Ecology of Pollen Storage in Honey Bees: Sugar Tolerant Yeast and the Aerobic Social Microbiota
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Monitoring Pollen Deposition and Age
2.2. Abiotic Factors Affecting Pollen Preservation
2.3. Culturing Beebread in Spring and Summer Dearth
2.4. Microscopic Identification
2.5. Sampling Long-Term Pollen Storage
2.5.1. Isolating Microbial DNA
2.5.2. Estimating Microbial Load of Long-Term Pollen Storage
3. Results
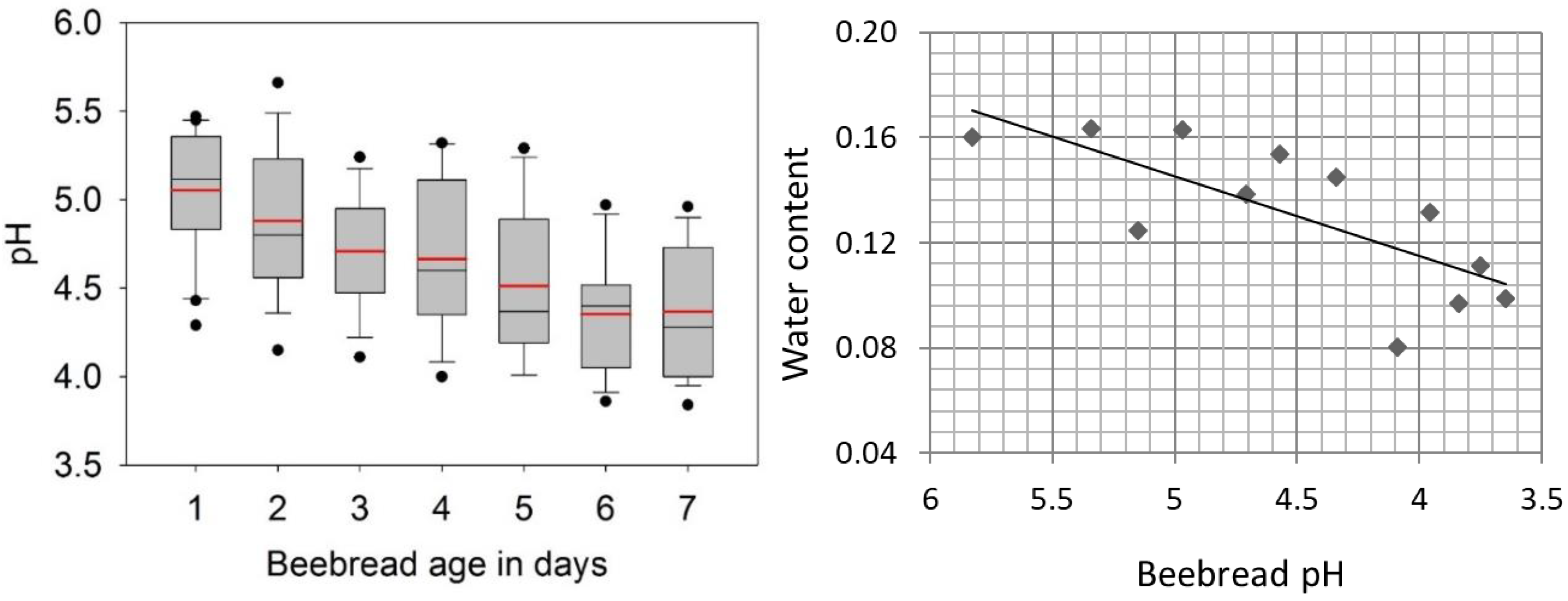
3.1. Abiotic Factors of Early Beebread Storage
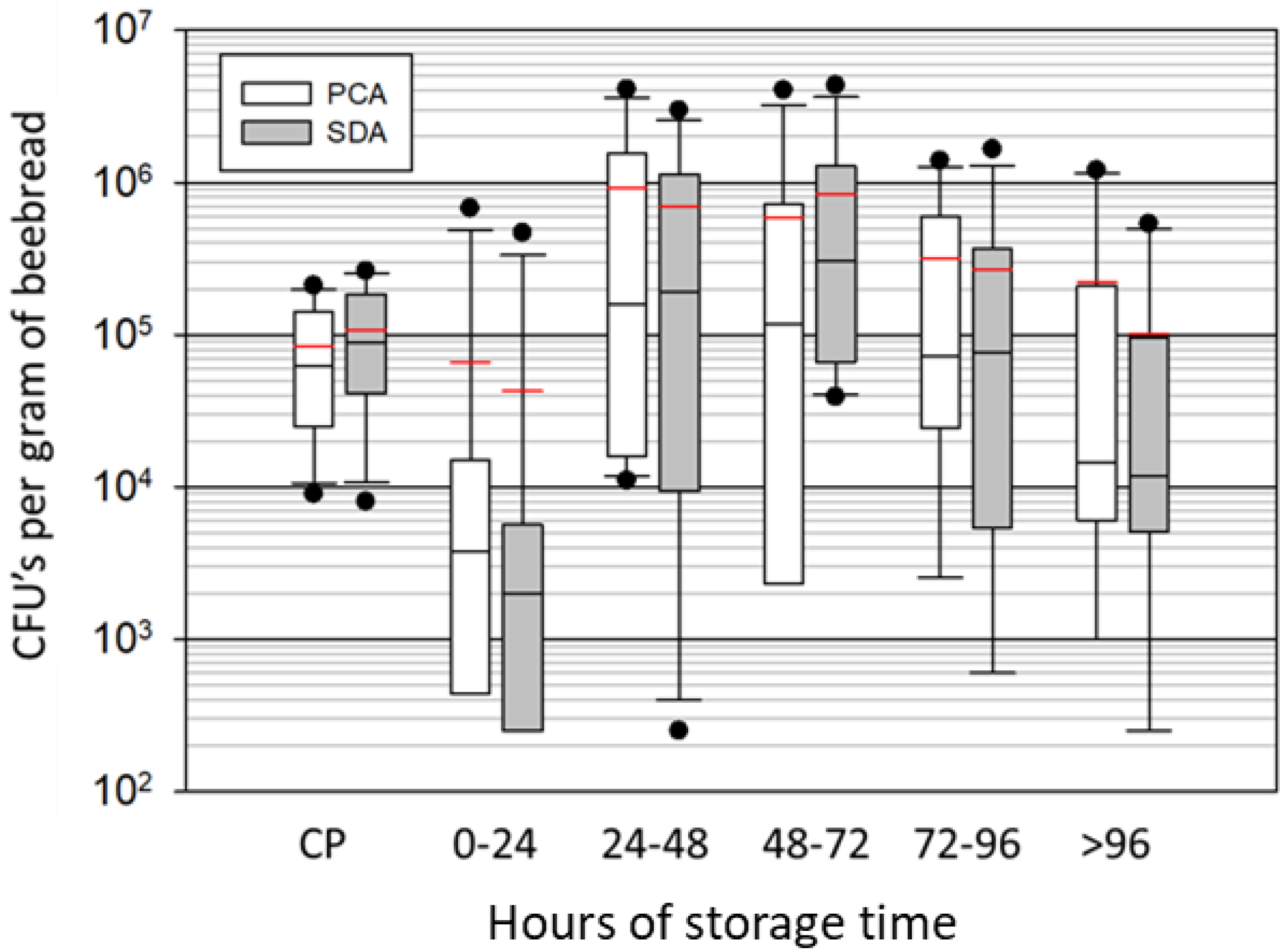
3.2. Microbial Growth in Beebread
3.3. Distinguishing Microbial Type
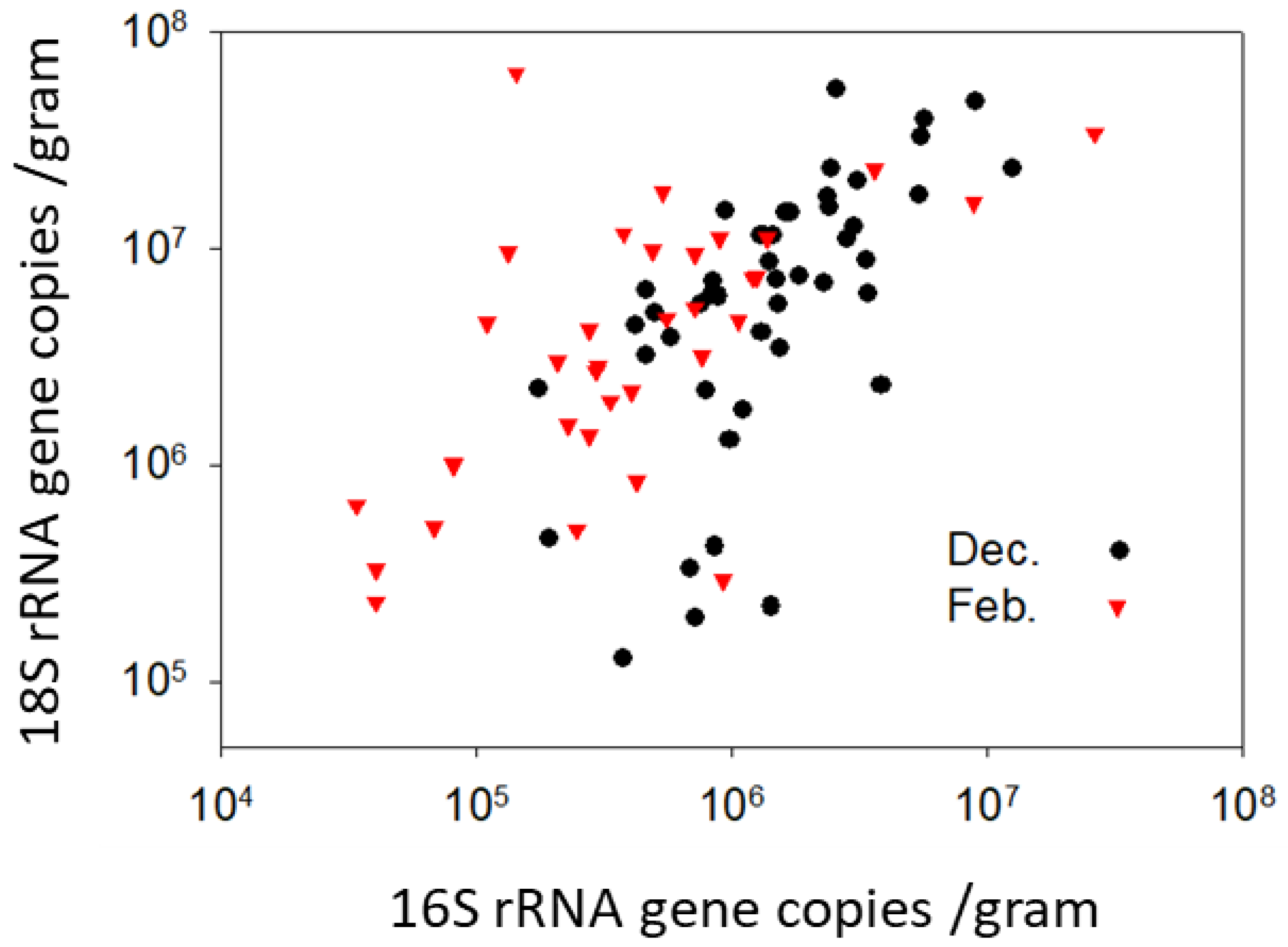
3.4. Long-Term Pollen Storage: FungiQuant and BactQuant
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwarz, R.S.; Huang, Q.; Evans, J.D. Hologenome theory and the honey bee pathosphere. Curr. Opin. Insect Sci. 2015, 10, 1–7. [Google Scholar] [CrossRef]
- Evans, J.D.; Spivak, M. Socialized medicine: Individual and communal disease barriers in honey bees. J. Invertebr. Pathol. 2010, 103, S62–S72. [Google Scholar] [CrossRef] [PubMed]
- Dalenberg, H.; Maes, P.; Mott, B.; Anderson, K.E.; Spivak, M. Propolis envelope promotes beneficial bacteria in the honey bee (Apis mellifera) mouthpart microbiome. Insects 2020, 11, 453. [Google Scholar] [CrossRef] [PubMed]
- Vojvodic, S.; Johnson, B.R.; Harpur, B.A.; Kent, C.F.; Zayed, A.; Anderson, K.E.; Linksvayer, T.A. The transcriptomic and evolutionary signature of social interactions regulating honey bee caste development. Ecol. Evol. 2015, 5, 4795–4807. [Google Scholar] [CrossRef] [Green Version]
- Grozinger, C.M.; Zayed, A. Improving bee health through genomics. Nat. Rev. Genet. 2020, 21, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Voulgari-Kokota, A.; McFrederick, Q.S.; Steffan-Dewenter, I.; Keller, A. Drivers, Diversity, and Functions of the Solitary-Bee Microbiota. Trends Microbiol. 2019, 27, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- McFrederick, Q.S.; Thomas, J.M.; Neff, J.L.; Vuong, H.Q.; Russell, K.A.; Hale, A.R.; Mueller, U.G. Flowers and Wild Megachilid Bees Share Microbes. Microb. Ecol. 2017, 73, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Carroll, M.J.; Sheehan, T.I.M.; Mott, B.M. Hive-stored pollen of honey bees: Many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014, 23, 5904–5917. [Google Scholar] [CrossRef]
