Application of Machine Learning Techniques to Discern Optimal Rearing Conditions for Improved Black Soldier Fly Farming
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection Procedure
2.2. Data Processing Techniques
2.2.1. Handling Missing Values
- (a)
- Missing completely at random (MCAR): MCAR happens when missing entries are randomly distributed across variables and are unrelated to other features. Kenyhercz and Passalacqua [17] came across the same challenge in their dataset. To solve this problem, they compared 4 different imputation techniques that included hot deck, iterative robust model, k-nearest neighbor (KNN), and variable means. Their results showed that KNN was the most accurate approach, as it had the lowest error between the imputed and actual values. Actual values refer to complete data entries collected during the data collection process, while imputed values refer to entries calculated to fill up missing values in the collected dataset. Technically, the cluster-based approach, using an appropriate algorithm such as KNN, entails identifying the nearest neighbors of the record with a missing entry, imputing the value of the nearest neighbors, and using the result to fill up the null record. It is for this reason that this research adopted this approach in imputing missing values. The actual variables evaluated using this approach were Phase 1 feed depth and total young larvae (YL) mass loaded (kg). This was implemented using sklearn’s KNNImputer [18].
- (b)
- Missing at random (MAR): This study also identified missing at random (MAR) data entries. In MAR, the missing data in one variable can be explained by another variable in the same dataset. For this study, the variables imputed using this approach include the feeding rate and the number of young larvae loaded at the first phase. To solve this, this research integrated the weight class adjustment approach. It was implemented using the group-by methodology [19] based on related records from other variables in the dataset to fill up the missing entries. This approach was also used by other researchers, such as [19], to solve similar problems.
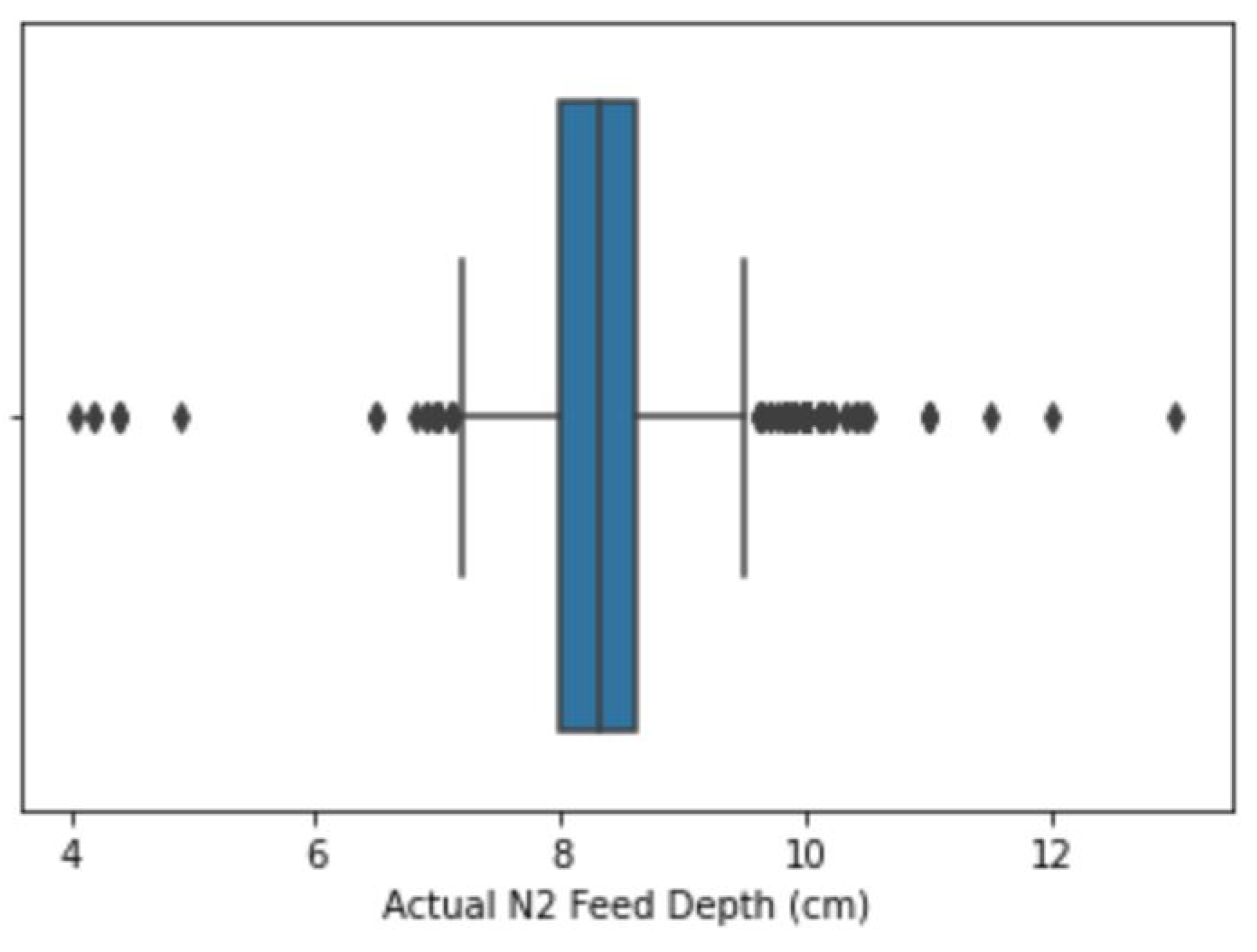
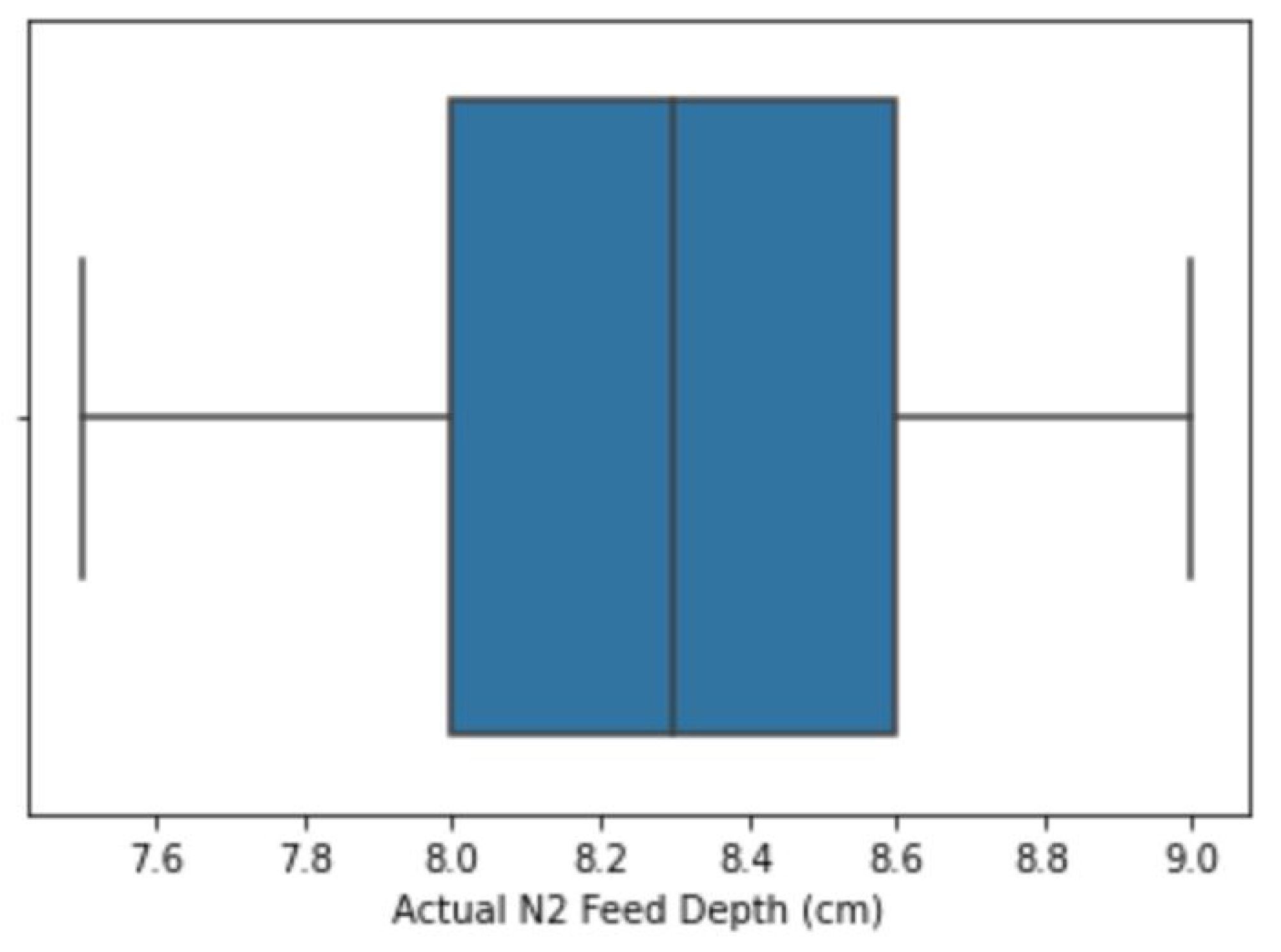
2.2.2. Outlier Treatment
2.3. Machine Learning Modeling
2.3.1. Feature Transformation
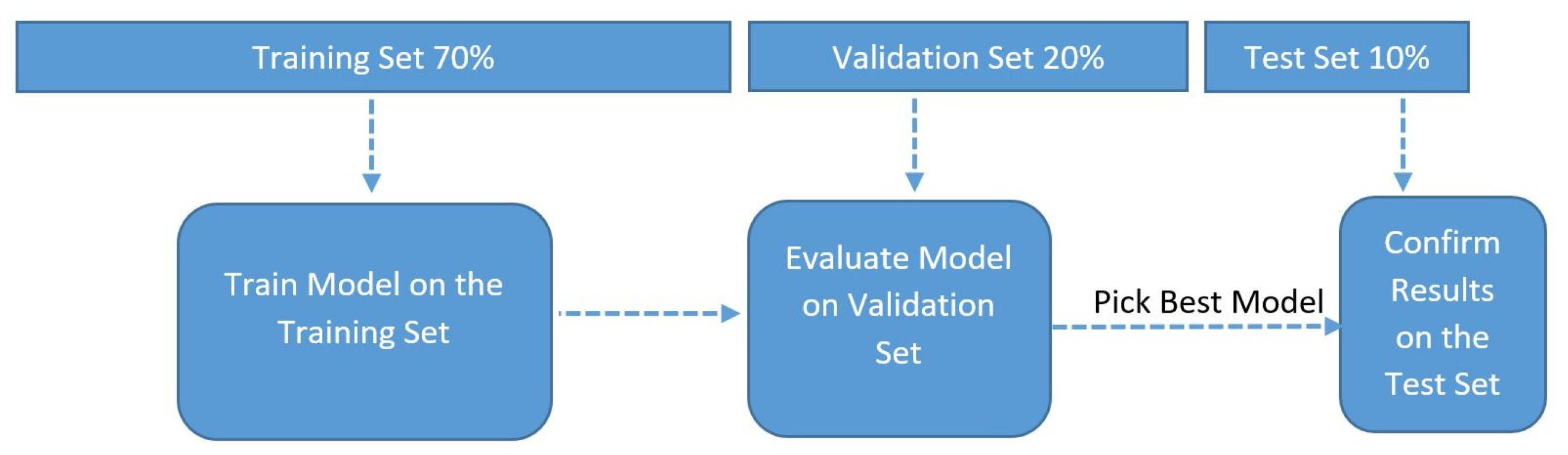
2.3.2. Data Splitting
2.3.3. Feature Scaling
2.3.4. Trained Machine Learning Models
2.3.5. Performance Evaluation Criteria
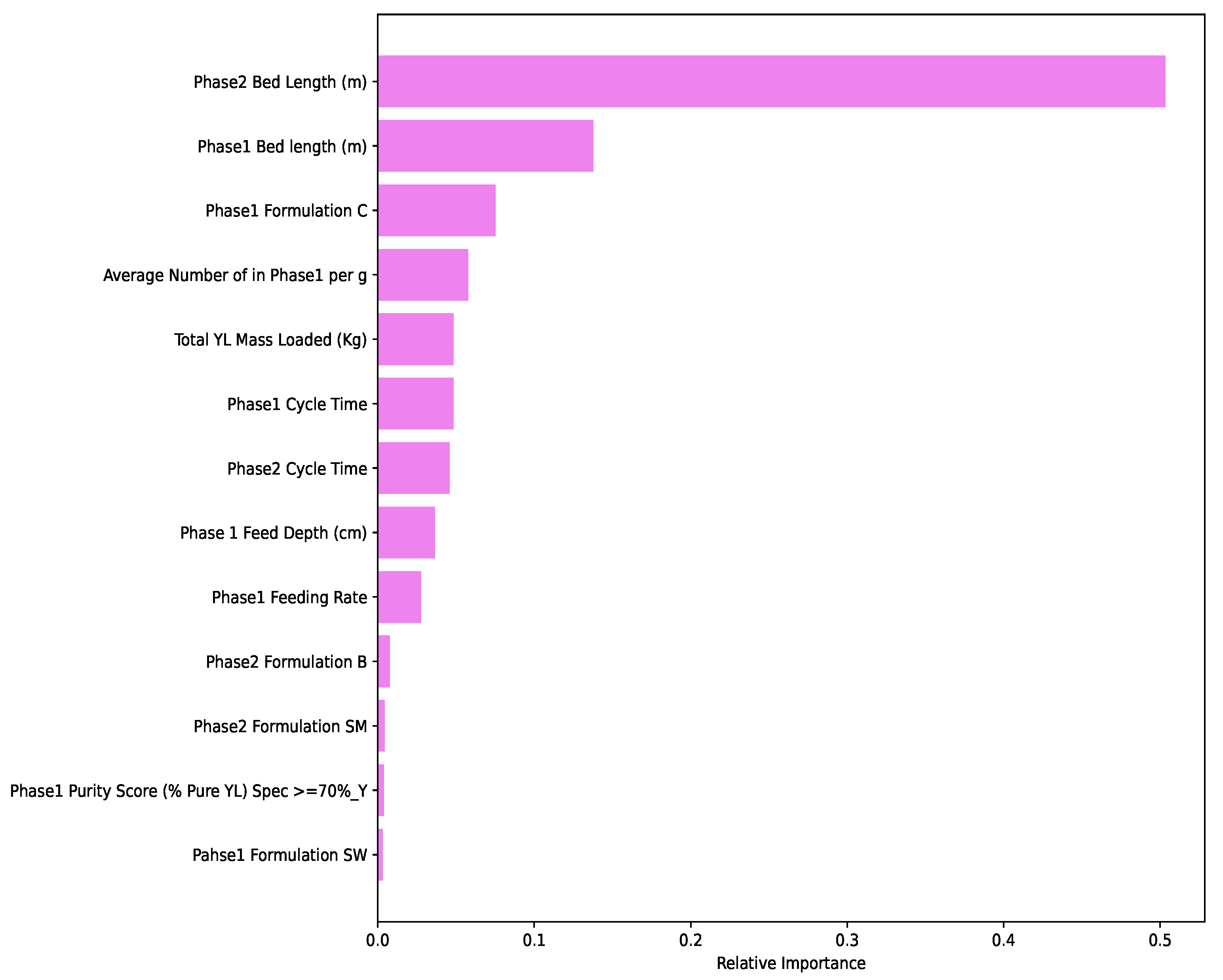
2.3.6. Ranking of Features
3. Results
3.1. Data Exploration
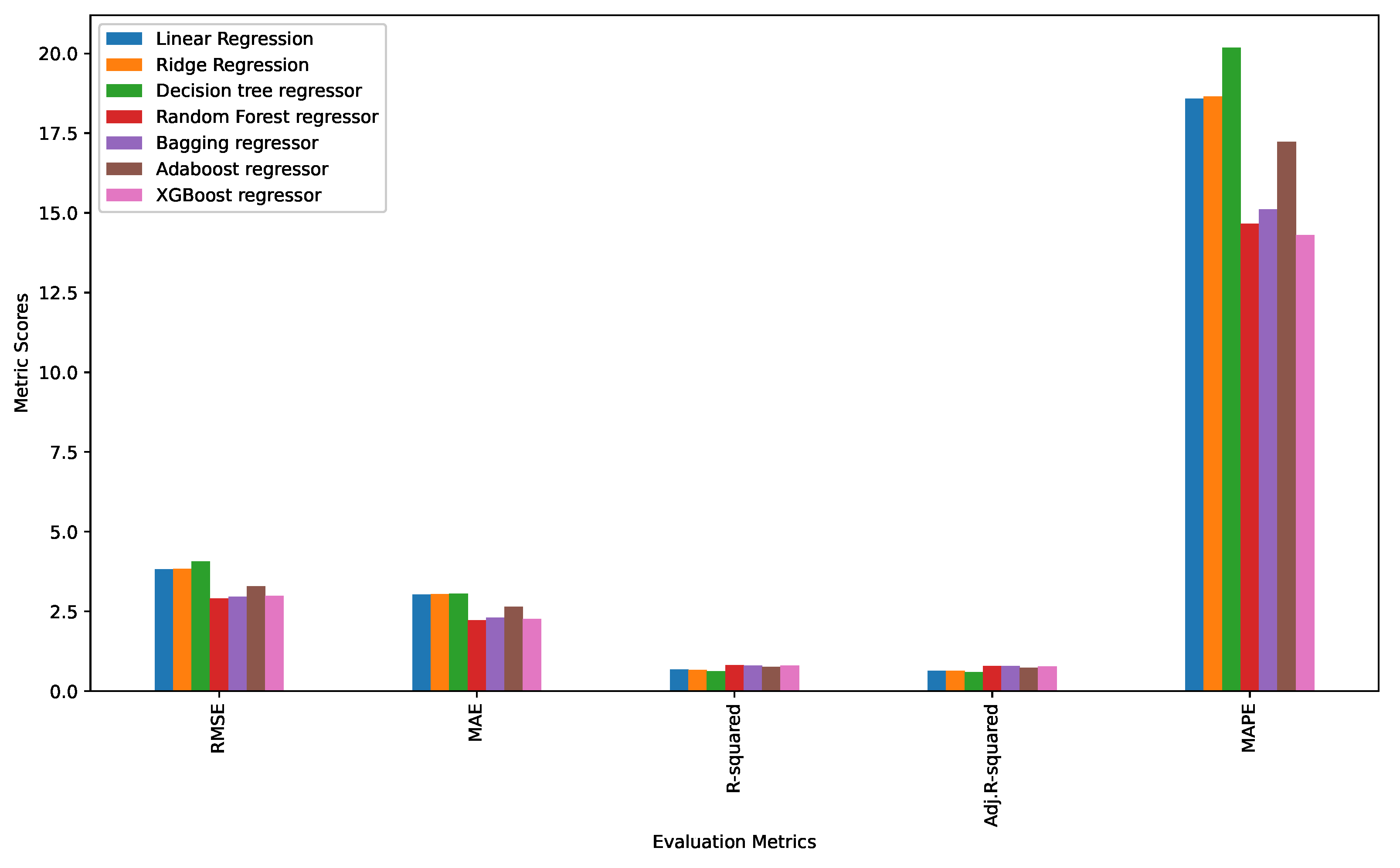
3.2. Performance Evaluation of Machine Learning Models and Ranking of Variables
4. Discussion
4.1. Data Analytics
