An Investigation of Burkholderia pseudomallei Seroprevalence in Market Pigs Slaughtered at Selected Pig Abattoirs in Uganda
Abstract
:1. Introduction
2. Materials and Methods
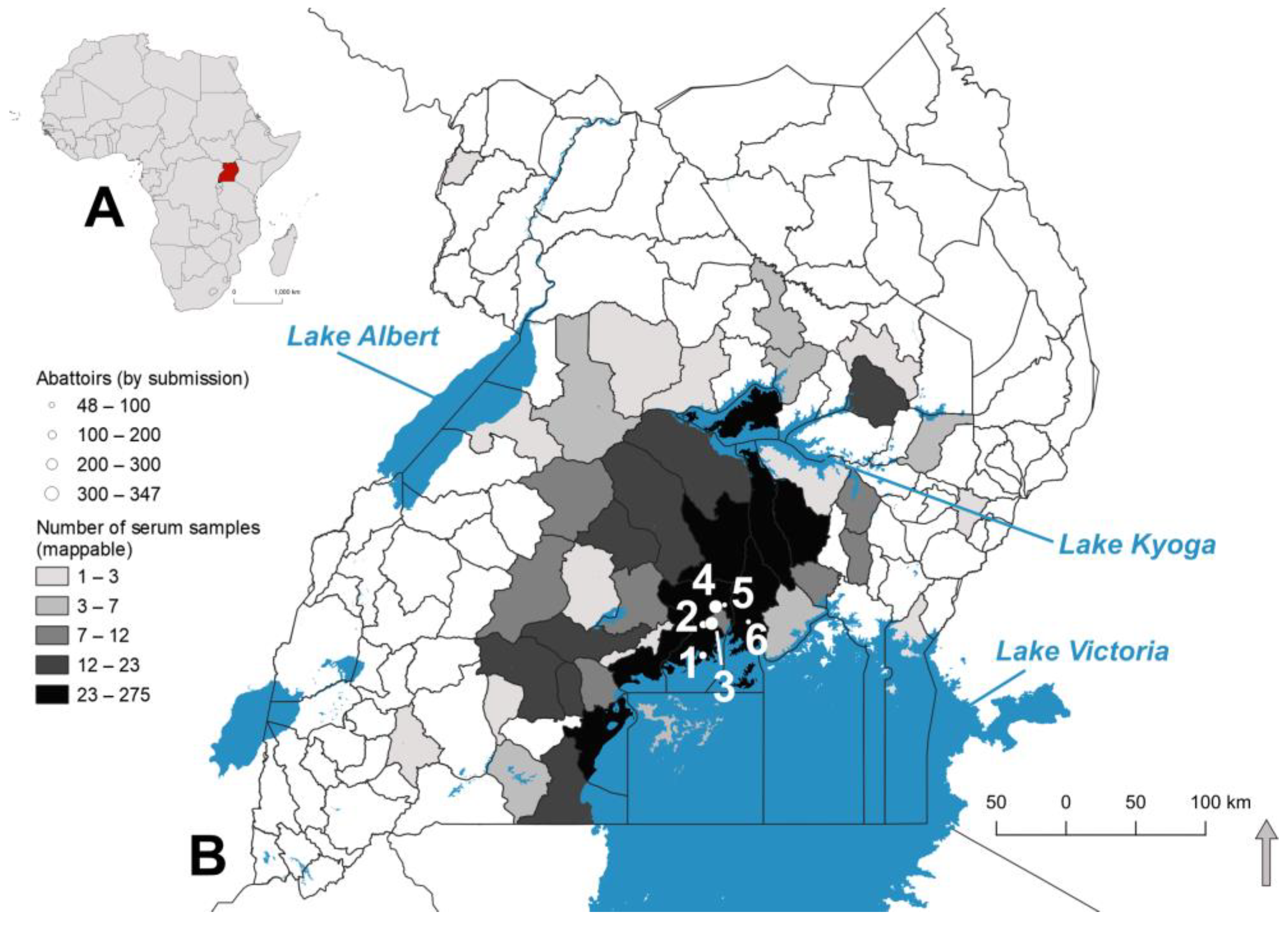
2.1. Sample Acquisition
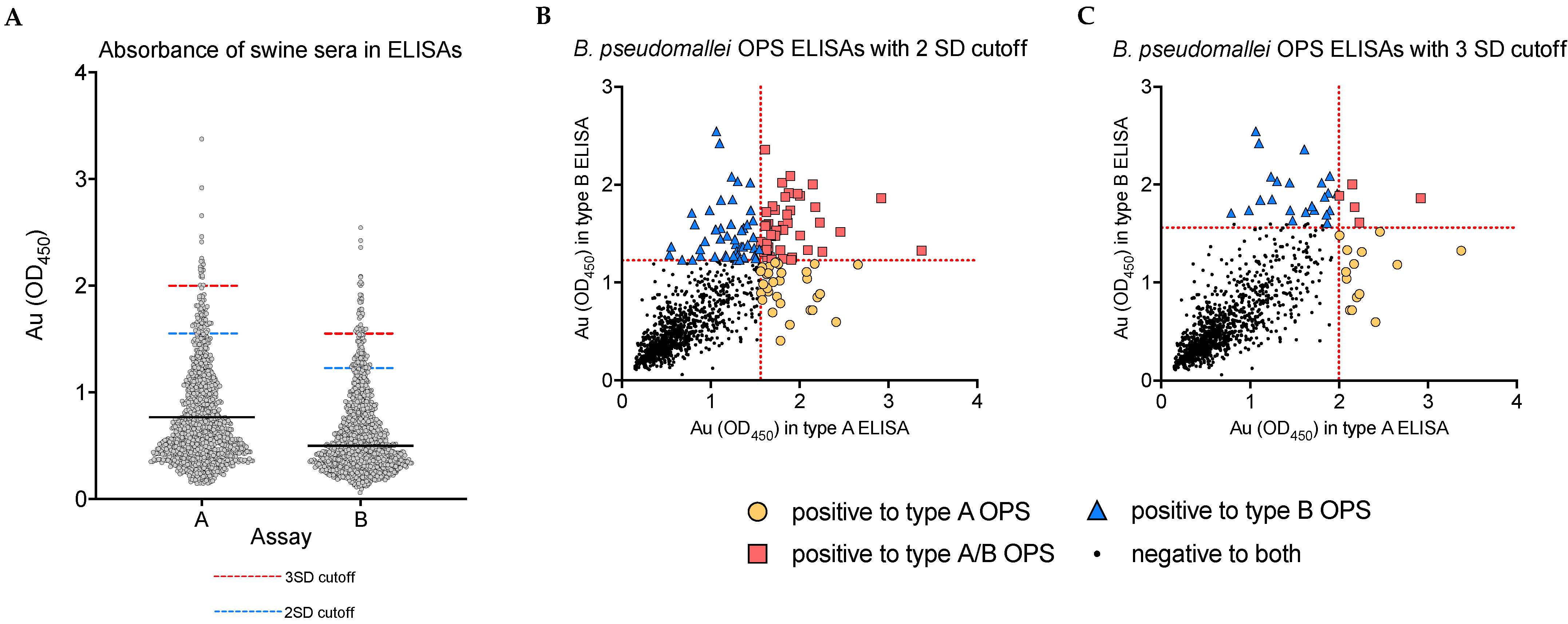
2.2. Serological Testing
2.3. Serological Data Analysis
2.4. Mapping Seroprevalence
2.5. Local Spatial Autocorrelation of Seroprevalence
3. Results
3.1. Pig Sample Characteristics
3.2. Analysis of Unadjusted Seropositivity
3.3. Distribution of Serological Findings by Abattoir, Sex, and Pig Breed
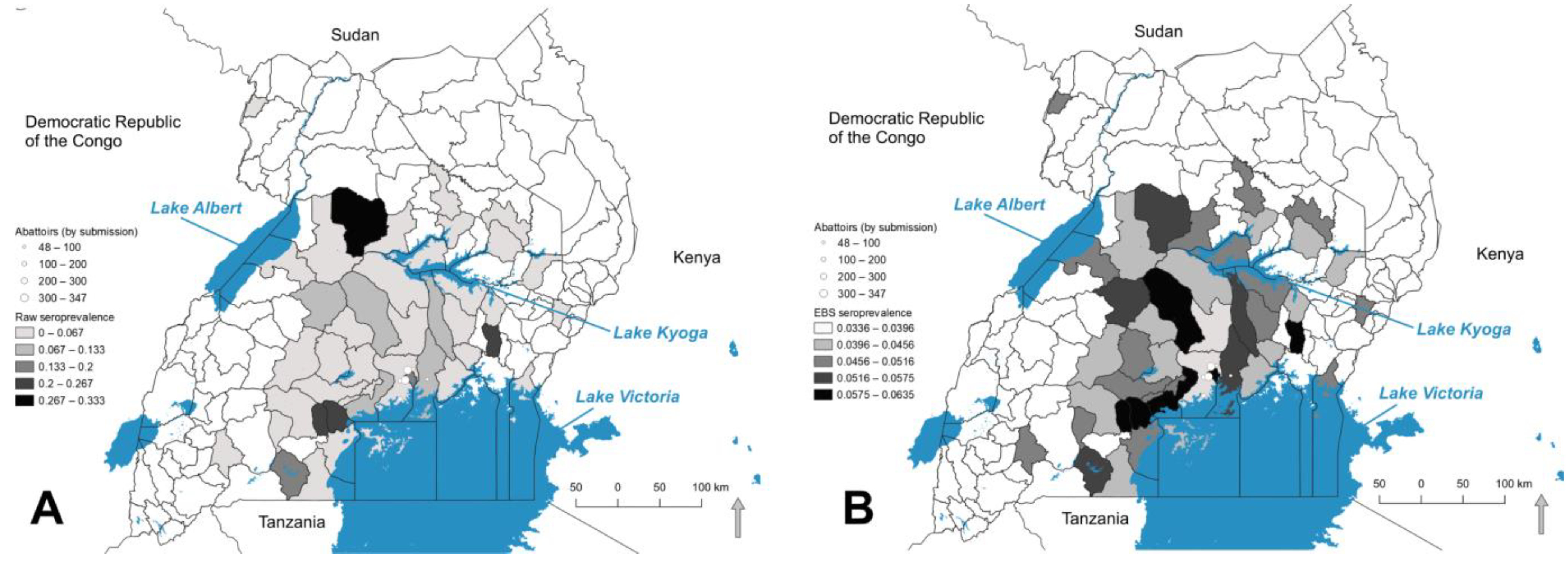
3.4. Seropositivity at the District Level
3.5. Local Spatial Autocorrelation of EBS Seroprevalence
3.6. Temporal and Seasonal Variation in Seropositivity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Narayanan, S. Burkholderia