Dual-Antigen Subunit Vaccine Nanoparticles for Scrub Typhus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of Recombinant Antigens
2.2. Rabbit Polyclonal Antibodies
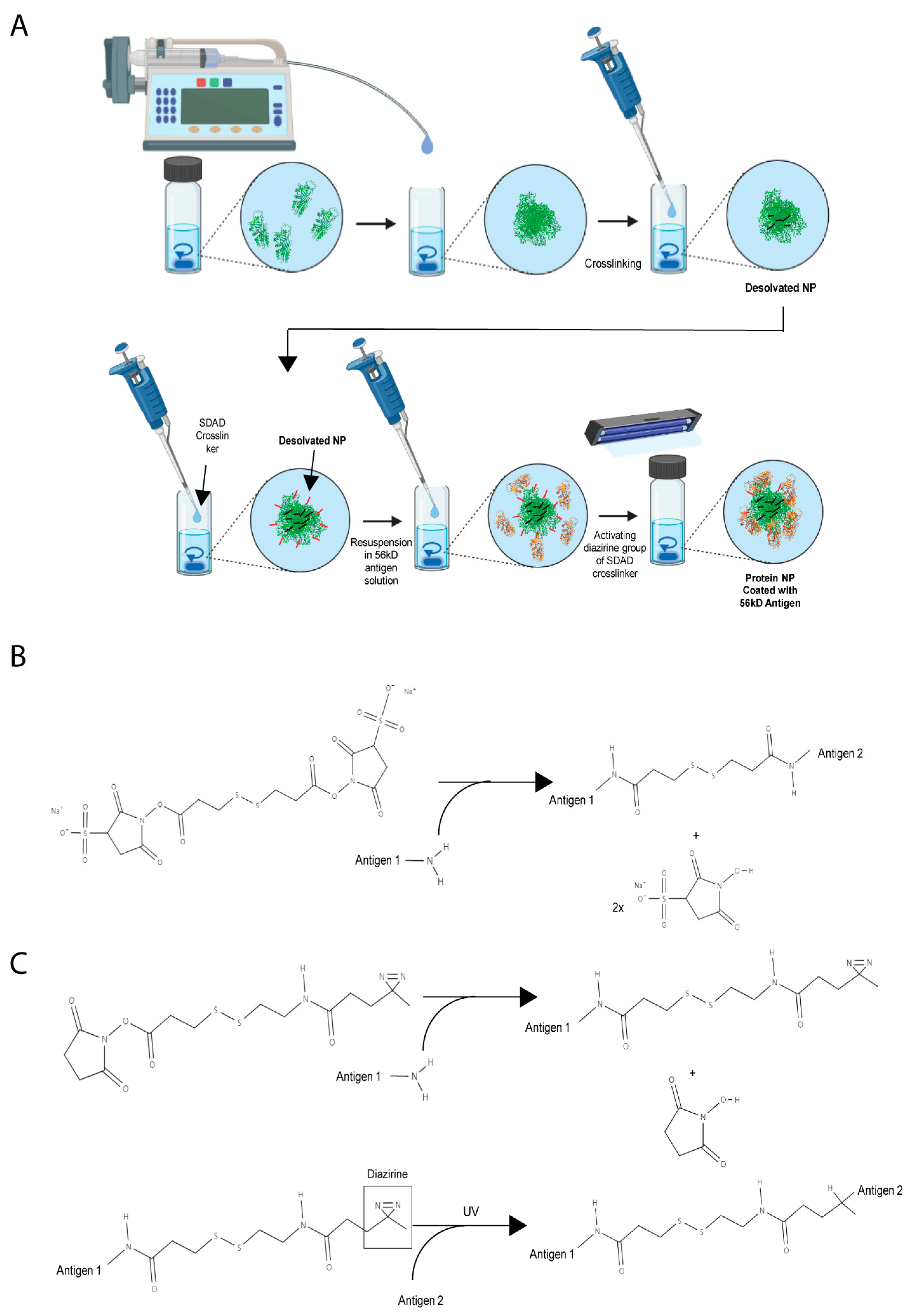
2.3. Synthesis of Orientia Subunit Vaccine Nanoparticles
2.4. Characterization of Nanoparticles
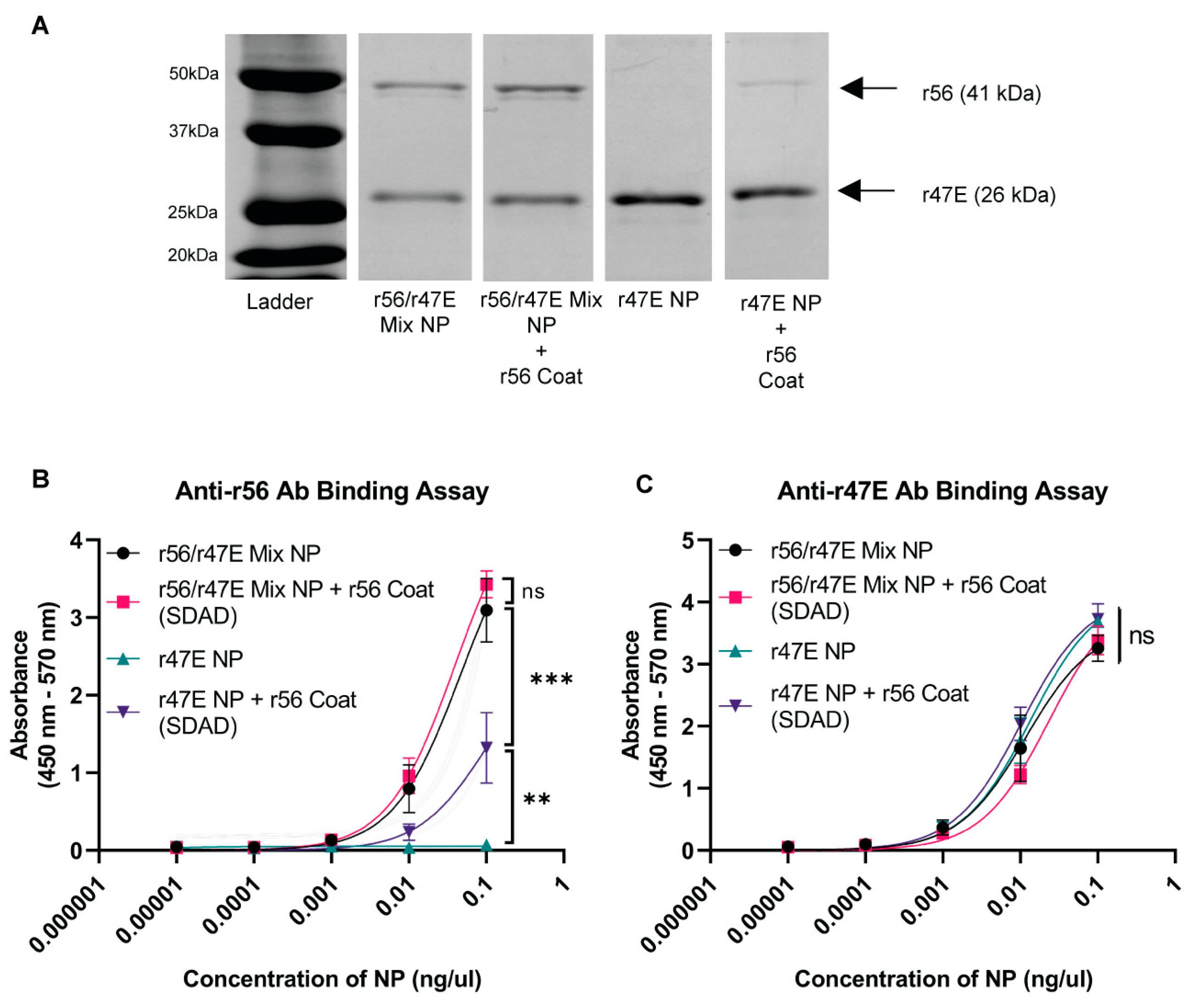
2.5. ELISA for Analyzing Antigen Composition of Nanoparticles
2.6. Immunization of Mice
2.7. ELISA for the Determination of Endpoint Titer
2.8. ELISPOT Assay for Cellular Immune Response
2.9. Statistical Analysis
3. Results and Discussion
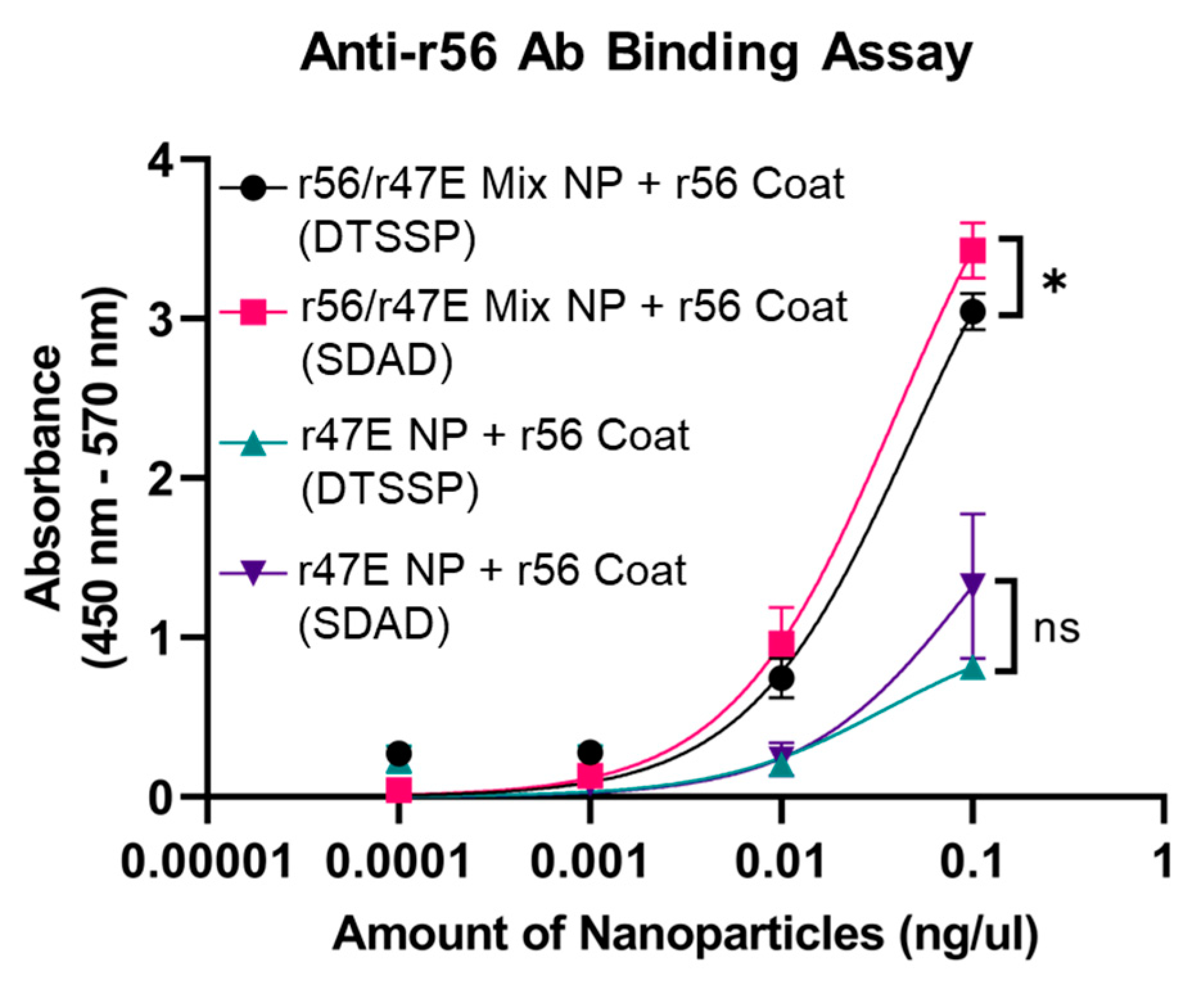
3.1. Optimization of Orientia Subunit Vaccine Nanoparticle Synthesis
3.2. In Vivo Assessment of Humoral Immunity of Orientia Subunit Vaccine Nanoparticles
3.3. In Vivo Assessment of Cellular Immunity of Orientia Subunit Vaccine Nanoparticles
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, J.; Richards, A.L. Scrub Typhus: No Longer Restricted to the Tsutsugamushi Triangle. Trop. Med. Infect. Dis. 2018, 3, 11. [Google Scholar] [CrossRef]
- Chakraborty, S.; Sarma, N. Scrub Typhus: An Emerging Threat. Indian J. Dermatol. 2017, 62, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Elliott, I.; Pearson, I.; Dahal, P.; Thomas, N.V.; Roberts, T.; Newton, P.N. Scrub typhus ecology: A systematic review of Orientia in vectors and hosts. Parasites Vectors 2019, 12, 513. [Google Scholar] [CrossRef] [PubMed]
- Paris, D.H.; Shelite, T.R.; Day, N.P.; Walker, D.H. Unresolved Problems Related to Scrub Typhus: A Seriously Neglected Life-Threatening Disease. Am. J. Trop. Med. Hyg. 2013, 89, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, G.; Walker, D.H. Approaches to vaccines against Orientia tsutsugamushi. Front. Cell. Infect. Microbiol. 2013, 2, 170. [Google Scholar] [CrossRef]
- Berge, T.O.; Gauld, R.L.; Kitaoka, M. A field trial of a vaccine prepared from the volner strain of rickettsia tsutsugamushi. Am. J. Hyg. 1949, 50, 337–342. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, K.-S.; Kim, T.-Y.; Cheong, H.-S.; Ahn, B.-Y. Long-term egg-yolk adaptation of the Orientia tsutsugamushi for preparation of a formalinized immunogen. Vaccine 2006, 24, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, G.H., Jr.; Osterman, J.V. Gamma-irradiated scrub typhus immunogens: Broad-spectrum immunity with combinations of rickettsial strains. Infect. Immun. 1979, 26, 131–136. [Google Scholar] [CrossRef]