4.2. Machine Learning Algorithms and Feature Ranking
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- United Nations Department of Economic and Social Affairs. World Population Prospects 2022: Summary of Results—UN DESA/POP/2022/TR/NO. 3; United Nations Department of Economic and Social Affairs: New York, NY, USA, 2022. [Google Scholar]
- FAO. The Future of Food and Agriculture: Alternative Pathways to 2050; Food and Agriculture Organization of the United Nations Rome: Rome, Italy, 2018. [Google Scholar]
- Hannah, F.; Munene, K.; Mohsen, M.; Lee-Anne, H.; Anna, K.S.; Elijah, G.K. Complete replacement of soybean meal with black soldier fly larvae meal in feeding program for broiler chickens from placement through to 49 days of age reduced growth performance and altered organs morphology. J. Poult. Sci. 2023, 102, 102293. [Google Scholar] [CrossRef]
- Veronica, C.; Kate, S.A.; Lee-Anne, H.; Elijah, K. Comparative protein quality in black soldier fly larvae meal vs. soybean meal and fish meal using classical protein efficiency ratio (PER) chick growth assay model. J. Poult. Sci. 2023, 102, 102255. [Google Scholar] [CrossRef]
- Sharvini, S.R.; Stringer, L.; Bruce, N.; Chun, S.C. Opportunities, challenges and solutions for black soldier fly larvae-based animal feed production. J. Clean. Prod. 2022, 373, 133802. [Google Scholar] [CrossRef]