mallei and Burkholderia pseudomallei. In Veterinary Microbiology; Wiley Online Library: Hoboken, NJ, USA, 2022; pp. 162–167. ISBN 978-1-119-65083-6. [Google Scholar]
- White, N.J. Melioidosis. Lancet 2003, 361, 1715–1722. [Google Scholar] [CrossRef]
- Dance, D.A. Ecology of Burkholderia pseudomallei and the Interactions between Environmental Burkholderia spp. and Human-Animal Hosts. Acta Trop. 2000, 74, 159–168. [Google Scholar] [CrossRef]
- Dance, D.A.B. Melioidosis: The Tip of the Iceberg? Clin. Microbiol. Rev. 1991, 4, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-S.; Chen, Y.-S.; Lin, H.-H.; Liu, P.-J.; Ni, W.-F.; Hsueh, P.-T.; Liang, S.-H.; Chen, C.; Chen, Y.-L. Airborne Transmission of Melioidosis to Humans from Environmental Aerosols Contaminated with B. pseudomallei. PLoS Negl. Trop. Dis. 2015, 9, e0003834. [Google Scholar] [CrossRef]
- Barnes, J.L.; Ketheesan, N. Route of Infection in Melioidosis. Emerg. Infect. Dis. 2005, 11, 638–639. [Google Scholar] [CrossRef]
- Limmathurotsakul, D.; Wuthiekanun, V.; Tuanyok, A.; Peacock, S.J. Presence and Sampling of Burkholderia pseudomallei in Soil. In Melioidosis—A Century of Observation and Research; Ketheesan, N., Ed.; Elsevier: San Diego, CA, USA, 2012; pp. 349–357. [Google Scholar]
- Hellebuyck, T.; Wattiau, P.; Boyen, F.; Moeremans, I.; Roosens, N.; Vanneste, K.; Garmyn, A.; Saey, V.; Pasmans, F.; Haesebrouck, F. Isolation of Burkholderia pseudomallei from a Pet Green Iguana, Belgium. Emerg. Infect. Dis. J. 2018, 24, 2331. [Google Scholar] [CrossRef] [Green Version]
- Sprague, L.D.; Neubauer, H. Melioidosis in Animals: A Review on Epizootiology, Diagnosis and Clinical Presentation. J. Vet. Med. Ser. B 2004, 51, 305–320. [Google Scholar] [CrossRef]
- Gassiep, I.; Armstrong, M.; Norton, R. Human Melioidosis. Clin. Microbiol. Rev. 2020, 33, e00006-19. [Google Scholar] [CrossRef]
- Titball, R.W.; Burtnick, M.N.; Bancroft, G.J.; Brett, P. Burkholderia pseudomallei and Burkholderia mallei Vaccines: Are We Close to Clinical Trials? Biosecurity 2017, 35, 5981–5989. [Google Scholar] [CrossRef]
- Karunanayake, P. Melioidosis: Clinical Aspects. Clin. Med. 2022, 22, 6–8. [Google Scholar] [CrossRef]
- Yee, K.C.; Lee, M.K.; Chua, C.T.; Puthucheary, S.D. Melioidosis, the Great Mimicker: A Report of 10 Cases from Malaysia. J. Trop. Med. Hyg. 1988, 91, 249–254. [Google Scholar] [PubMed]
- Melioidosis-Infectious Diseases. Available online: https://www.merckmanuals.com/professional/infectious-diseases/gram-negative-bacilli/melioidosis (accessed on 6 September 2022).
