Concatenated ScaA and TSA56 Surface Antigen Sequences Reflect Genome-Scale Phylogeny of Orientia tsutsugamushi: An Analysis Including Two Genomes from Taiwan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation, Purification, and Genomic DNA Isolation
2.2. Quantitative PCR
2.3. Whole Genome Sequencing, Assembly, and Annotation
2.4. Phylogenetic Analysis
3. Results
3.1. Complete Genomes of O. tsutsugamushi Strains TW-1 and TW-22
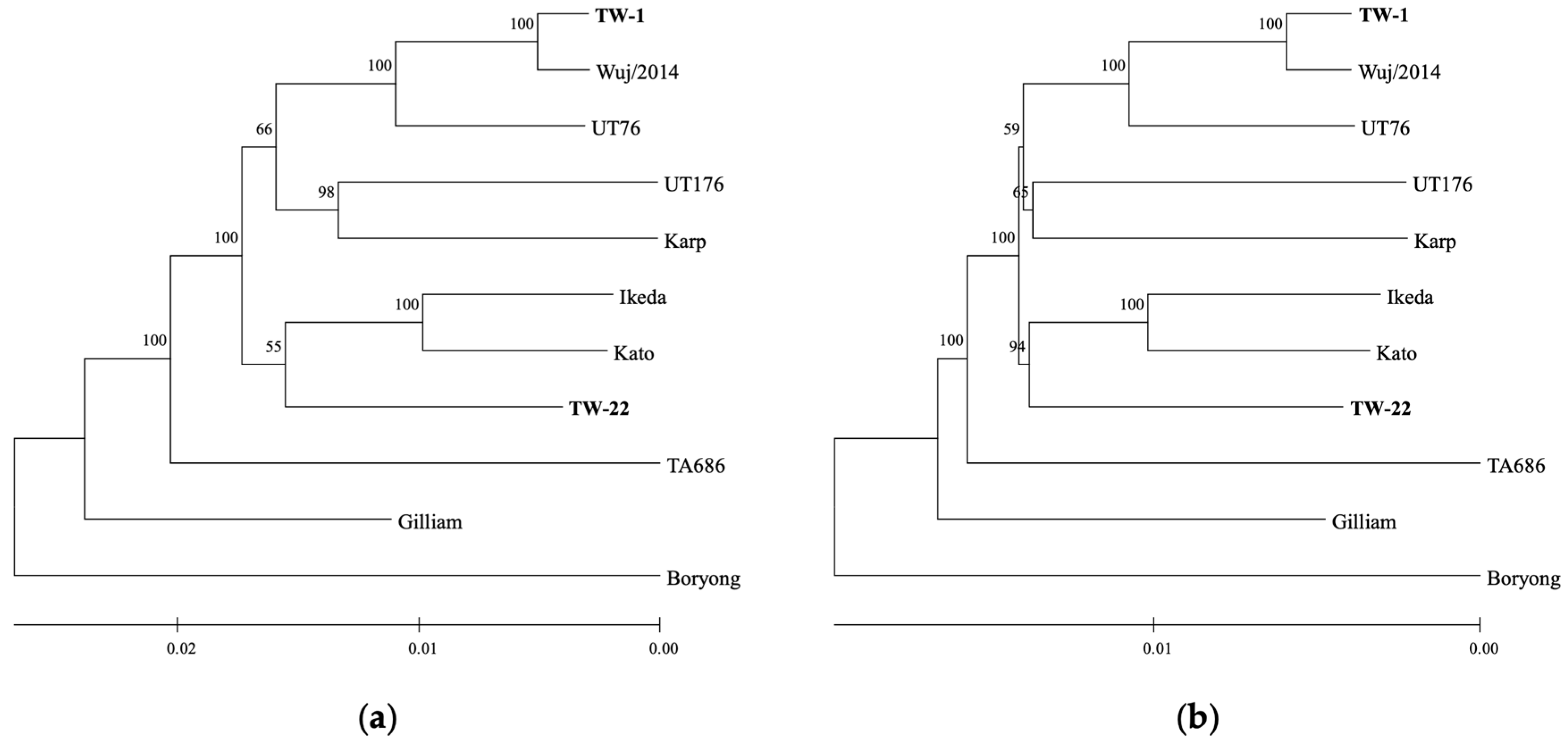
3.2. Core Genome Phylogeny of O. tsutsugamushi
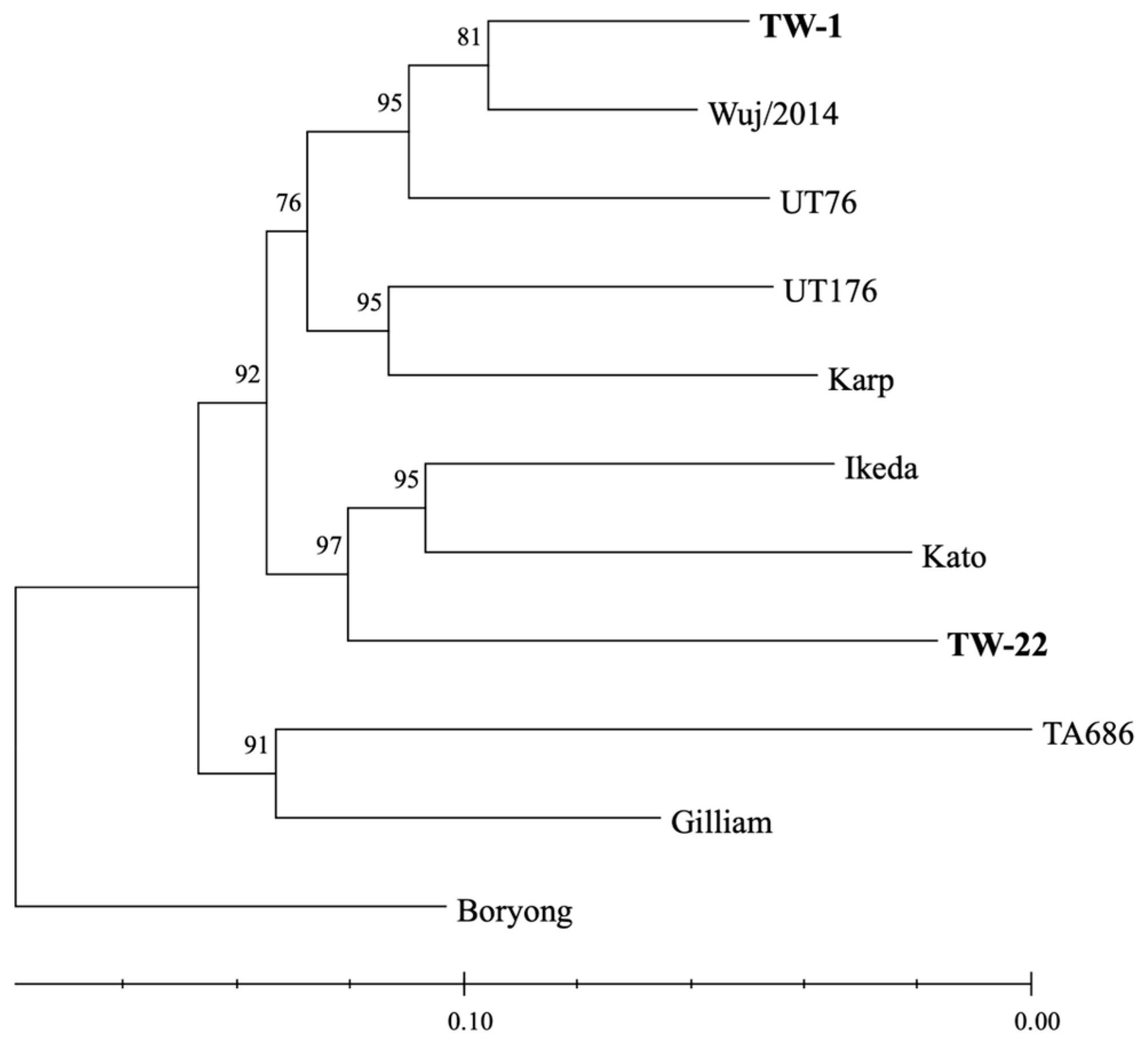
3.3. Surface Antigen-Based Phylogeny of O. tsutsugamushi
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Traub, R.; Wisseman, C.L., Jr. Ecological considerations in scrub typhus. 2. Vector species. Bull. World Health Organ. 1968, 39, 219–230. [Google Scholar] [PubMed]
- Paris, D.H.; Shelite, T.R.; Day, N.P.; Walker, D.H. Unresolved problems related to scrub typhus: A seriously neglected life-threatening disease. Am. J. Trop. Med. Hyg. 2013, 89, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Richards, A.L. Scrub typhus: No longer restricted to the Tsutsugamushi Triangle. Trop. Med. Infect. Dis. 2018, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Minahan, N.T.; Chao, C.C.; Tsai, K.H. The re-emergence and emergence of vector-borne rickettsioses in Taiwan. Trop. Med. Infect. Dis. 2018, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- El Sayed, I.; Liu, Q.; Wee, I.; Hine, P. Antibiotics for treating scrub typhus. Cochrane Database Syst. Rev. 2018, 9, CD002150. [Google Scholar] [CrossRef] [PubMed]
- Minahan, N.T.; Davis, R.J.; Graves, S.R.; Tsai, K.H. Australian case of scrub typhus contracted while hiking in northern Taiwan. J. Formos. Med. Assoc. 2020, 119, 662–663. [Google Scholar] [CrossRef] [PubMed]
- Devasagayam, E.; Dayanand, D.; Kundu, D.; Kamath, M.S.; Kirubakaran, R.; Varghese, G.M. The burden of scrub typhus in India: A systematic review. PLoS Negl. Trop. Dis. 2021, 15, e0009619. [Google Scholar] [CrossRef] [PubMed]
- Bonell, A.; Lubell, Y.; Newton, P.N.; Crump, J.A.; Paris, D.H. Estimating the burden of scrub typhus: A systematic review. PLoS Negl. Trop. Dis. 2017, 11, e0005838. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, G.; Walker, D.H. Approaches to vaccines against Orientia tsutsugamushi. Front. Cell Infect. Microbiol. 2012, 2, 170. [Google Scholar] [CrossRef]
- Stover, C.K.; Marana, D.P.; Carter, J.M.; Roe, B.A.; Mardis, E.; Oaks, E.V. The 56-kilodalton major protein antigen of Rickettsia tsutsugamushi: Molecular cloning and sequence analysis of the sta56 gene and precise identification of a strain-specific epitope. Infect. Immun. 1990, 58, 2076–2084. [Google Scholar] [CrossRef]