- Beyers, M.; Coudron, C.; Ravi, R.; Meers, E.; Bruun, S. Black soldier fly larvae as an alternative feed source and agro-waste disposal route—A life cycle perspective. Resour. Conserv. Recycl. 2023, 192, 106917. [Google Scholar] [CrossRef]
- Astuti, D.A.; Wiryawan, K.G. Black Soldier Fly as feed ingredient for ruminants. Anim. Biosci. 2022, 35, 356. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.S.; Shelomi, M. Review of Black Soldier Fly (Hermetia illucens) as animal feed and human food. Foods 2017, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Tschirner, M.; Simon, A. Influence of different growing substrates and processing on the nutrient composition of black soldier fly larvae destined for animal feed. J. Insects Food Feed. 2015, 1, 249–259. [Google Scholar] [CrossRef]
- Lydia, P.M.; Fernandez-Bayo, J.; Putri, F.; Niemeier, D.; Bischel, H.; VanderGheynst, J.S. Predicting black soldier fly larvae biomass and methionine accumulation using a kinetic model for batch cultivation and improving system performance using semi-batch cultivation. Bioprocess Biosyst. Eng. 2022, 45, 333–344. [Google Scholar] [CrossRef]
- Chia, S.Y. Black Soldier Fly Larvae as a Sustainable Animal Feed Ingredient in Kenya. Ph.D. Thesis, Wageningen University and Research, Wageningen, The Netherlands, 2019. [Google Scholar]