- Constable, P.D.; Hinchcliff, K.W.; Done, S.H.; Grünberg, W. Veterinary Medicine: A Textbook of the Diseases of Cattle, Horses, Sheep, Pigs and Goats, 11th ed.; Elsevier: St. Louis, MO, USA, 2017; pp. 2029–2031. [Google Scholar]
- Choy, J.L.; Mayo, M.; Janmaat, A.; Currie, B.J. Animal Melioidosis in Australia. Acta Trop. 2000, 74, 153–158. [Google Scholar] [CrossRef]
- Hang, N.T.T.; Hang, T.T.T.; Trung, T.T.; Khong, N.V. Initial Investigation of Burkholderia pseudomallei in Pigs in Nghe an Province, Vietnam in 2016 and 2017. In Proceedings of the World Melioidosis Congress, Hanoi, Vietnam, 15–18 October 2019; Volume WMC 2019, p. 116. [Google Scholar]
- Inglis, T.J.J.; Sousa, A.Q. The Public Health Implications of Melioidosis. Braz. J. Infect. Dis. 2009, 13, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birnie, E.; Virk, H.S.; Savelkoel, J.; Spijker, R.; Bertherat, E.; Dance, D.A.B.; Limmathurotsakul, D.; Devleesschauwer, B.; Haagsma, J.A.; Wiersinga, W.J. Global Burden of Melioidosis in 2015: A Systematic Review and Data Synthesis. Lancet Infect. Dis. 2019, 19, 892–902. [Google Scholar] [CrossRef] [Green Version]
- Dance, D. Treatment and Prophylaxis of Melioidosis. Int. J. Antimicrob. Agents 2014, 43, 310–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limmathurotsakul, D.; Golding, N.; Dance, D.A.B.; Messina, J.P.; Pigott, D.M.; Moyes, C.L.; Rolim, D.B.; Bertherat, E.; Day, N.P.J.; Peacock, S.J.; et al. Predicted Global Distribution of Burkholderia pseudomallei and Burden of Melioidosis. Nat. Microbiol. 2016, 1, 15008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiersinga, W.J.; Currie, B.J.; Peacock, S.J. Melioidosis. N. Engl. J. Med. 2012, 367, 1035–1044. [Google Scholar] [CrossRef]
- Hatcher, C.L.; Muruato, L.A.; Torres, A.G. Recent Advances in Burkholderia mallei and B. pseudomallei Research. Curr. Trop. Med. Rep. 2015, 2, 62–69. [Google Scholar] [CrossRef] [Green Version]
- Steinmetz, I.; Wagner, G.E.; Kanyala, E.; Sawadogo, M.; Soumeya, H.; Teferi, M.; Andargie, E.; Yeshitela, B.; Yaba Atsé-Achi, L.; Sanogo, M.; et al. Melioidosis in Africa: Time to Uncover the True Disease Load. Trop. Med. Infect. Dis. 2018, 3, 62. [Google Scholar] [CrossRef] [Green Version]
- Savelkoel, J.; Dance, D.A.B.; Currie, B.J.; Limmathurotsakul, D.; Wiersinga, W.J. A Call to Action: Time to Recognise Melioidosis as a Neglected Tropical Disease. Lancet Infect. Dis. 2022, 22, e176–e182. [Google Scholar] [CrossRef]
- Birnie, E.; Wiersinga, W.J.; Limmathurotsakul, D.; Grobusch, M.P. Melioidosis in Africa: Should We Be Looking More Closely? Future Microbiol. 2015, 10, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Frazer, D.N. Melioidosis. J. R. Army Med. Corps 1982, 128, 123. [Google Scholar] [CrossRef] [Green Version]
- Currie, B.J.; Dance, D.A.B.; Cheng, A.C. The Global Distribution of Burkholderia pseudomallei and Melioidosis: An Update. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, S1–S4. [Google Scholar] [CrossRef]
- Norris, M.H.; Tran, H.T.T.; Walker, M.A.; Bluhm, A.P.; Zincke, D.; Trung, T.T.; Thi, N.V.; Thi, N.P.; Schweizer, H.P.; Unger, F.; et al. Distribution of Serological Response to Burkholderia pseudomallei in Swine from Three Provinces of Vietnam. Int. J. Environ. Res. Public. Health 2020, 17, 5203. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, R.H. Validation of serological assays for diagnosis of infectious diseases. Rev. Sci. Tech. Int. Off. Epizoot. 1998, 17, 469–526. [Google Scholar] [CrossRef]
- Nsubuga, F.N.W.; Olwoch, J.M.; Rautenbach, C.J.d.W.; Botai, O.J. Analysis of Mid-Twentieth Century Rainfall Trends and Variability over Southwestern Uganda. Theor. Appl. Climatol. 2014, 115, 53–71. [Google Scholar] [CrossRef] [Green Version]
