A Comparison of Methods for Identifying Enterobacterales Isolates from Fish and Prawns
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Strain Isolation
4.2. EnteroTest 24N
4.3. MALDI-TOF Identification
4.4. Sequencing
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Meier, M.; Hamprecht, A. Systematic comparison of four methods for detection of carbapenemase-producing enterobacterales directly from blood cultures. J. Clin. Microbiol. 2019, 57, e00709-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, N.M.B.; Bronzato, G.F.; Santiago, G.S.; Botelho, L.A.B.; Moreira, B.M.; Coelho, I.D.S.; de Souza, M.M.S.; Coelho, S.D.M.D.O. The Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS) identification versus biochemical tests: A study with enterobacteria from a dairy cattle environment. Braz. J. Microbiol. 2017, 48, 132–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lydon, K.A.; Kinsey, T.; Le, C.; Gulig, P.A.; Jones, J.L. Biochemical and Virulence Characterization of Vibrio vulnificus Isolates From Clinical and Environmental Sources. Front. Cell. Infect. Microbiol. 2021, 11, 64. [Google Scholar] [CrossRef]
- Jesumirhewe, C.; Ogunlowo, P.O.; Olley, M.; Springer, B.; Allerberger, F.; Ruppitsch, W. Accuracy of conventional identification methods used for Enterobacteriaceae isolates in three Nigerian hospitals. PeerJ 2016, 4, e2511. [Google Scholar] [CrossRef] [Green Version]
- Strejcek, M.; Smrhova, T.; Junkova, P.; Uhlik, O. Whole-Cell MALDI-TOF MS Versus 16S rRNA Gene Analysis for Identification and Dereplication of Recurrent Bacterial Isolates. Front. Microbiol. 2018, 9, 1294. [Google Scholar] [CrossRef]
- Piamsomboon, P.; Jaresitthikunchai, J.; Hung, T.Q.; Roytrakul, S.; Wongtavatchai, J. Identification of bacterial pathogens in cultured fish with a custom peptide database constructed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). BMC Vet. Res. 2020, 16, 52. [Google Scholar] [CrossRef]
- Delmas, J.; Breysse, F.; Devulder, G.; Flandrois, J.P.; Chomarat, M. Rapid identification of Enterobacteriaceae by sequencing DNA gyrase subunit B encoding gene. Diagn. Microbiol. Infect. Dis. 2006, 55, 263–268. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Tse, H.; Yuen, K.Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934. [Google Scholar] [CrossRef] [Green Version]
- Awong-Taylor, J.; Craven, K.S.; Griffiths, L.; Bass, C.; Muscarella, M. Comparison of biochemical and molecular methods for the identification of bacterial isolates associated with failed loggerhead sea turtle eggs. J. Appl. Microbiol. 2008, 104, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Montero, D.; Bodero, M.; Riveros, G.; Lapierre, L.; Gaggero, A.; Vidal, R.M.; Vidal, M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015, 6, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Liu, H.; Pan, Y.; Xie, J.; Zhao, Y. Comparison of the effects of environmental parameters on the growth variability of Vibrio parahaemolyticus coupled with strain sources and genotypes analyses. Front. Microbiol. 2016, 7, 994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buszewski, B.; Rogowska, A.; Pomastowski, P.; Złoch, M.; Railean-Plugaru, V. Identification of Microorganisms by Modern Analytical Techniques. J. AOAC Int. 2017, 100, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Popper, M.; Gancarčíková, S.; Maďar, M.; Mudroňová, D.; Hrčková, G.; Nemcová, R. Amoxicillin-clavulanic acid and ciprofloxacin-treated SPF mice as gnotobiotic model. Appl. Microbiol. Biotechnol. 2016, 100, 9671–9682. [Google Scholar] [CrossRef]
- Navrátilová, L.; Šafářová, D.; Raclavský, V. Usefulness of PCR-HRMA in identification of non-fermentative Gram-negative rods recovered from patients suffering from cystic fibrosis or chronic obstructive pulmonary disease. Folia Microbiol. 2014, 59, 17–21. [Google Scholar] [CrossRef]
- MacHorowska-Pieniazek, A.; Mertas, A.; Skucha-Nowak, M.; Tanasiewicz, M.; Morawiec, T. A Comparative Study of Oral Microbiota in Infants with Complete Cleft Lip and Palate or Cleft Soft Palate. Biomed. Res. Int. 2017, 2017, 1460243. [Google Scholar] [CrossRef] [Green Version]