- Corby-Harris, V.; Snyder, L.A.; Schwan, M.R.; Maes, P.; McFrederick, Q.S.; Anderson, K.E. Origin and Effect of Alpha 2.2 Acetobacteraceae in Honey Bee Larvae and Description of Parasaccharibacter apium gen. nov., sp. nov. Appl. Environ. Microbiol. 2014, 80, 7460–7472. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.L.; Smith, E.A.; Newton, I.L.G. A bacterial symbiont protects honey bees from fungal disease. MBio 2021, 12, e0050321. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batra, S.W.T.; Bohart, G.E. Alkali bees: Response of adults to pathogenic fungi in brood cells. Science 1969, 165, 607–608. [Google Scholar] [CrossRef] [PubMed]
- Vannette, R.L.; Mohamed, A.; Johnson, B.R. Forager bees (Apis mellifera) highly express immune and detoxification genes in tissues associated with nectar processing. Sci. Rep. 2015, 5, 16224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannette, R.L.; Gauthier, M.-P.L.; Fukami, T. Nectar bacteria, but not yeast, weaken a plant-pollinator mutualism. Proc. Biol. Sci. 2013, 280, 20122601. [Google Scholar] [CrossRef] [Green Version]
- Nicolson, S.W. Bee food: The chemistry and nutritional value of nectar, pollen and mixtures of the two. Afr. Zool. 2011, 46, 197–204. [Google Scholar] [CrossRef]
- Graystock, P.; Rehan, S.M.; McFrederick, Q.S. Hunting for healthy microbiomes: Determining the core microbiomes of Ceratina, Megalopta, and Apis bees and how they associate with microbes in bee collected pollen. Conserv. Genet. 2017, 18, 701–711. [Google Scholar] [CrossRef]
- Mcfrederick, Q.S.; Mueller, U.G.; James, R.R. Interactions between fungi and bacteria influence microbial community structure in the Megachile rotundata larval gut. Proc. Biol. Sci. 2014, 281, 20132653. [Google Scholar] [CrossRef] [Green Version]