- Cho, B.A.; Cho, N.H.; Seong, S.Y.; Choi, M.S.; Kim, I.S. Intracellular invasion by Orientia tsutsugamushi is mediated by integrin signaling and actin cytoskeleton rearrangements. Infect. Immun. 2010, 78, 1915–1923. [Google Scholar] [CrossRef] [PubMed]
- Rights, F.L.; Smadel, J.E. Studies on scrub typhus (tsutsugamushi disease). III. Heterogenicity of strains of R. tsutsugamushi as demonstrated by cross-vaccination studies. J. Exp. Med. 1948, 87, 339–351. [Google Scholar] [CrossRef]
- Bozeman, F.M.; Elisberg, B.L. Serological diagnosis of scrub typhus by indirect immunofluorescence. Proc. Soc. Exp. Biol. Med. 1963, 112, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Blacksell, S.D.; Bryant, N.J.; Paris, D.H.; Doust, J.A.; Sakoda, Y.; Day, N.P.J. Scrub typhus serologic testing with the indirect immunofluorescence method as a diagnostic gold standard: A lack of consensus leads to a lot of confusion. Clin. Infect. Dis. 2007, 44, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.J.; Fuerst, P.A.; Richards, A.L. Origins, importance and genetic stability of the prototype strains Gilliam, Karp and Kato of Orientia tsutsugamushi. Trop. Med. Infect. Dis. 2019, 4, 75. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Yoshida, Y.; Osono, M.; Ohashi, N.; Oyanagi, M.; Urakami, H.; Tamura, A.; Nogami, S.; Tanaka, H.; Kawamura, A., Jr. Production and characterization of monoclonal strain-specific antibodies against prototype strains of Rickettsia tsutsugamushi. Microbiol. Immunol. 1986, 30, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Kawabata, N.; Tamura, A.; Urakami, H.; Ohashi, N.; Murata, M.; Yoshida, Y.; Kawamura, A., Jr. Immunological properties of Rickettsia tsutsugamushi, Kawasaki strain, isolated from a patient in Kyushu. Microbiol. Immunol. 1986, 30, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, N.; Tamura, A.; Sakurai, H.; Yamamoto, S. Characterization of a new antigenic type, Kuroki, of Rickettsia tsutsugamushi isolated from a patient in Japan. J. Clin. Microbiol. 1990, 28, 2111–2113. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.H.; Kang, J.S.; Lee, W.K.; Choi, M.S.; Lee, J.H. Serological classification by monoclonal antibodies of Rickettsia tsutsugamushi isolated in Korea. J. Clin. Microbiol. 1990, 28, 685–688. [Google Scholar] [CrossRef]
