility Study of Radiomic Features on a Phantom and Human Cohort. Sci. Rep. 2021, 11, 2055. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Refaee, T.; Leijenaar, R.T.H.; Primakov, S.; Hustinx, R.; Mottaghy, F.M.; Woodruff, H.C.; Maidment, A.D.A.; Lambin, P. The Application of a Workflow Integrating the Variable Reproducibility and Harmonizability of Radiomic Features on a Phantom Dataset. PLoS ONE 2021, 16, e0251147. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Leger, S.; Agolli, L.; Pilz, K.; Troost, E.G.C.; Richter, C.; Löck, S. Assessing Robustness of Radiomic Features by Image Perturbation. Sci. Rep. 2019, 9, 614. [Google Scholar] [CrossRef] [Green Version]



| Specifications | (a) Internal Dataset | (b) External Dataset |
|---|---|---|
| No. of Patients | 100 | 15 |
| Manufacturer | Ingenia (Philips Medical System, Best, The Netherlands) | GE Signa HDxt platform and GE Discovery MR750w (General Electric Healthcare, Milwaukee, WI) machines. |
| Magnetic Field Strength | 1.5 T | 3.0 T |
| Endorectal Coil | Yes | Yes |
| PIRADSv2 Compliant | Yes | Yes |
| Acquisition Protocol | T2w (TR/TE = 4910/110 ms, slice thickness = 3 mm, pixel spacing = 0.297 mm); DWI (b-values = 0, 1500 and 2000 s/mm2, TR/TE = 3320/106 ms, slice thickness = 3 mm, pixel spacing = 1.250 mm); DCE (TR/TE = 4.03/1.88 ms, slice thickness = 3 mm, pixel spacing = 1.136 mm, acquired with high temporal resolution < 10 s). | T2w (TR/TE = 3350–5109/84–107 ms, slice thickness = 3 mm, pixel spacing = 0.273–0.312 mm); DWI (b-values of 0 and 1400 s/mm2, TR/TE = 2500–8150/76.7–80.6 ms, slice thickness = 3–4 mm, pixel spacing = 0.625–0.703 mm); DCE (TR/TE = 3.68–4.1/1.3–1.42 ms, slice thickness = 5–6 mm, pixel spacing = 0.547–1.015 mm). |
| GT Segmentation | Whole prostate gland segmentation on T2w | Whole prostate gland segmentation on T2w, ADC, and SUB |
| (a) Internal | ||||||||
| aug config | T2w | ADC | SUBwin | SUBwout | ||||
| mean | std | mean | std | mean | std | mean | std | |
| InP-R | 0.95 | 0.01 | 0.95 | 0.01 | 0.95 | 0.01 | 0.95 | 0.01 |
| InP-S | 0.95 | 0.02 | 0.95 | 0.02 | 0.95 | 0.02 | 0.95 | 0.02 |
| OutP | 0.99 | 0.01 | 0.99 | 0.01 | 0.99 | 0.01 | 0.99 | 0.01 |
| In&OutP-R | 0.95 | 0.01 | 0.95 | 0.01 | 0.95 | 0.02 | 0.95 | 0.01 |
| In&OutP-S | 0.94 | 0.03 | 0.95 | 0.03 | 0.94 | 0.02 | 0.94 | 0.03 |
| (b) External | ||||||||
| aug config | T2w | ADC | SUB | |||||
| mean | std | mean | std | mean | std | |||