- Carroll, M.J.; Brown, N.; Goodall, C.; Downs, A.M.; Sheenan, T.H.; Anderson, K.E. Honey bees preferentially consume freshly stored pollen. PLoS ONE 2017, 12, e0175933. [Google Scholar] [CrossRef] [Green Version]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef]
- Floyd, A.S.; Mott, B.M.; Maes, P.; Copeland, D.C.; McFrederick, Q.S.; Anderson, K.E. Microbial ecology of European foulbrood disease in the honey bee (Apis mellifera): Towards a microbiome understanding of disease susceptibility. Insects 2020, 11, 555. [Google Scholar] [CrossRef]
- Kwakman, P.H.S.; te Velde, A.A.; de Boer, L.; Speijer, D.; Vandenbroucke-Grauls, M.J.C.; Zaat, S.A.J. How honey kills bacteria. FASEB J. 2010, 24, 2576–2582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, R.; Mendes, M.A.; De Souza, B.M.; Konno, K.; César, L.M.M.; Malaspina, O.; Palma, M.S. Jelleines: A family of antimicrobial peptides from the royal jelly of honeybees (Apis mellifera). Peptides 2004, 25, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Ricigliano, V.A. Honey bee gut dysbiosis: A novel context of disease ecology. Curr. Opin. Insect Sci. 2017, 22, 125–132. [Google Scholar] [CrossRef]
- Gilliam, M. Identification and roles of non-pathogenic microflora associated with honey bees. FEMS Microbiol. Lett. 1997, 155, 1–10. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Eckholm, B.J.; Mott, B.M.; DeGrandi-Hoffman, G. An emerging paradigm of colony health: Microbial balance of the honey bee and hive (Apis mellifera). Insectes Soc. 2011, 58, 431–444. [Google Scholar] [CrossRef]
- Roessink, I.; van der Steen, J.J.M. Beebread consumption by honey bees is fast: Results of a six-week field study. J. Apic. Res. 2021, 60, 659–664. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Jia, E.; Soh, Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callegari, M.; Crotti, E.; Fusi, M.; Marasco, R.; Gonella, E.; De Noni, I.; Romano, D.; Borin, S.; Tsiamis, G.; Cherif, A.; et al. Compartmentalization of bacterial and fungal microbiomes in the gut of adult honeybees. NPJ Biofilms Microbiomes 2021, 7, 42. [Google Scholar] [CrossRef]