- Ohashi, N.; Koyama, Y.; Urakami, H.; Fukuhara, M.; Tamura, A.; Kawamori, F.; Yamamoto, S.; Kasuya, S.; Yoshimura, K. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol. Immunol. 1996, 40, 627–638. [Google Scholar] [CrossRef]
- Enatsu, T.; Urakami, H.; Tamura, A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol. Lett. 1999, 180, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Blacksell, S.D.; Luksameetanasan, R.; Kalambaheti, T.; Aukkanit, N.; Paris, D.H.; McGready, R.; Nosten, F.; Peacock, S.J.; Day, N.P.J. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol. Med. Microbiol. 2008, 52, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.Y.; Tsai, K.H.; Yu, S.K.; Cheng, C.H.; Yang, J.S.; Su, C.L.; Hu, H.C.; Wang, H.C.; Huang, J.H.; Shu, P.Y. Phylogenetic analysis of 56-kDa type-specific antigen gene of Orientia tsutsugamushi isolates in Taiwan. Am. J. Trop. Med. Hyg. 2010, 83, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.R.; Tsai, H.P.; Tsui, P.Y.; Weng, M.H.; Kuo, M.D.; Lin, H.C.; Chen, K.C.; Ji, D.D.; Chu, D.M.; Liu, W.T. Genetic typing, based on the 56-kilodalton type-specific antigen gene, of Orientia tsutsugamushi strains isolated from chiggers collected from wild-caught rodents in Taiwan. Appl. Environ. Microbiol. 2011, 77, 3398–3405. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.F.; Peng, S.H.; Tsai, K.H.; Yang, C.F.; Chang, M.C.; Hsueh, Y.L.; Su, C.L.; Wang, R.Y.; Shu, P.Y.; Yang, S.L. Molecular epidemiology of scrub typhus in Taiwan during 2006–2016. PLoS Negl. Trop. Dis. 2022, 16, e0010369. [Google Scholar] [CrossRef]
- Mahajan, S.K.; Rolain, J.M.; Kashyap, R.; Bakshi, D.; Sharma, V.; Prasher, B.S.; Pal, L.S.; Raoult, D. Scrub typhus in Himalayas. Emerg. Infect. Dis. 2006, 12, 1590–1592. [Google Scholar] [CrossRef] [PubMed]
- Trung, N.V.; Hoi, L.T.; Cuong, D.D.; Ha, D.T.; Hoa, T.M.; Lien, V.N.; Hoa, N.T.; Hoa, L.N.M.; Huong, D.T.; Bich, V.T.N.; et al. Analysis of the 56-kDa type specific antigen gene of Orientia tsutsugamushi from northern Vietnam. PLoS ONE 2019, 14, e0221588. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Ha, N.Y.; Min, C.K.; Kim, H.I.; Yen, N.T.H.; Lee, K.H.; Oh, I.; Kang, J.S.; Choi, M.S.; Kim, I.S.; et al. Diversification of Orientia tsutsugamushi genotypes by intragenic recombination and their potential expansion in endemic areas. PLoS Negl. Trop. Dis. 2017, 11, e0005408. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.H.; Kim, H.R.; Lee, J.H.; Kim, S.Y.; Kim, J.; Cha, S.; Kim, S.Y.; Darby, A.C.; Fuxelius, H.H.; Yin, J.; et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. USA 2007, 104, 7981–7986. [Google Scholar] [CrossRef]