- Koscova, J.; Hurnikova, Z.; Pistl, J. Degree of Bacterial Contamination of Mobile Phone and Computer Keyboard Surfaces and Efficacy of Disinfection with Chlorhexidine Digluconate and Triclosan to Its Reduction. Int. J. Environ. Res. Public Health 2018, 15, 2238. [Google Scholar] [CrossRef] [Green Version]
- Maina, D.; Okinda, N.; Mulwa, E.; Revathi, G. A Five Year Review of API20E Bacteria Identification System’s Performance at A Teaching Hospital. East Afr. Med. 2014, 91, 73–76. [Google Scholar]
- Vithanage, N.R.; Yeager, T.R.; Jadhav, S.R.; Palombo, E.A.; Datta, N. Comparison of identification systems for psychrotrophic bacteria isolated from raw bovine milk. Int. J. Food Microbiol. 2014, 189, 26–38. [Google Scholar] [CrossRef]
- Carbonnelle, E.; Grohs, P.; Jacquier, H.; Day, N.; Tenza, S.; Dewailly, A.; Vissouarn, O.; Rottman, M.; Herrmann, J.L.; Podglajen, I.; et al. Robustness of two MALDI-TOF mass spectrometry systems for bacterial identification. J. Microbiol. Methods 2012, 89, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Arnold, T.; Neubauer, H.; Nikolaou, K.; Roesler, U.; Hensel, A. Identification of Yersinia enterocolitica in Minced Meat: A Comparative Analysis of API 20E, Yersinia Identification Kit and a 16S rRNA-based PCR Method. J. Vet. Med. Ser. B 2004, 51, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Cernela, N.; Ziegler, D.; Pflüger, V.; Tonolla, M.; Ravasi, D.; Fredriksson-Ahomaa, M.; Hächler, H. Rapid species specific identification and subtyping of Yersinia enterocolitica by MALDI-TOF Mass spectrometry. J. Microbiol. Methods 2011, 87, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Ayyadurai, S.; Flaudrops, C.; Raoult, D.; Drancourt, M. Rapid identification and typing of Yersinia pestis and other Yersinia species by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. BMC Microbiol. 2010, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Wanger, A.; Chavez, V.; Huang, R.S.P.; Wahed, A.; Actor, J.K.; Dasgupta, A. Overview of Bacteria. In Microbiology and Molecular Diagnosis in Pathology; Elsevier: Amsterdam, The Netherlands, 2017; pp. 75–117. [Google Scholar]
- Kolínská, R.; Španělová, P.; Dřevínek, M.; Hrabák, J.; Žemličková, H. Species identification of strains belonging to genus Citrobacter using the biochemical method and MALDI-TOF mass spectrometry. Folia Microbiol. 2015, 60, 53–59. [Google Scholar] [CrossRef]
- Pavlovic, M.; Konrad, R.; Iwobi, A.N.; Sing, A.; Busch, U.; Huber, I. A dual approach employing MALDI-TOF MS and real-time PCR for fast species identification within the Enterobacter cloacae complex. FEMS Microbiol. Lett. 2012, 328, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Osman, E.A.; El-Amin, N.; Adrees, E.A.E.; Al-Hassan, L.; Mukhtar, M. Comparing conventional, biochemical and genotypic methods for accurate identification of Klebsiella pneumoniae in Sudan. Access Microbiol. 2020, 2, e000096. [Google Scholar] [CrossRef]
- Guo, L.; Ye, L.; Zhao, Q.; Ma, Y.; Yang, J.; Luo, Y. Comparative study of MALDI-TOF MS and VITEK 2 in bacteria identification. J. Thorac. Dis. 2014, 6, 534–538. [Google Scholar] [CrossRef]
- Dhiman, N.; Hall, L.; Wohlfiel, S.L.; Buckwalter, S.P.; Wengenack, N.L. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 2011, 49, 1614–1616. [Google Scholar] [CrossRef] [Green Version]
- ISO 6887-1:2017; Microbiology of the Food Chain—Preparation of Test Samples, Initial Suspension and Decimal Dilutions for Microbiological Examination—Part 1: General Rules for the Preparation of the Initial Suspension and Decimal Dilutions (ISO 6887-1:2017). International Organization for Standardization: Geneva, Switzerland, 2017.
- Przemieniecki, S.W.; Kurowski, T.P.; Kotlarz, K.; Krawczyk, K.; Damszel, M.; Karwowska, A. Plant growth promoting properties of Serratia fonticola ART-8 and Pseudomonas putida ART-9 and their effect on the growth of spring wheat (Triticum aestivum L.). Environ. Biotechnol. 2016, 12, 35–39. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saffert, R.T.; Cunningham, S.A.; Ihde, S.M.; Monson Jobe, K.E.; Mandrekar, J.; Patel, R. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometer to BD phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol. 2011, 49, 887–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graphical Abstract. Available online: BioRender.com (accessed on 22 February 2022).