- Endo, A.; Irisawa, T.; Futagawa-Endo, Y.; Sonomoto, K.; Itoh, K.; Takano, K.; Okada, S.; Dicks, L.M.T. Fructobacillus tropaeoli sp. nov., a fructophilic lactic acid bacterium isolated from a flower. Int. J. Syst. Evol. Microbiol. 2011, 61, 898–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neveling, D.P.; Endo, A.; Dicks, L.M.T. Fructophilic Lactobacillus kunkeei and Lactobacillus brevis Isolated from Fresh Flowers, Bees and Bee-hives. Curr. Microbiol. 2012, 65, 507–515. [Google Scholar] [CrossRef]
- Jones, B.; Shipley, E.; Arnold, K.E. Social immunity in honeybees—Density dependence, diet, and body mass trade-offs. Ecol. Evol. 2018, 8, 4852–4859. [Google Scholar] [CrossRef] [PubMed]
- López-Uribe, M.M.; Fitzgerald, A.; Simone-Finstrom, M. Inducible versus constitutive social immunity: Examining effects of colony infection on glucose oxidase and defensin-1 production in honeybees. R. Soc. Open Sci. 2017, 4, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolson, S.W. Water homeostasis in bees, with the emphasis on sociality. J. Exp. Biol. 2009, 212, 429–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pain, J.; Maugenet, J. Recherches biochimiques et physiologiques sur le pollen emmagsiné par les abeilles. Ann. Abeille 1966, 9, 209–236. [Google Scholar] [CrossRef] [Green Version]
- Oliver, R. Reevaluating Beebread: Part 3- For Preservation or Digestion? Am. Bee J. 2015, 155, 1311–1316. [Google Scholar]
- SAS Institute Inc. SAS/ACCESS® 9.4 Interface to ADABAS: Reference; SAS Institute Inc.: Cary, NC, USA, 2013. [Google Scholar]
- Liu, C.M.; Aziz, M.; Kachur, S.; Hsueh, P.-R.; Huang, Y.-T.; Keim, P.; Price, L.B. BactQuant: An enhanced broad-coverage bacterial quantitative real-time PCR assay. BMC Microbiol. 2012, 12, 56. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.M.; Kachur, S.; Dwan, M.G.; Abraham, A.G.; Aziz, M.; Hsueh, P.R.; Huang, Y.T.; Busch, J.D.; Lamit, L.J.; Gehring, C.A.; et al. FungiQuant: A broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 2012, 12, 1. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.P.; Pierce, N.E.; Boomsma, J.J. Social insect symbionts: Evolution in homeostatic fortresses. Trends Ecol. Evol. 2008, 23, 672–677. [Google Scholar] [CrossRef]