- Choi, S.; Jeong, H.J.; Hwang, K.-J.; Gill, B.; Ju, Y.R.; Lee, Y.S.; Lee, J. A Recombinant 47-kDa Outer Membrane Protein Induces an Immune Response against Orientia tsutsugamushi Strain Boryong. Am. J. Trop. Med. Hyg. 2017, 97, 30–37. [Google Scholar] [CrossRef]
- NI, Y.-S.; Chan, T.-C.; Dasch, G.A.; Chao, C.-C.; Richards, A.L.; Ching, W.-M. Protection against scrub typhus by a plasmid vaccine encoding the 56-kd outer membrane protein antigen gene. Am. J. Trop. Med. Hyg. 2005, 73, 936–941. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Jiang, J.; Chan, T.-C.; Manetz, T.S.; Chao, C.-C.; Ching, W.-M.; Richards, A.L. Scrub Typhus Vaccine Candidate Kp r56 Induces Humoral and Cellular Immune Responses in Cynomolgus Monkeys. Infect. Immun. 2005, 73, 5039–5047. [Google Scholar] [CrossRef]
- Park, S.M.; Gu, M.J.; Ju, Y.J.; Cheon, I.S.; Hwang, K.J.; Gill, B.; Shim, B.-S.; Jeong, H.-J.; Son, Y.M.; Choi, S.; et al. Intranasal Vaccination with Outer-Membrane Protein of Orientia Tsutsugamushi Induces Protective Immunity against Scrub Typhus. Immune Netw. 2021, 21, e14. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chen, M.; Qiu, L.; Wen, B.; Niu, D.; Wen, B. Induction of protective immunity against scrub typhus with a 56-kilodalton recombinant antigen fused with a 47-kilodalton antigen of orientia tsutsugamushi karp. Am. J. Trop. Med. Hyg. 2005, 72, 458–464. [Google Scholar] [CrossRef]
- Seong, S.Y.; Huh, M.S.; Jang, W.J.; Park, S.G.; Kim, J.G.; Woo, S.G.; Choi, M.S.; Kim, I.S.; Chang, W.H. Induction of homologous immune response to Rickettsia tsutsugamushi Boryong with a partial 56-kilodalton recombinant antigen fused with the maltose-binding protein MBP-Bor56. Infect. Immun. 1997, 65, 1541–1545. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B. Identification and partial characterization of Rickettsia tsutsugamushi major protein immunogens. Infect. Immun. 1985, 50, 603–609. [Google Scholar] [CrossRef]