- Jung, S.; Jung, J.M.; Tsang, Y.F.; Bhatnagar, A.; Chen, W.H.; Lin, K.Y.A.; Kwon, E.E. Biodiesel production from black soldier fly larvae derived from food waste by non-catalytic transesterification. Energy 2022, 238, 121700. [Google Scholar] [CrossRef]
- Moritz, G.; Jeffery, T.; Stefan, D.; Christian, Z.; Mathys, A. Decomposition of biowaste macronutrients, microbes, and chemicals in black soldier fly larval treatment: A review. Int. J. Mol. Sci. 2020, 21, 4955. [Google Scholar] [CrossRef]
- Cuncheng, L.; Cunwen, W.; Huaiying, Y. Comprehensive Resource Utilization of Waste Using the Black Soldier Fly (Hermetia illucens (L.)) (Diptera: Stratiomyidae). Animals 2019, 9, 349. [Google Scholar] [CrossRef]
- Rena, M.; Masami, S.; Seiichi, F.; Takuya, U. Inoculation with black soldier fly larvae alters the microbiome and volatile organic compound profile of decomposing food waste. Sci. Rep. 2023, 13, 4297. [Google Scholar] [CrossRef]
- Jianwei, H.; Shuang, L.; Aiguo, L.; Jia, Z.; Shengli, S.; Yun, Z.; Chujun, L. Assessing Nursery-Finishing Pig Manures on Growth of Black Soldier Fly Larvae. Animals 2023, 13, 452. [Google Scholar] [CrossRef]
- Kenyhercz, M.; Passalacqua, N. Missing data imputation methods and their performance with biodistance analyses. In Biological Distance Analysis; Elsevier: Amsterdam, The Netherlands, 2016; pp. 181–194. [Google Scholar] [CrossRef]