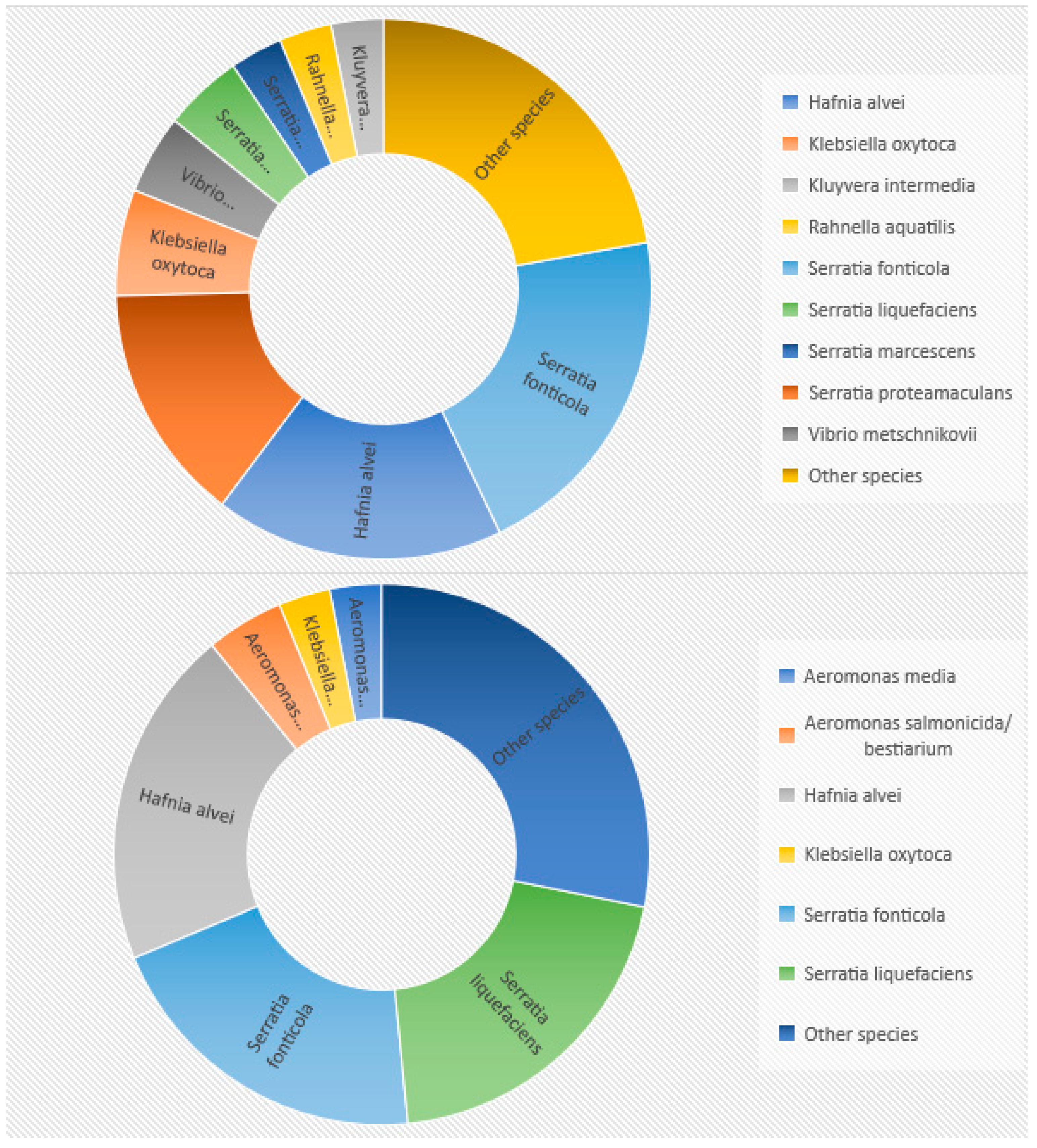
| No. | Strain | Source | MALDI-TOF | EnteroTest 24N | ||
|---|---|---|---|---|---|---|
| ID | Confidence Level | ID | Confidence Level | |||
| 1 | Escherichia coli (ATCC® 8739™) | feces | Escherichia coli | 99.8% | Escherichia coli | excellent |
| 2 | Escherichia coli (ATCC® 25922™) | clinical isolate | Escherichia coli | 99.8% | Escherichia coli | excellent |
| 3 | Hafnia alvei (ATCC® 51815™) | milk | Hafnia alvei | 99.8% | Hafnia alvei | good |
| 4 | Serratia marcescens subsp. marcescens (ATCC® 13880™) | pond water | Serratia marcescens | 99.8% | Serratia marcescens | very good |
| 5 | Enterobacter cloacae subsp. cloacae (ATCC® BAA-1143™) | ND | Enterobacter cloacae | 99.8% | Enterobacter cloacae subsp. cloacae | very good |
| 6 | Klebsiella pneumoniae subsp. pneumoniae (ATCC® 700603™) | clinical isolate | Klebsiella pneumoniae | 99.8% | Klebsiella oxytoca | genus |
| 7 | Salmonella enterica subsp. arizonae (ATCC® 13314™) | ND | Salmonella enterica subsp. arizonae | 99.8% | Salmonella enterica subsp. arizonae | species |
| 8 | Salmonella enterica subsp. enterica (ATCC® 14028™) | tissue | Salmonella enterica subsp. enterica | 99.8% | Salmonella serovar Enteritidis | genus |
| 9 | Salmonella enterica subsp. enterica (ATCC® BAA-664™) | ND | Salmonella enterica subsp. enterica | 99.8% | Salmonella serovar Enteritidis | excellent |