- Nakayama, K.; Yamashita, A.; Kurokawa, K.; Morimoto, T.; Ogawa, M.; Fukuhara, M.; Urakami, H.; Ohnishi, M.; Uchiyama, I.; Ogura, Y.; et al. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008, 15, 185–199. [Google Scholar] [CrossRef]
- Batty, E.M.; Chaemchuen, S.; Blacksell, S.; Richards, A.L.; Paris, D.; Bowden, R.; Chan, C.; Lachumanan, R.; Day, N.; Donnelly, P.; et al. Long-read whole genome sequencing and comparative analysis of six strains of the human pathogen Orientia tsutsugamushi. PLoS Negl. Trop. Dis. 2018, 12, e0006566. [Google Scholar] [CrossRef] [PubMed]
- Fleshman, A.; Mullins, K.; Sahl, J.; Hepp, C.; Nieto, N.; Wiggins, K.; Hornstra, H.; Kelly, D.; Chan, T.C.; Phetsouvanh, R.; et al. Comparative pan-genomic analyses of Orientia tsutsugamushi reveal an exceptional model of bacterial evolution driving genomic diversity. Microb. Genom. 2018, 4, e000199. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; O’Neill, K.; Li, W.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef] [PubMed]
- Giengkam, S.; Kullapanich, C.; Wongsantichon, J.; Adcox, H.E.; Gillespie, J.J.; Salje, J. Orientia tsutsugamushi: Comprehensive analysis of the mobilome of a highly fragmented and repetitive genome reveals the capacity for ongoing lateral gene transfer in an obligate intracellular bacterium. mSphere 2023, 8, e00268-23. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, N.; Fukuhara, M.; Shimada, M.; Tamura, A. Phylogenetic position of Rickettsia tsutsugamushi and the relationship among its antigenic variants by analyses of 16S rRNA gene sequences. FEMS Microbiol. Lett. 1995, 125, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Tamura, A.; Ohashi, N.; Urakami, H.; Miyamura, S. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int. J. Syst. Bacteriol. 1995, 45, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Salje, J. Orientia and Rickettsia: Different flowers from the same garden. Curr. Opin. Microbiol. 2023, 74, 102318. [Google Scholar] [CrossRef] [PubMed]
- Nicolay, T.; Vanderleyden, J.; Spaepen, S. Autotransporter-based cell surface display in Gram-negative bacteria. Crit. Rev. Microbiol. 2015, 41, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Diop, A.; El Karkouri, K.; Raoult, D.; Fournier, P.E. Genome sequence-based criteria for demarcation and definition of species in the genus Rickettsia. Int. J. Syst. Evol. Microbiol. 2020, 70, 1738–1750. [Google Scholar] [CrossRef]