- Fadlil, A.; Praseptian, M.D. K Nearest Neighbor imputation performance on missing value data graduate user satisfaction. J. RESTI (Rekayasa Sist. Teknol. Inf.) 2022, 6, 570–576. [Google Scholar] [CrossRef]
- Harrell, F.E. Missing data. In Regression Modeling Strategies; Springer: Berlin/Heidelberg, Germany, 2015; pp. 45–61. [Google Scholar] [CrossRef]
- Tiwari, K.; Mehta, K.; Jain, N.; Tiwari, R.; Kanda, G. Selecting the appropriate outlier treatment for common industry applications. In Proceedings of the NESUG Conference Proceedings on Statistics and Data Analysis, Baltimore, MA, USA, 11–14 November 2007; pp. 1–5. [Google Scholar]
- Mahalingam, P.; Kalpana, D.; Sendhilkumar, S.; Thyagarajan, T. Prefatory data analysis approach to synthetically generated pneumatic actuator data set. Proc. Inst. Mech. Eng. Part I J. Syst. Control Eng. 2022, 236, 1807–1818. [Google Scholar] [CrossRef]
- Seger, C. An investigation of categorical variable encoding techniques in machine learning: Binary versus one-hot and feature hashing. Int. J. Comput. Appl. 2018, 175, 7–9. [Google Scholar]
- Alshaher, H. Studying the Effects of Feature Scaling in Machine Learning. Ph.D. Thesis, North Carolina Agricultural and Technical State University, Greensboro, NC, USA, 2021. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. Available online: http://jmlr.org/papers/v12/pedregosa11a.html (accessed on 1 May 2023).