- Diem, J.E.; Hartter, J.; Ryan, S.J.; Palace, M.W. Validation of Satellite Rainfall Products for Western Uganda. J. Hydrometeorol. 2014, 15, 2030–2038. [Google Scholar] [CrossRef]
- Anselin, L.; Lozano, N.; Koschinsky, J. Rate Transformation and Smoothing; University of Illinois: Champaign, IL, USA, 2006. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020. [Google Scholar]
- Anselin, L. Local Indicators of Spatial Association—LISA. Geogr. Anal. 1995, 27, 93–115. [Google Scholar] [CrossRef]
- Kracalik, I.T.; Malania, L.; Tsertsvadze, N.; Manvelyan, J.; Bakanidze, L.; Imnadze, P.; Tsanava, S.; Blackburn, J.K. Evidence of Local Persistence of Human Anthrax in the Country of Georgia Associated with Environmental and Anthropogenic Factors. PLoS Negl. Trop. Dis. 2013, 7, e2388. [Google Scholar] [CrossRef] [Green Version]
- Reif, J.S. Animal Sentinels for Environmental and Public Health. Public Health Rep. 2011, 126, 50–57. [Google Scholar] [CrossRef]
- Nugent, G.; Whitford, J.; Young, N. Use of Released Pigs as Sentinels for Mycobacterium bovis. J. Wildl. Dis. 2002, 38, 665–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwanhian, W.; Jiranantasak, T.; Kessler, A.T.; Tolchinsky, B.E.; Parker, S.; Songsri, J.; Wisessombat, S.; Pukanha, K.; Testamenti, V.A.; Khrongsee, P.; et al. Investigation of Melioidosis Outbreak in Pig Farms in Southern Thailand. Vet. Sci. 2020, 7, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Villamil, J.I.; Tapia, D.; Borlee, G.I.; Borlee, B.R.; Walker, D.H.; Torres, A.G. Burkholderia pseudomallei as an Enteric Pathogen: Identification of Virulence Factors Mediating Gastrointestinal Infection. Infect. Immun. 2020, 89, e00654-20. [Google Scholar] [CrossRef]
- Norris, M.H.; Schweizer, H.P.; Tuanyok, A. Structural Diversity of Burkholderia pseudomallei Lipopolysaccharides Affects Innate Immune Signaling. PLoS Negl. Trop. Dis. 2017, 11, e0005571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norris, M.H.; Rahman Khan, M.S.; Schweizer, H.P.; Tuanyok, A. An Avirulent Burkholderia pseudomallei ΔpurM Strain with Atypical Type B LPS: Expansion of the Toolkit for Biosafe Studies of Melioidosis. BMC Microbiol. 2017, 17, 132. [Google Scholar] [CrossRef] [Green Version]
- Sarovich, D.S.; Garin, B.; De Smet, B.; Kaestli, M.; Mayo, M.; Vandamme, P.; Jacobs, J.; Lompo, P.; Tahita, M.C.; Tinto, H.; et al. Phylogenomic Analysis Reveals an Asian Origin for African Burkholderia pseudomallei and Further Supports Melioidosis Endemicity in Africa. mSphere 2016, 1, e000089-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vongphayloth, K.; Rattanavong, S.; Moore, C.E.; Phetsouvanh, R.; Wuthiekanun, V.; Sengdouangphachanh, A.; Phouminh, P.; Newton, P.N.; Buisson, Y. Burkholderia pseudomallei Detection in Surface Water in Southern Laos Using Moore’s Swabs. Am. Soc. Trop. Med. Hyg. 2012, 86, 872–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaestli, M.; Grist, E.P.M.; Ward, L.; Hill, A.; Mayo, M.; Currie, B.J. The Association of Melioidosis with Climatic Factors in Darwin, Australia: A 23-Year Time-Series Analysis. J. Infect. 2016, 72, 687–697. [Google Scholar] [CrossRef]
- Currie, B.J.; Jacups, S.P. Intensity of Rainfall and Severity of Melioidosis, Australia. Emerg. Infect. Dis. 2003, 9, 1538. [Google Scholar] [CrossRef]
- Ministry of Agriculture, Animal Industry, and Fisheries; Uganda Bureau of Statistics. The National Livestock Census Report 2008; Ministry of Agriculture, Animal Industry, and Fisheries: Entebbe, Uganda; Uganda Bureau of Statistics: Kampala, Uganda, 2009.