- Oaks, E.V.; Rice, R.M.; Kelly, D.J.; Stover, C.K. Antigenic and genetic relatedness of eight Rickettsia tsutsugamushi antigens. Infect. Immun. 1989, 57, 3116–3122. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, N.; Tamura, A.; Suto, T. Immunoblotting Analysis of Anti-Rickettsial Antibodies Produced in Patients of Tsutsugamushi Disease. Microbiol. Immunol. 1988, 32, 1085–1092. [Google Scholar] [CrossRef]
- Tamura, A.; Ohashi, N.; Urakami, H.; Takahashi, K.; Oyanagi, M. Analysis of polypeptide composition and antigenic components of Rickettsia tsutsugamushi by polyacrylamide gel electrophoresis and immunoblotting. Infect. Immun. 1985, 48, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-W.; Zhang, Z.; Huber, E.; Mutumanje, E.; Chao, C.-C.; Ching, W.-M. Kinetics and Magnitude of Antibody Responses against the Conserved 47-Kilodalton Antigen and the Variable 56-Kilodalton Antigen in Scrub Typhus Patients. Clin. Vaccine Immunol. 2011, 18, 1021–1027. [Google Scholar] [CrossRef]
- Chen, H.W.; Zhang, Z.; Huber, E.; Chao, C.C.; Wang, H.; Dasch, G.A.; Ching, W.M. Identification of Cross-Reactive Epitopes on the Conserved 47-Kilodalton Antigen of Orientia Tsutsugamushi and Human Serine Protease. Infect. Immun. 2009, 77, 2311–2319. [Google Scholar] [CrossRef]
- Pati, R.; Shevtsov, M.; Sonawane, A. Nanoparticle Vaccines Against Infectious Diseases. Front. Immunol. 2018, 9, 2224. [Google Scholar] [CrossRef] [PubMed]
- Taki, A.; Smooker, P. Small Wonders—The Use of Nanoparticles for Delivering Antigen. Vaccines 2015, 3, 638–661. [Google Scholar] [CrossRef] [PubMed]
- Bezbaruah, R.; Chavda, V.P.; Nongrang, L.; Alom, S.; Deka, K.; Kalita, T.; Ali, F.; Bhattacharjee, B.; Vora, L. Nanoparticle-Based Delivery Systems for Vaccines. Vaccines 2022, 10, 1946. [Google Scholar] [CrossRef] [PubMed]
- Chamundeeswari, M.; Jeslin, J.; Verma, M.L. Nanocarriers for drug delivery applications. Environ. Chem. Lett. 2018, 17, 849–865. [Google Scholar] [CrossRef]