| InP-R | 0.95 | 0.01 | 0.96 | 0.01 | 0.95 | 0.02 | ||
| InP-S | 0.95 | 0.03 | 0.95 | 0.02 | 0.95 | 0.03 | ||
| OutP | 0.99 | 0.01 | 0.99 | 0.01 | 0.99 | 0.01 | ||
| In&OutP-R | 0.94 | 0.02 | 0.95 | 0.01 | 0.94 | 0.02 | ||
| In&OutP-S | 0.94 | 0.03 | 0.95 | 0.03 | 0.94 | 0.03 | ||
| (a) T2w | ||||||||||||||||||||
| aug config | firstorder | glcm | glrlm | glszm | Overall | |||||||||||||||
| O | BF | O | BF | O | BF | O | BF | O | BF | |||||||||||
| S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | |
| InP-R | 0.72 | 0.72 | 1 | 1 | 0.95 | 0.86 | 1 | 1 | 0.94 | 0.75 | 1 | 1 | 0.81 | 0.62 | 1 | 0.88 | 0.86 | 0.75 | 1 | 0.97 |
| InP-S | 0.44 | 0.22 | 1 | 0.94 | 0.73 | 0.41 | 1 | 0.86 | 0.94 | 0.44 | 1 | 0.62 | 0.75 | 0.44 | 1 | 0.81 | 0.71 | 0.38 | 1 | 0.82 |
| OutP | 0.83 | 0.83 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.94 | 0.81 | 1 | 1 | 0.94 | 0.92 | 1 | 1 |
| In&OutP-R | 0.72 | 0.61 | 1 | 1 | 0.95 | 0.82 | 1 | 1 | 0.94 | 0.56 | 1 | 1 | 0.81 | 0.56 | 1 | 0.88 | 0.86 | 0.65 | 1 | 0.97 |
| In&OutP-S | 0.44 | 0.22 | 1 | 0.94 | 0.73 | 0.41 | 1 | 0.86 | 0.94 | 0.44 | 1 | 0.62 | 0.69 | 0.38 | 0.88 | 0.75 | 0.69 | 0.36 | 0.97 | 0.81 |
| (b) ADC | ||||||||||||||||||||
| aug config | firstorder | glcm | glrlm | glszm | Overall | |||||||||||||||
| O | BF | O | BF | O | BF | O | BF | O | BF | |||||||||||
| S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | |
| InP-R | 0.94 | 0.89 | 1 | 1 | 1 | 0.91 | 1 | 1 | 1 | 0.94 | 1 | 1 | 0.81 | 0.69 | 0.94 | 0.94 | 0.94 | 0.86 | 0.99 | 0.99 |
| InP-S | 0.5 | 0.5 | 1 | 1 | 0.55 | 0.41 | 1 | 1 | 0.69 | 0.5 | 1 | 1 | 0.44 | 0.31 | 0.88 | 0.88 | 0.54 | 0.43 | 0.97 | 0.97 |
| OutP | 1 | 0.89 | 1 | 1 | 1 | 0.95 | 1 | 1 | 0.94 | 0.94 | 1 | 1 | 0.81 | 0.81 | 1 | 1 | 0.94 | 0.9 | 1 | 1 |
| In&OutP-R | 0.94 | 0.89 | 1 | 0.94 | 1 | 0.91 | 1 | 1 | 0.94 | 0.88 | 1 | 1 | 0.75 | 0.56 | 0.94 | 0.94 | 0.92 | 0.82 | 0.99 | 0.97 |
| In&OutP-S | 0.56 | 0.56 | 1 | 0.94 | 0.5 | 0.36 | 1 | 1 | 0.5 | 0.31 | 1 | 1 | 0.31 | 0.19 | 0.88 | 0.81 | 0.47 | 0.36 | 0.97 | 0.94 |
| (c) SUBwin | ||||||||||||||||||||
| aug config | firstorder | glcm | glrlm | glszm | Overall | |||||||||||||||
| O | BF | O | BF | O | BF | O | BF | O | BF | |||||||||||
| S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | |
| InP-R | 0.72 | 0.67 | 1 | 1 | 0.91 | 0.91 | 1 | 1 | 0.81 | 0.62 | 1 | 1 | 0.81 | 0.56 | 1 | 0.88 | 0.82 | 0.71 | 1 | 0.97 |