| 10 | Salmonella enterica subsp. enterica (ATCC® 7001™) | ND | Salmonella enterica subsp. enterica | 99.8% | Salmonella enterica subsp. arizonae | species |
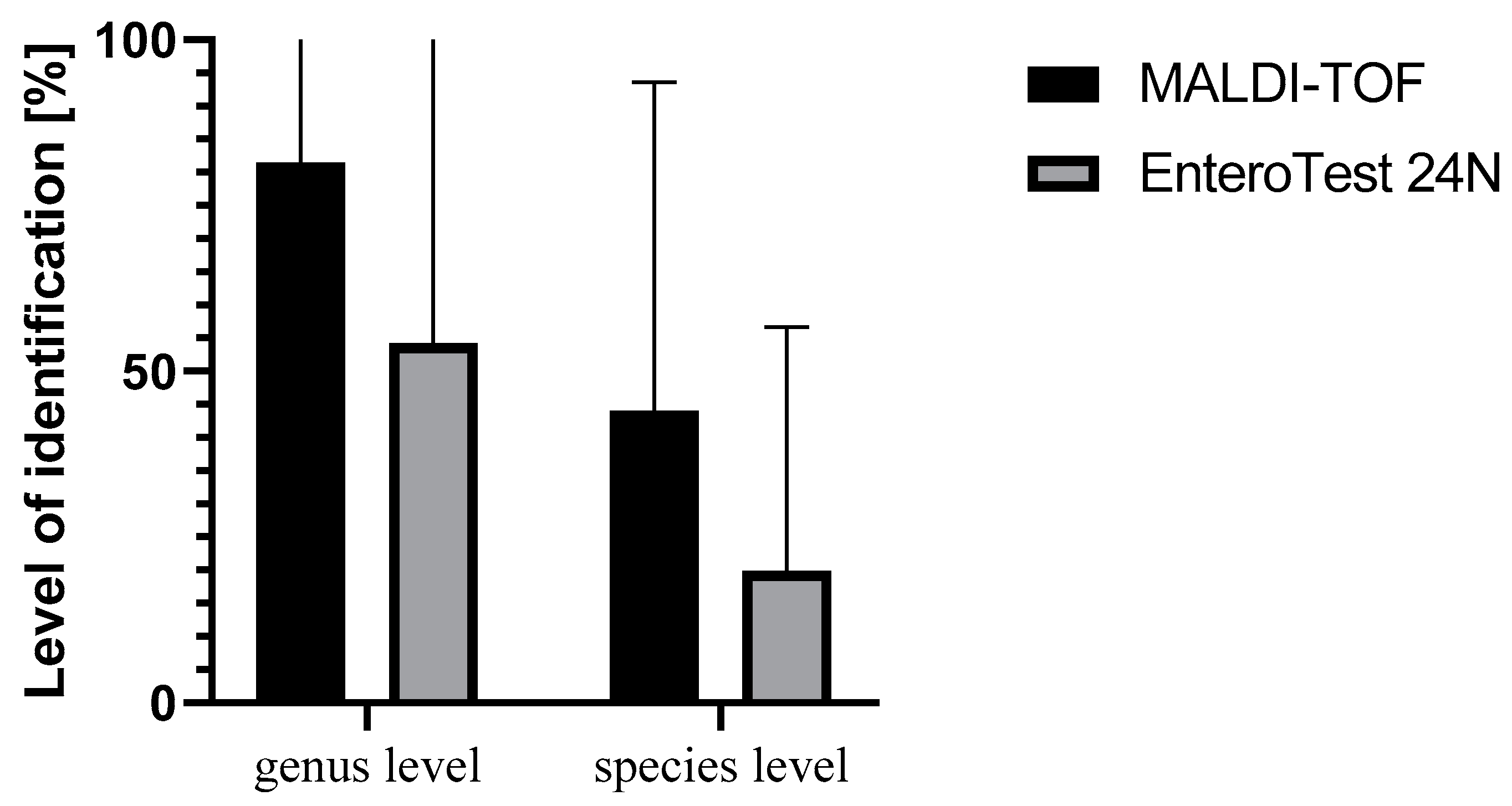
| Method | SE (%) | SP (%) | PPV (%) | NPV (%) | AC (%) | PLR | NLR |
|---|---|---|---|---|---|---|---|
| MALDI-TOF MS | 83.33 | 30.76 | 68.96 | 72.72 | 69.56 | 1.20 | 0.54 |
| EnteroTest 24N | 100 | 0 | 0 | 0 | 37.68 | 1 | - |
| Family: | Genus: | Species (Identified by 16S RNA Sequencing) | No. of Isolates | MALDI-TOF Identification (%) | EnteroTest 24N (%) | ||
|---|---|---|---|---|---|---|---|
| Genus | Species | Genus | Species | ||||
| Aeromonadaceae | Aeromonas | A. salmonicida | 4 | 75.00 | 75.00 | 0.00 | 0.00 |
| A. rivipollensis | 1 | 100.00 | 0.00 | 0.00 | 0.00 | ||
| A. hydrophila | 1 | 100.00 | 0.00 | 0.00 | 0.00 | ||
| Enterobcteriaceae | Buttiauxella | B. agrestis | 1 | 100.00 | 100.00 | 0.00 | 0.00 |
| Citrobacter | C. koseri | 1 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Enterobacter | E. cloacae | 1 | 100.00 | 100.00 | 100.00 | 100.00 | |
| E. kobei | 2 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| E. ludwigii | 2 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Klebsiella | K. oxytoca | 2 | 100.00 | 100.00 | 100.00 | 50.00 | |
| Klebsiella | K. variicola | 1 | 100.00 | 0.00 | 100.00 | 0.00 | |
| Klebsiella | K. pneumoniae | 1 | 100.00 | 0.00 | 100.00 | 0.00 | |
| Leclercia | L. adecarboxylata | 1 | 100.00 | 0.00 | 0.00 | 0.00 | |