- Park, J.; Pho, T.; Champion, J.A. Chemical and biological conjugation strategies for the development of multivalent protein vaccine nanoparticles. Biopolymers 2023, 114, e23563. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Chen, Z.; Wang, Y.; Chen, C. Design and application of nanoparticles as vaccine adjuvants against human corona virus infection. J. Inorg. Biochem. 2021, 219, 111454. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Zhang, Q.; Qie, L.; Baker, G.L. The Adjuvant Effect of Emerging Nanomaterials: A Double-Edged Sword. In Interactions of Nanomaterials with Emerging Environmental Contaminants; American Chemical Society: Washington, DC, USA, 2013; pp. 3–21. [Google Scholar]
- Zhu, M.; Wang, R.; Nie, G. Applications of nanomaterials as vaccine adjuvants. Hum. Vaccines Immunother. 2014, 10, 2761–2774. [Google Scholar] [CrossRef]
- Guo, S.; Fu, D.; Utupova, A.; Sun, D.; Zhou, M.; Jin, Z.; Zhao, K. Applications of polymer-based nanoparticles in vaccine field. Nanotechnol. Rev. 2019, 8, 143–155. [Google Scholar] [CrossRef]
- Singh, R.P.; Ramarao, P. Accumulated Polymer Degradation Products as Effector Molecules in Cytotoxicity of Polymeric Nanoparticles. Toxicol. Sci. 2013, 136, 131–143. [Google Scholar] [CrossRef]
- Soenen, S.J.; Montenegro, J.M.; Montenegro, A.M.; Abdelmonem, B.B.; Manshian, S.H.; Doak, W.J.; Parak, S.C.; Smedt, D.; Braeckmans, K. The Effect of Nanoparticle Degradation on Amphiphilic Polymer-Coated Quantum Dot Toxicity: The Importance of Particle Functionality Assessment in Toxicology [Corrected]. Acta Biomater. 2014, 10, 732–741. [Google Scholar] [CrossRef]
- Voigt, N.; Henrich-Noack, P.; Kockentiedt, S.; Hintz, W.; Tomas, J.; Sabel, B.A. Toxicity of Polymeric Nanoparticles in vivo and in vitro. J. Nanopart. Res. 2014, 16, 2379. [Google Scholar] [CrossRef] [PubMed]
- Cox, F.; Khalib, K.; Conlon, N. PEG That Reaction: A Case Series of Allergy to Polyethylene Glycol. J. Clin. Pharmacol. 2021, 61, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Sellaturay, P.; Nasser, S.; Ewan, P. Polyethylene Glycol–Induced Systemic Allergic Reactions (Anaphylaxis). J. Allergy Clin. Immunol. Pract. 2020, 9, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Sellaturay, P.; Nasser, S.; Islam, S.; Gurugama, P.; Ewan, P.W. Polyethylene glycol (PEG) is a cause of anaphylaxis to the Pfizer/BioNTech mRNA COVID-19 vaccine. Clin. Exp. Allergy 2021, 51, 861–863. [Google Scholar] [CrossRef] [PubMed]