| InP-S | 0.5 | 0.33 | 1 | 1 | 0.82 | 0.5 | 1 | 1 | 0.56 | 0.56 | 0.94 | 0.81 | 0.75 | 0.56 | 0.88 | 0.88 | 0.67 | 0.49 | 0.96 | 0.93 |
| OutP | 0.83 | 0.78 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.88 | 1 | 1 | 0.96 | 0.92 | 1 | 1 |
| In&OutP-R | 0.72 | 0.61 | 1 | 1 | 0.91 | 0.82 | 1 | 1 | 0.81 | 0.62 | 1 | 0.88 | 0.81 | 0.5 | 1 | 0.88 | 0.82 | 0.65 | 1 | 0.94 |
| In&OutP-S | 0.5 | 0.33 | 1 | 1 | 0.82 | 0.5 | 1 | 1 | 0.56 | 0.56 | 0.94 | 0.81 | 0.69 | 0.44 | 0.88 | 0.88 | 0.65 | 0.46 | 0.96 | 0.93 |
| (d) SUBwout | ||||||||||||||||||||
| aug config | firstorder | glcm | glrlm | glszm | Overall | |||||||||||||||
| O | BF | O | BF | O | BF | O | BF | O | BF | |||||||||||
| S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | |
| InP-R | 0.78 | 0.67 | 1 | 1 | 0.95 | 0.91 | 1 | 1 | 0.94 | 0.62 | 1 | 1 | 0.88 | 0.62 | 0.94 | 0.88 | 0.89 | 0.72 | 0.99 | 0.97 |
| InP-S | 0.33 | 0.33 | 1 | 1 | 0.68 | 0.41 | 1 | 1 | 0.56 | 0.56 | 1 | 0.81 | 0.56 | 0.5 | 0.94 | 0.88 | 0.54 | 0.44 | 0.99 | 0.93 |
| OutP | 0.89 | 0.78 | 1 | 1 | 1 | 1 | 1 | 1 | 0.94 | 0.94 | 1 | 1 | 1 | 0.88 | 1 | 1 | 0.96 | 0.9 | 1 | 1 |
| In&OutP-R | 0.72 | 0.61 | 1 | 1 | 0.91 | 0.82 | 1 | 1 | 0.81 | 0.62 | 1 | 0.88 | 0.81 | 0.56 | 0.88 | 0.88 | 0.82 | 0.67 | 0.97 | 0.94 |
| In&OutP-S | 0.33 | 0.33 | 1 | 1 | 0.64 | 0.41 | 1 | 1 | 0.56 | 0.56 | 0.88 | 0.81 | 0.56 | 0.44 | 0.88 | 0.88 | 0.53 | 0.43 | 0.94 | 0.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thulasi Seetha, S.; Garanzini, E.; Tenconi, C.; Marenghi, C.; Avuzzi, B.; Catanzaro, M.; Stagni, S.; Villa, S.; Chiorda, B.N.; Badenchini, F.; et al. Stability of Multi-Parametric Prostate MRI Radiomic Features to Variations in Segmentation. J. Pers. Med. 2023, 13, 1172. https://doi.org/10.3390/jpm13071172
Thulasi Seetha S, Garanzini E, Tenconi C, Marenghi C, Avuzzi B, Catanzaro M, Stagni S, Villa S, Chiorda BN, Badenchini F, et al. Stability of Multi-Parametric Prostate MRI Radiomic Features to Variations in Segmentation. Journal of Personalized Medicine. 2023; 13(7):1172. https://doi.org/10.3390/jpm13071172
Chicago/Turabian StyleThulasi Seetha, Sithin, Enrico Garanzini, Chiara Tenconi, Cristina Marenghi, Barbara Avuzzi, Mario Catanzaro, Silvia Stagni, Sergio Villa, Barbara Noris Chiorda, Fabio Badenchini, and et al. 2023. "Stability of Multi-Parametric Prostate MRI Radiomic Features to Variations in Segmentation" Journal of Personalized Medicine 13, no. 7: 1172. https://doi.org/10.3390/jpm13071172