- Fournier, P.E.; Dumler, J.S.; Greub, G.; Zhang, J.; Wu, Y.; Raoult, D. Gene sequence-based criteria for identification of new Rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J. Clin. Microbiol. 2003, 41, 5456–5465. [Google Scholar] [CrossRef]
- Koralur, M.C.; Ramaiah, A.; Dasch, G.A. Detection and distribution of Sca autotransporter protein antigens in diverse isolates of Orientia tsutsugamushi. PLoS Negl. Trop. Dis. 2018, 12, e0006784. [Google Scholar] [CrossRef]
- Nguyen, Y.T.H.; Kim, C.; Kim, H.I.; Kim, Y.; Lee, S.E.; Chang, S.; Ha, N.Y.; Cho, N.H. An alternative splicing variant of the Mixed-Lineage Leukemia 5 protein is a cellular adhesion receptor for ScaA of Orientia tsutsugamushi. mBio 2023, 14, e0154322. [Google Scholar] [CrossRef]
- Ha, N.Y.; Cho, N.H.; Kim, Y.S.; Choi, M.S.; Kim, I.S. An autotransporter protein from Orientia tsutsugamushi mediates adherence to nonphagocytic host cells. Infect. Immun. 2011, 79, 1718–1727. [Google Scholar] [CrossRef]
- Ha, N.Y.; Sharma, P.; Kim, G.; Kim, Y.; Min, C.K.; Choi, M.S.; Kim, I.S.; Cho, N.H. Immunization with an autotransporter protein of Orientia tsutsugamushi provides protective immunity against scrub typhus. PLoS Negl. Trop. Dis. 2015, 9, e0003585. [Google Scholar] [CrossRef]
- Giengkam, S.; Blakes, A.; Utsahajit, P.; Chaemchuen, S.; Atwal, S.; Blacksell, S.D.; Paris, D.H.; Day, N.P.J.; Salje, J. Improved quantification, propagation, purification and storage of the obligate intracellular human pathogen Orientia tsutsugamushi. PLoS Negl. Trop. Dis. 2015, 9, e0004009. [Google Scholar] [CrossRef]
- Sunyakumthorn, P.; Paris, D.H.; Chan, T.C.; Jones, M.; Luce-Fedrow, A.; Chattopadhyay, S.; Jiang, J.; Anantatat, T.; Turner, G.D.H.; Day, N.P.J.; et al. An intradermal inoculation model of scrub typhus in Swiss CD-1 mice demonstrates more rapid dissemination of virulent strains of Orientia tsutsugamushi. PLoS ONE 2013, 8, e54570. [Google Scholar] [CrossRef]
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. 2023. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 25 April 2023).
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Anders, S.; Lawrence, M.; Aboyoun, P.; Pagès, H.; Gentleman, R. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 2009, 25, 2607–2608. [Google Scholar] [CrossRef] [PubMed]
- Pagès, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: Efficient manipulation of biological strings. R Package Version 2023, 2, 10–18129. [Google Scholar] [CrossRef]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2020, 48, D84–D86. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Avram, O.; Rapoport, D.; Portugez, S.; Pupko, T. M1CR0B1AL1Z3R—A user-friendly web server for the analysis of large-scale microbial genomics data. Nucleic Acids Res. 2019, 47, W88–W92. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Smith, M.R. Quartet: Comparison of Phylogenetic Trees Using Quartet and Split Measures; CERN: Geneva, Switzerland, 2022. [Google Scholar] [CrossRef]