- Wylon, K.; Dölle, S.; Worm, M. Polyethylene glycol as a cause of anaphylaxis. Allergy Asthma Clin. Immunol. 2016, 12, 67. [Google Scholar] [CrossRef]
- Fu, K.; Pack, D.W.; Klibanov, A.M.; Langer, R. Visual Evidence of Acidic Environment Within Degrading Poly(lactic-co-glycolic acid) (PLGA) Microspheres. Pharm. Res. 2000, 17, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Martin, A.; Han, X.; Shirihai, O.S.; Grinstaff, M.W. Biodegradable PLGA Nanoparticles Restore Lysosomal Acidity and Protect Neural PC-12 Cells against Mitochondrial Toxicity. Ind. Eng. Chem. Res. 2019, 58, 13910–13917. [Google Scholar] [CrossRef]
- Chang, T.Z.; Champion, J.A. Protein and Peptide Nanocluster Vaccines. In Nanoparticles for Rational Vaccine Design; Gill, H.S., Compans, R.W., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 107–130. [Google Scholar]
- Park, J.; Champion, J.A. Effect of Antigen Structure in Subunit Vaccine Nanoparticles on Humoral Immune Responses. ACS Biomater. Sci. Eng. 2023, 9, 1296–1306. [Google Scholar] [CrossRef]
- Deng, L.; Chang, T.Z.; Wang, Y.; Li, S.; Wang, S.; Matsuyama, S.; Yu, G.; Compans, R.W.; Li, J.-D.; Prausnitz, M.R.; et al. Heterosubtypic influenza protection elicited by double-layered polypeptide nanoparticles in mice. Proc. Natl. Acad. Sci. USA 2018, 115, E7758–E7767. [Google Scholar] [CrossRef]
- Deng, L.; Mohan, T.; Chang, T.Z.; Gonzalez, G.X.; Wang, Y.; Kwon, Y.-M.; Kang, S.-M.; Compans, R.W.; Champion, J.A.; Wang, B.-Z. Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nat. Commun. 2018, 9, 359. [Google Scholar] [CrossRef]
- Zhu, W.; Park, J.; Pho, T.; Wei, L.; Dong, C.; Kim, J.; Ma, Y.; Champion, J.A.; Wang, B. ISCOMs/MPLA-Adjuvanted SDAD Protein Nanoparticles Induce Improved Mucosal Immune Responses and Cross-Protection in Mice. Small 2023, 19, e2301801. [Google Scholar] [CrossRef]