- Cheng, A.C.; Jacups, S.P.; Gal, D.; Mayo, M.; Currie, B.J. Extreme Weather Events and Environmental Contamination Are Associated with Case-Clusters of Melioidosis in the Northern Territory of Australia. Int. J. Epidemiol. 2006, 35, 323–329. [Google Scholar] [CrossRef]
- Currie, B.J.; Fisher, D.A.; Howard, D.M.; Burrow, J.N.; Lo, D.; Selva-Nayagam, S.; Anstey, N.M.; Huffam, S.E.; Snelling, P.L.; Marks, P.J.; et al. Endemic Melioidosis in Tropical Northern Australia: A 10-Year Prospective Study and Review of the Literature. Clin. Infect. Dis. 2000, 31, 981–986. [Google Scholar] [CrossRef]


| 2 Standard Deviation Cutoff | 3 Standard Deviation Cutoff | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abattoir (District) | # Positive A | % (95% CI) Positive A | # Positive B | % (95% CI) Positive B | # Positive Both A & B | % (95% CI) Positive Both A & B | # Positive A | % (95% CI) Positive A | # Positive B | % (95% CI) Positive B | # Positive Both A & B | % (95% CI) Positive Both A & B | Total |
| Budo (Wakiso) | 4 | 3.74% (1.2–9.5%) | 8 | 7.48% (3.6–14.3%) | 3 | 2.80% (1.6–8.3%) | 0 | 0.00% | 2 | 1.87% (0.1–7%) | 0 | 0.00% | 107 |
| Buwate (Wakiso) | 4 | 5.13% (1.6–12.8%) | 3 | 3.85% (0.9–11.2%) | 1 | 1.28% (0–7.6%) | 1 | 1.28% (0–7.6%) | 0 | 0.00% | 0 | 0.00% | 78 |
| Katabi (Wakiso) | 8 | 5.52% (2.7–10.7%) | 13 | 8.97% (5.2–14.8%) | 7 | 4.83% (2.2–9.8%) | 1 | 0.69% (0–4.2%) | 4 | 2.76% (0.8–7.1%) | 0 | 0.00% | 145 |
| Lusanja (Wakiso) | 32 | 9.22% (6.6–12.8%) | 27 | 7.78% (5.4–11.1%) | 16 | 4.61% (2.8–7.4%) | 7 | 2.02% (0.9–4.2%) | 5 | 1.44% (0.5–3.4%) | 0 | 0.00% | 347 |
| Kyetume (Mukono) | 2 | 4.17% (0.4–14.8%) | 3 | 6.25% (1.5–17.5%) | 2 | 4.17% (0.4–14.8%) | 1 | 2.08% (0–11.9%) | 2 | 4.17% (0.4–14.8%) | 1 | 2.08% (0–11.9%) | 48 |
| Wambizi (Kampala) | 25 | 8.06% (5.5–11.7%) | 39 | 12.58% (9.3–16.8%) | 15 | 4.84% (2.9–7.9%) | 9 | 2.90% (1.5–5.5%) | 15 | 4.84% (2.9–7.9%) | 4 | 1.29% (0.4–3.4%) | 310 |
| Total | 75 | 7.25% (5.8–9%) | 93 | 8.99% (7.4–10.9%) | 44 | 4.25% (3.2–5.7%) | 19 | 1.84% (1.2–2.9%) | 28 | 2.71% (1.9–3.9%) | 5 | 0.48% (0.2–1.2%) | 1035 |
| Sex of Pigs | |||||||||||||
| Female | 57 | 10.07% (7.8–12.8%) | 57 | 10.07% (7.8–12.8%) | 33 | 5.83% (4.2–8.1%) | 17 | 3.00% (1.8–4.8%) | 20 | 3.53% (2.3–5.4%) | 4 | 0.71% (0.2–1.9%) | 566 |