- Shumo, M.; Khamis, F.M.; Tanga, C.M.; Fiaboe, K.K.; Subramanian, S.; Ekesi, S.; Van Huis, A.; Borgemeister, C. Influence of temperature on selected life-history traits of black soldier fly (Hermetia illucens) reared on two common urban organic waste streams in Kenya. Animals 2019, 9, 79. [Google Scholar] [CrossRef]
- Barbi, S.; Macavei, L.I.; Fuso, A.; Luparelli, A.V.; Caligiani, A.; Ferrari, A.M.; Maistrello, L.; Montorsi, M. Valorization of seasonal agri-food leftovers through insects. Sci. Total Environ. 2020, 709, 136209. [Google Scholar] [CrossRef]
- Schneider, P.; Xhafa, F. Anomaly Detection and Complex Event Processing Over IoT Data Streams: With Application to EHealth and Patient Data Monitoring; Academic Press: London, UK, 2022. [Google Scholar] [CrossRef]
- Hackeling, G. Mastering Machine Learning with Scikit-Learn; Packt Publishing Ltd.: Birmingham, UK, 2017. [Google Scholar]
- De Myttenaere, A.; Golden, B.; Le Grand, B.; Rossi, F. Mean absolute percentage error for regression models. Neurocomputing 2016, 192, 38–48. [Google Scholar] [CrossRef]
- Miles, J. R-squared, adjusted R-squared. In Encyclopedia of Statistics in Behavioral Science; John Wiley & Sons Ltd.: London, UK, 2005. [Google Scholar] [CrossRef]
- Broeckx, L.; Frooninckx, L.; Slegers, L.; Berrens, S.; Noyens, I.; Goossens, S.; Verheyen, G.; Wuyts, A.; Van Miert, S. Growth of Black Soldier Fly larvae reared on organic side-streams. Sustainability 2021, 13, 12953. [Google Scholar] [CrossRef]
- Eriksen, N.T. Dynamic modelling of feed assimilation, growth, lipid accumulation, and CO2 production in black soldier fly larvae. PLoS ONE 2022, 17, e0276605. [Google Scholar] [CrossRef] [PubMed]
- Barragán-Fonseca, K.; Pineda-Mejia, J.; Dicke, M.; Van Loon, J.J. Performance of the black soldier fly (Diptera: Stratiomyidae) on vegetable residue-based diets formulated based on protein and carbohydrate contents. J. Econ. Entomol. 2018, 111, 2676–2683. [Google Scholar] [CrossRef]
- Barragan-Fonseca, K.B.; Gort, G.; Dicke, M.; van Loon, J.J. Effects of dietary protein and carbohydrate on life-history traits and body protein and fat contents of the black soldier fly Hermetia illucens. Physiol. Entomol. 2019, 44, 148–159. [Google Scholar] [CrossRef]
- ICIPE. Machine Learning to Discern Optimal Conditions BSF; International Centre of Insect Physiology and Ecology (ICIPE): Nairobi, Kenya, 2022; Available online: https://github.com/icipe-official/Machine-Learning-to-Discern-Optimal-Conditions-BSF (accessed on 1 May 2023).


| No. | Variable Name | Description |
|---|---|---|
| 1 | Phase 1 Cycle Time (days) | Time taken to rear the larvae in the first phase. |
| 2 | Phase 1 Formulation C | Coded name of feed type used in Phase 1. |
| 3 | Phase 1 Bed Length | length of the rearing area at Phase1. |
| 4 | Phase 1 Purity Score | percentage purity recorded after separating the larvae from the substrate. |
| 5 | Phase 1 Formulation SW | Coded name of a feed used in Phase 1. |
| 6 | Average Number of Larvae in Phase 1 in 1 g | Estimated number of young larvae per 1 g. |
| 7 | Phase 1 Feed Depth (in) | Measurement of the feed depth. |
| 8 | Phase 1 Feeding Rate in milligrams (mg) per young larvae (YL) in a day, i.e., mg/YL-Day. | This refers to the estimated mass of feed given to each larva per day. The young larvae refer to those that were 2–3 days old. |
| 9 | Phase 2 Formulation SM | Coded name of feed type used in Phase 2. |
| 10 | Phase 2 Bed Length (m) | Length of the beds at the second phase. |
| 11 | Phase 2 Formulation B | Coded name of the feed type used. |
| 12 | Phase 2 Cycle Time (days) | Rearing time under Phase 2. |
| 13 | Total Young Larvae (YL) Mass Loaded (kg) | Amount of larvae added in mass at each bed. |