- Deng, L.; Kim, J.R.; Chang, T.Z.; Zhang, H.; Mohan, T.; Champion, J.A.; Wang, B.-Z. Protein nanoparticle vaccine based on flagellin carrier fused to influenza conserved epitopes confers full protection against influenza A virus challenge. Virology 2017, 509, 82–89. [Google Scholar] [CrossRef]
- Wang, L.; Chang, T.Z.; He, Y.; Kim, J.R.; Wang, S.; Mohan, T.; Berman, Z.; Tompkins, S.M.; Tripp, R.A.; Compans, R.W.; et al. Coated protein nanoclusters from influenza H7N9 HA are highly immunogenic and induce robust protective immunity. Nanomed. Nanotechnol. Biol. Med. 2016, 13, 253–262. [Google Scholar] [CrossRef]
- Ching, W.-M.; Wang, H.; Eamsila, C.; Kelly, D.J.; Dasch, G.A. Expression and Refolding of Truncated Recombinant Major Outer Membrane Protein Antigen (r56) of Orientia tsutsugamushi and Its Use in Enzyme-Linked Immunosorbent Assays. Clin. Diagn. Lab. Immunol. 1998, 5, 519–526. [Google Scholar] [CrossRef]
- Amanat, F.D.; Stadlbauer, S.; Strohmeier, T.H.O.; Nguyen, V.; Chromikova, M.; McMahon, K.; Jiang, G.A.; Arunkumar, D.; Jurczyszak, J.; Polanco, J.; et al. A Serological Assay to Detect Sars-Cov-2 Seroconversion in Humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef]
- Majji, S.; Wijayalath, W.; Shashikumar, S.; Pow-Sang, L.; Villasante, E.; Brumeanu, T.D.; Casares, S. Differential effect of HLA class-I versus class-II transgenes on human T and B cell reconstitution and function in NRG mice. Sci. Rep. 2016, 6, 28093. [Google Scholar] [CrossRef] [PubMed]
- Estrada, L.H.; Chu, S.; Champion, J.A. Protein Nanoparticles for Intracellular Delivery of Therapeutic Enzymes. J. Pharm. Sci. 2014, 103, 1863–1871. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Siggers, K.; Zhang, S.; Jiang, H.; Xu, Z.; Zernicke, R.F.; Matyas, J.; Uludağ, H. Preparation of BMP-2 Containing Bovine Serum Albumin (BSA) Nanoparticles Stabilized by Polymer Coating. Pharm. Res. 2008, 25, 2896–2909. [Google Scholar] [CrossRef] [PubMed]
- Tarhini, M.; Benlyamani, I.; Hamdani, S.; Agusti, G.; Fessi, H.; Greige-Gerges, H.; Bentaher, A.; Elaissari, A. Protein-Based Nanoparticle Preparation via Nanoprecipitation Method. Materials 2018, 11, 394. [Google Scholar] [CrossRef] [PubMed]
- Cyster, J.G. B cell follicles and antigen encounters of the third kind. Nat. Immunol. 2010, 11, 989–996. [Google Scholar] [CrossRef]