- Ueno, T.; Nakaoka, T.; Takeuchi, H.; Kubo, T. Differential Gene Expression in the Hypopharyngeal Glands of Worker Honeybees (Apis mellifera L.) Associated with an Age-Dependent Role Change. Zoolog. Sci. 2009, 26, 557–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leschine, S.B. Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 1995, 49, 399–426. [Google Scholar] [CrossRef]
- Anderson, K.E.; Ricigliano, V.A.; Copeland, D.C.; Mott, B.M.; Maes, P. Social Interaction is Unnecessary for Hindgut Microbiome Transmission in Honey Bees: The Effect of Diet and Social Exposure on Tissue-Specific Microbiome Assembly. Microb. Ecol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Vojvodic, S.; Rehan, S.M.; Anderson, K.E. Microbial Gut Diversity of Africanized and European Honey Bee Larval Instars. PLoS ONE 2013, 8, 0072106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Detry, R.; Simon-Delso, N.; Bruneau, E.; Daniel, H.M. Specialisation of yeast genera in different phases of bee bread maturation. Microorganisms 2020, 8, 1789. [Google Scholar] [CrossRef]
- González, G.; Hinojo, M.J.; Mateo, R.; Medina, A.; Jiménez, M. Occurrence of mycotoxin producing fungi in bee pollen. Int. J. Food Microbiol. 2005, 105, 1–9. [Google Scholar] [CrossRef]
- Tauber, J.P.; McMahon, D.; Ryabov, E.V.; Kunat, M.; Ptaszynska, A.; Evans, J.D. Honeybee intestines retain low yeast titers, but no bacterial mutualists, at emergence. Yeast 2021, 39, 95–107. [Google Scholar] [CrossRef]
- Kacániová, M.; Chlebo, R.; Kopernický, M.; Trakovická, A. Microflora of the honeybee gastrointestinal tract. Folia Microbiol. 2004, 49, 169–171. [Google Scholar] [CrossRef]
- Brown, A.D.; Simpson, J.R. Water relations of sugar-tolerant yeasts: The role of intracellular polyols. J. Gen. Microbiol. 1972, 72, 589–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrålstad, T. ITS, OTUs and beyond—Fungal hyperdiversity calls for supplementary solutions. Mol. Ecol. 2011, 20, 2873–2875. [Google Scholar] [CrossRef]