| 14 | Mass Harvested | Refers to the amount of larvae harvested in kilograms (kg) at the end of the rearing cycle comprising of phase 1 and phase 2. |
| No. | Variable Name | Outlier Treatment |
|---|---|---|
| 1 | Phase 1 Actual Cycle Time (days) | Outliers were corrected using the flooring and capping approach. Percentile values are shown in Table 3. |
| 2 | Phase 1 Formulation Coke | Categorical variable; no outlier score. |
| 3 | Phase 1 Bed Length | Outliers were corrected using the flooring and capping approach. Percentile values are shown in Table 3. |
| 4 | Total Young Larvae (YL) Mass Loaded (kg) | No presence of outliers. |
| 5 | Phase 1 Purity Score | Boolean (Yes/No) variable; no outlier score. |
| 6 | Phase 1 Formulation SW | Categorical variable; no outlier score. |
| 7 | Average Number of Larvae Phase 1 in 1 g | Outliers were corrected using the flooring and capping approach. Percentile values are shown in Table 3. |
| 8 | Phase 1 Feed Depth (in) | Outliers were corrected using the flooring and capping approach. Percentile values are shown in Table 3. |
| 9 | Actual Phase 1 Feeding rate (mg/YL-Day) | Anomaly outliers not detected. |
| 10 | Phase 2 Bed Length (m) | No presence of outliers. |
| 11 | Phase 2 Formulation B | Categorical variable; no outlier score. |
| 12 | Phase 2 Formulation SM | Categorical variable; no outlier score. |
| 13 | Phase 2 Cycle Time (days) | Outliers were corrected using the flooring and capping approach. Percentile values are shown in Table 3. |
| 14 | Mass Harvested | This variable was not treated for outliers to avoid altering the distribution of the target variable. |
| Variable Name | Lower Percentile | Upper Percentile |
|---|---|---|
| Phase 1 Cycle Time (days) | 0.01 | 0.99 |
| Phase 1 Bed Length | 0.05 | 0.95 |
| Phase 1 Average Number of Larvae in 1 g | 0.02 | 0.98 |
| Phase 2 Cycle Time (days) | 0.01 | 0.99 |
| RMSE | MAE | R | Adjusted R | MAPE | |
|---|---|---|---|---|---|
| Linear Regression | 3.818960 | 3.027252 | 0.671549 | 0.638467 | 18.587188 |
| Ridge regression | 3.832344 | 3.043502 | 0.669242 | 0.638529 | 18.645440 |
| Decision tree regressor | 4.073082 | 3.048241 | 0.626382 | 0.591689 | 20.185357 |
| Random forest regressor | 2.912162 | 2.221066 | 0.809009 | 0.791275 | 14.661590 |
| Bagging regressor | 2.961843 | 2.298820 | 0.802437 | 0.784092 | 15.113145 |
| Adaboost regressor | 3.29355 | 2.647179 | 0.755707 | 0.733023 | 17.228926 |
| XGBoost regressor | 2.990458 | 2.255420 | 0.798601 | 0.779900 | 14.305270 |
| RMSE | MAE | R | Adjusted R | MAPE | |
|---|---|---|---|---|---|
| Statistical model (ordinary least square) | 3.818960 | 3.027252 | 0.671549 | 0.638467 | 18.587188 |
| Generalized linear model | 3.855807 | 3.087771 | 0.665180 | 0.634089 | 18.712122 |
| Random forest regressor | 2.912162 | 2.221066 | 0.809009 | 0.791275 | 14.661590 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muinde, J.; Tanga, C.M.; Olukuru, J.; Odhiambo, C.; Tonnang, H.E.Z.; Senagi, K. Application of Machine Learning Techniques to Discern Optimal Rearing Conditions for Improved Black Soldier Fly Farming. Insects 2023, 14, 479. https://doi.org/10.3390/insects14050479
Muinde J, Tanga CM, Olukuru J, Odhiambo C, Tonnang HEZ, Senagi K. Application of Machine Learning Techniques to Discern Optimal Rearing Conditions for Improved Black Soldier Fly Farming. Insects. 2023; 14(5):479. https://doi.org/10.3390/insects14050479
Chicago/Turabian StyleMuinde, John, Chrysantus M. Tanga, John Olukuru, Clifford Odhiambo, Henri E. Z. Tonnang, and Kennedy Senagi. 2023. "Application of Machine Learning Techniques to Discern Optimal Rearing Conditions for Improved Black Soldier Fly Farming" Insects 14, no. 5: 479. https://doi.org/10.3390/insects14050479