- Foged, C.; Brodin, B.; Frokjaer, S.; Sundblad, A. Particle size and surface charge affect particle uptake by human dendritic cells in an in vitro model. Int. J. Pharm. 2005, 298, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-H.; Jang, J.-H.; Cho, H.-Y.; Lee, Y.-B. Soft- and hard-lipid nanoparticles: A novel approach to lymphatic drug delivery. Arch. Pharmacal Res. 2018, 41, 797–814. [Google Scholar] [CrossRef] [PubMed]
- LeBien, T.W.; Tedder, T.F. B lymphocytes: How they develop and function. Blood 2008, 112, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Porter, C.J.H.; Trevaskis, N.L. Targeting immune cells within lymph nodes. Nat. Nanotechnol. 2020, 15, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Schudel, A.; Francis, D.M.; Thomas, S.N. Material design for lymph node drug delivery. Nat. Rev. Mater. 2019, 4, 415–428. [Google Scholar] [CrossRef]
- Linna, D.; Mukherjee, E. Janeway’s Immunobiology, Ninth Edition. Yale J. Biol. Med. 2016, 89, 424–425. [Google Scholar]
- Manolova, V.; Flace, A.; Bauer, M.; Schwarz, K.; Saudan, P.; Bachmann, M.F. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 2008, 38, 1404–1413. [Google Scholar] [CrossRef]
- Hickman, C.J.; Stover, C.K.; Joseph, S.W.; Oaks, E.V. Murine T-cell response to native and recombinant protein antigens of Rickettsia tsutsugamushi. Infect. Immun. 1993, 61, 1674–1681. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Zhang, Z.; Belinskaya, T.; Tsoras, A.N.; Chao, C.-C.; Jiang, L.; Champion, J.A. Dual-Antigen Subunit Vaccine Nanoparticles for Scrub Typhus. Pathogens 2023, 12, 1390. https://doi.org/10.3390/pathogens12121390
Park J, Zhang Z, Belinskaya T, Tsoras AN, Chao C-C, Jiang L, Champion JA. Dual-Antigen Subunit Vaccine Nanoparticles for Scrub Typhus. Pathogens. 2023; 12(12):1390. https://doi.org/10.3390/pathogens12121390
Chicago/Turabian StylePark, Jaeyoung, Zhiwen Zhang, Tatyana Belinskaya, Alexandra N. Tsoras, Chien-Chung Chao, Le Jiang, and Julie A. Champion. 2023. "Dual-Antigen Subunit Vaccine Nanoparticles for Scrub Typhus" Pathogens 12, no. 12: 1390. https://doi.org/10.3390/pathogens12121390