- Vásquez, A.; Olofsson, T.C. The lactic acid bacteria involved in the production of bee pollen and bee bread. J. Apic. Res. 2009, 48, 189–195. [Google Scholar] [CrossRef]
- Meyer, T.; Rosa, C.A.; Lachance, M. The yeast genus Starmerella gen. nov. and Starmerella bombicola sp. nov., the teleomorph of Candida bombicola. Int. J. Syst. Bacteriol. 1998, 48, 1413–1417. [Google Scholar]
- Anderson, K.E.; Maes, P. Social microbiota and social gland gene expression of worker honey bees by age and climate. Sci. Rep. 2022, 12, 10690. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Ricigliano, V.A.; Mott, B.M.; Copeland, D.C.; Floyd, A.S.; Maes, P. The Queen Gut Refines with Age: Longevity Phenotypes in a Social Insect Model. Microbiome 2018, 6, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, P.W.; Floyd, A.S.; Mott, B.M.; Anderson, K.E. Overwintering honey bee colonies: Effect of worker age and climate on the hindgut microbiota. Insects 2021, 12, 224. [Google Scholar] [CrossRef] [PubMed]
- Tauber, J.P.; Nguyen, V.; Lopez, D.; Evans, J.D. Effects of a Resident Yeast from the Honeybee Gut on Immunity, Microbiota, and Nosema Disease. Insects 2019, 10, 296. [Google Scholar] [CrossRef] [Green Version]
- Ellegaard, K.M.; Engel, P. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 2019, 10, 446. [Google Scholar] [CrossRef] [Green Version]
- Bae, Y.S.; Choi, M.K.; Lee, W.J. Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 2010, 31, 278–287. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anderson, K.E.; Mott, B.M. Ecology of Pollen Storage in Honey Bees: Sugar Tolerant Yeast and the Aerobic Social Microbiota. Insects 2023, 14, 265. https://doi.org/10.3390/insects14030265
Anderson KE, Mott BM. Ecology of Pollen Storage in Honey Bees: Sugar Tolerant Yeast and the Aerobic Social Microbiota. Insects. 2023; 14(3):265. https://doi.org/10.3390/insects14030265
Chicago/Turabian StyleAnderson, Kirk E., and Brendon M. Mott. 2023. "Ecology of Pollen Storage in Honey Bees: Sugar Tolerant Yeast and the Aerobic Social Microbiota" Insects 14, no. 3: 265. https://doi.org/10.3390/insects14030265