- Asher, R.J.; Smith, M.R. Phylogenetic signal and bias in paleontology. Syst. Biol. 2022, 71, 986–1008. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Kaur, S.J.; Rahman, M.S.; Rennoll-Bankert, K.; Sears, K.T.; Beier-Sexton, M.; Azad, A.F. Secretome of obligate intracellular Rickettsia. FEMS Microbiol. Rev. 2015, 39, 47–80. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.I.; Ha, N.Y.; Kim, G.; Min, C.K.; Kim, Y.; Yen, N.T.H.; Choi, M.S.; Cho, N.H. Immunization with a recombinant antigen composed of conserved blocks from TSA56 provides broad genotype protection against scrub typhus. Emerg. Microbes Infect. 2019, 8, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Hickman, C.J.; Stover, C.K.; Joseph, S.W.; Oaks, E.V. Murine T-cell response to native and recombinant protein antigens of Rickettsia tsutsugamushi. Infect. Immun. 1993, 61, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Seong, S.Y.; Kim, M.K.; Lee, S.M.; Odgerel, Z.; Choi, M.S.; Han, T.H.; Kim, I.S.; Kang, J.S.; Lim, B.U. Neutralization epitopes on the antigenic domain II of the Orientia tsutsugamushi 56-kDa protein revealed by monoclonal antibodies. Vaccine 2000, 19, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.L.; Tsai, K.H.; Chen, H.F.; Luo, J.Y.; Shu, P.Y. Evaluation of enzyme-linked immunosorbent assay using recombinant 56-kDa type-specific antigens derived from multiple Orientia tsutsugamushi strains for detection of scrub typhus infection. Am. J. Trop. Med. Hyg. 2019, 100, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.W.; Zhang, Z.; Huber, E.; Mutumanje, E.; Chao, C.C.; Ching, W.M. Kinetics and magnitude of antibody responses against the conserved 47-kilodalton antigen and the variable 56-kilodalton antigen in scrub typhus patients. Clin. Vaccine Immunol. 2011, 18, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Blacksell, S.D.; Tanganuchitcharnchai, A.; Nawtaisong, P.; Kantipong, P.; Laongnualpanich, A.; Day, N.P.; Paris, D.H. Diagnostic accuracy of the InBios scrub typhus detect enzyme-linked immunoassay for the detection of IgM antibodies in northern Thailand. Clin. Vaccine Immunol. 2016, 23, 148–154. [Google Scholar] [CrossRef]