| Male | 18 | 3.89% (2.4–6.1%) | 36 | 7.78% (5.6–10.6%) | 11 | 2.38% (1.3–4.3%) | 2 | 0.43% (0–1.7%) | 8 | 1.73% (0.8–3.4%) | 1 | 0.22% (0–1.3%) | 463 |
| Unknown | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% | 6 |
| Total | 75 | 7.25% (5.8–9%) | 93 | 8.99% (7.4–10.9%) | 44 | 4.25% (3.2–5.7%) | 19 | 1.84% (1.2–2.9%) | 28 | 2.71% (1.9–3.9%) | 5 | 0.48% (0.2–1.2%) | 1035 |
| Breed of Pigs | |||||||||||||
| Mixed | 30 | 10.53% (7.4–14.7%) | 41 | 14.39% (10.8–19%) | 17 | 5.96% (3.7–9.4%) | 7 | 2.46% (1.1–5.1%) | 14 | 4.91% (2.9–8.1%) | 1 | 0.35% (0–2.2%) | 285 |
| Exotic | 33 | 5.81% (4.1–8.1%) | 39 | 6.87% (5–9.3%) | 22 | 3.87% (2.5–5.8%) | 7 | 1.23% (0.5–2.6%) | 12 | 2.11% (1.2–3.7%) | 3 | 0.53% (0.1–1.6%) | 568 |
| Local | 10 | 6.17% (3.3–11.1%) | 13 | 8.02% (4.6–13.4%) | 5 | 3.09% (1.1–7.2%) | 4 | 2.47% (0.7–6.4%) | 2 | 1.23% (0.1–4.7%) | 1 | 0.62% (0–3.8%) | 162 |
| Unknown | 2 | 10.00% (1.6–31.3%) | 0 | 0.00% | 0 | 0.00% | 1 | 5.00% (0–25.4%) | 0 | 0.00% | 0 | 0.00% | 20 |
| Total | 75 | 7.25% (5.8–9%) | 93 | 8.99% (7.4–10.9%) | 44 | 4.25% (3.2–5.7%) | 19 | 1.84% (1.2–2.9%) | 28 | 2.71% (1.9–3.9%) | 5 | 0.48% (0.2–1.2%) | 1035 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ekakoro, J.E.; Lubega, A.; Kayaga, E.B.; Ndoboli, D.; Bluhm, A.P.; Wampande, E.M.; Blackburn, J.K.; Havas, K.A.; Norris, M.H. An Investigation of Burkholderia pseudomallei Seroprevalence in Market Pigs Slaughtered at Selected Pig Abattoirs in Uganda. Pathogens 2022, 11, 1363. https://doi.org/10.3390/pathogens11111363
Ekakoro JE, Lubega A, Kayaga EB, Ndoboli D, Bluhm AP, Wampande EM, Blackburn JK, Havas KA, Norris MH. An Investigation of Burkholderia pseudomallei Seroprevalence in Market Pigs Slaughtered at Selected Pig Abattoirs in Uganda. Pathogens. 2022; 11(11):1363. https://doi.org/10.3390/pathogens11111363
Chicago/Turabian StyleEkakoro, John E., Arnold Lubega, Edrine B. Kayaga, Dickson Ndoboli, Andrew P. Bluhm, Eddie M. Wampande, Jason K. Blackburn, Karyn A. Havas, and Michael H. Norris. 2022. "An Investigation of Burkholderia pseudomallei Seroprevalence in Market Pigs Slaughtered at Selected Pig Abattoirs in Uganda" Pathogens 11, no. 11: 1363. https://doi.org/10.3390/pathogens11111363