| Lelliottia | L. nimipressuralis | 2 | 0.00 | 0.00 | 0.00 | 0.00 | |
| L. amnigena | 1 | 100.00 | 100.00 | 0.00 | 0.00 | ||
| Raoultella | R. ornithinolytica | 1 | 100.00 | 100.00 | 0.00 | 0.00 | |
| Erwiniaceae | Pantoea | P. agglomerans | 1 | 100.00 | 100.00 | 0.00 | 0.00 |
| Hafniaceae | Hafnia | H. alvei | 13 | 92.86 | 92.86 | 78.57 | 78.57 |
| Yersiniaceae | Rahnella | R. aquatilis | 1 | 100.00 | 100.00 | 100.00 | 100.00 |
| Serratia | S. fonticola | 13 | 92.30 | 92.30 | 92.30 | 92.30 | |
| S. proteamaculans | 1 | 100.00 | 0.00 | 100.00 | 0.00 | ||
| S. liquefaciens | 10 | 100.00 | 100.00 | 100.00 | 20.20 | ||
| Yersinia | Y. enterocolitica | 1 | 100.00 | 100.00 | 100.00 | 0.00 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakrzewski, A.J.; Zarzecka, U.; Chajęcka-Wierzchowska, W.; Zadernowska, A. A Comparison of Methods for Identifying Enterobacterales Isolates from Fish and Prawns. Pathogens 2022, 11, 410. https://doi.org/10.3390/pathogens11040410
Zakrzewski AJ, Zarzecka U, Chajęcka-Wierzchowska W, Zadernowska A. A Comparison of Methods for Identifying Enterobacterales Isolates from Fish and Prawns. Pathogens. 2022; 11(4):410. https://doi.org/10.3390/pathogens11040410
Chicago/Turabian StyleZakrzewski, Arkadiusz Józef, Urszula Zarzecka, Wioleta Chajęcka-Wierzchowska, and Anna Zadernowska. 2022. "A Comparison of Methods for Identifying Enterobacterales Isolates from Fish and Prawns" Pathogens 11, no. 4: 410. https://doi.org/10.3390/pathogens11040410