- Ha, N.Y.; Kim, Y.; Min, C.K.; Kim, H.I.; Yen, N.T.H.; Choi, M.S.; Kang, J.S.; Kim, Y.S.; Cho, N.H. Longevity of antibody and T-cell responses against outer membrane antigens of Orientia tsutsugamushi in scrub typhus patients. Emerg. Microbes Infect. 2017, 6, e116. [Google Scholar] [CrossRef] [PubMed]
- Hickman, C.J.; Stover, C.K.; Joseph, S.W.; Oaks, E.V. Molecular cloning and sequence analysis of a Rickettsia tsutsugamushi 22 kDa antigen containing B- and T-cell epitopes. Microb. Pathog. 1991, 11, 19–31. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, M.K.; Kang, J.S. Involvement of lipid rafts in the budding-like exit of Orientia tsutsugamushi. Microb. Pathog. 2013, 63, 37–43. [Google Scholar] [CrossRef]
- Kim, C.G.; Kim, W.K.; Kim, N.; Pyung, Y.J.; Park, D.J.; Lee, J.C.; Cho, C.S.; Chu, H.; Yun, C.H. Intranasal immunization with nanoparticles containing an Orientia tsutsugamushi protein vaccine candidate and a polysorbitol transporter adjuvant enhances both humoral and cellular immune responses. Immune Netw. 2023, 23, e47. [Google Scholar] [CrossRef]
- Park, J.; Zhang, Z.; Belinskaya, T.; Tsoras, A.N.; Chao, C.C.; Jiang, L.; Champion, J.A. Dual-antigen subunit vaccine nanoparticles for scrub typhus. Pathogens 2023, 12, 1390. [Google Scholar] [CrossRef]
- Boyoglu-Barnum, S.; Ellis, D.; Gillespie, R.A.; Hutchinson, G.B.; Park, Y.J.; Moin, S.M.; Acton, O.J.; Ravichandran, R.; Murphy, M.; Pettie, D.; et al. Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 2021, 592, 623–628. [Google Scholar] [CrossRef]
- Nguyen, Y.T.H.; Kim, C.; Kim, Y.; Jeon, K.; Kim, H.I.; Ha, N.Y.; Cho, N.H. The Orientia tsutsugamushi ScaB autotransporter protein is required for adhesion and invasion of mammalian cells. Front. Microbiol. 2021, 12, 626298. [Google Scholar] [CrossRef]
- Mika-Gospodorz, B.; Giengkam, S.; Westermann, A.J.; Wongsantichon, J.; Kion-Crosby, W.; Chuenklin, S.; Wang, L.C.; Sunyakumthorn, P.; Sobota, R.M.; Subbian, S.; et al. Dual RNA-seq of Orientia tsutsugamushi informs on host-pathogen interactions for this neglected intracellular human pathogen. Nat. Commun. 2020, 11, 3363. [Google Scholar] [CrossRef]
- Paris, D.H.; Phetsouvanh, R.; Tanganuchitcharnchai, A.; Jones, M.; Jenjaroen, K.; Vongsouvath, M.; Ferguson, D.P.J.; Blacksell, S.D.; Newton, P.N.; Day, N.P.J.; et al. Orientia tsutsugamushi in human scrub typhus eschars shows tropism for dendritic cells and monocytes rather than endothelium. PLoS Negl. Trop. Dis. 2012, 6, e1466. [Google Scholar] [CrossRef]
- Inthawong, M.; Sunyakumthorn, P.; Wongwairot, S.; Anantatat, T.; Dunachie, S.J.; Im-Erbsin, R.; Jones, J.W.; Mason, C.J.; Lugo, L.A.; Blacksell, S.D.; et al. A time-course comparative clinical and immune response evaluation study between the human pathogenic Orientia tsutsugamushi strains: Karp and Gilliam in a rhesus macaque (Macaca mulatta) model. PLoS Negl. Trop. Dis. 2022, 16, e0010611. [Google Scholar] [CrossRef]
- Jiang, L.; Morris, E.K.; Aguilera-Olvera, R.; Zhang, Z.; Chan, T.C.; Shashikumar, S.; Chao, C.C.; Casares, S.A.; Ching, W.M. Dissemination of Orientia tsutsugamushi, a causative agent of scrub typhus, and immunological responses in the humanized DRAGA mouse. Front. Immunol. 2018, 9, 816. [Google Scholar] [CrossRef]
- Fang, C.T.; Ferng, W.F.; Hwang, J.J.; Yu, C.J.; Chen, Y.C.; Wang, M.H.; Chang, S.C.; Hsieh, W.C. Life-threatening scrub typhus with meningoencephalitis and acute respiratory distress syndrome. J. Formos. Med. Assoc. 1997, 96, 213–216. [Google Scholar]
- Lee, H.C.; Ko, W.C.; Lee, H.L.; Chen, H.Y. Clinical manifestations and complications of rickettsiosis in southern Taiwan. J. Formos. Med. Assoc. 2002, 101, 385–392. [Google Scholar]
- Fisher, J.; Gonzales, C.; Chroust, Z.; Liang, Y.; Soong, L. Orientia tsutsugamushi infection stimulates Syk-dependent responses and innate cytosolic defenses in macrophages. Pathogens 2023, 12, 53. [Google Scholar] [CrossRef]
- Traub, R.; Wisseman, C.L., Jr. The ecology of chigger-borne rickettsiosis (scrub typhus). J. Med. Entomol. 1974, 11, 237–303. [Google Scholar] [CrossRef]
- Kuo, C.C.; Lin, Y.F.; Yao, C.T.; Shih, H.C.; Chung, L.H.; Liao, H.C.; Hsu, Y.C.; Wang, H.C. Tick-borne pathogens in ticks collected from birds in Taiwan. Parasit. Vectors 2017, 10, 587. [Google Scholar] [CrossRef]
- Chung, L.H.; Wu, W.J.; Kuo, C.C.; Wang, H.C. A checklist of chigger mites (Acari: Trombiculidae and Leeuwenhökiidae) from Taiwan, with descriptions of three new species. J. Med. Entomol. 2015, 52, 1241–1253. [Google Scholar] [CrossRef]
- Takhampunya, R.; Tippayachai, B.; Korkusol, A.; Promsathaporn, S.; Leepitakrat, S.; Sinwat, W.; Schuster, A.L.; Richards, A.L. Transovarial transmission of co-existing Orientia tsutsugamushi genotypes in laboratory-reared Leptotrombidium imphalum. Vector Borne Zoonotic Dis. 2016, 16, 33–41. [Google Scholar] [CrossRef]
- Minahan, N.T.; Wu, W.J.; Tsai, K.H. Rickettsia felis is an emerging human pathogen associated with cat fleas: A review of findings in Taiwan. J. Microbiol. Immunol. Infect. 2023, 56, 10–19. [Google Scholar] [CrossRef]
- Izzard, L.; Fuller, A.; Blacksell, S.D.; Paris, D.H.; Richards, A.L.; Aukkanit, N.; Nguyen, C.; Jiang, J.; Fenwick, S.; Day, N.P.J.; et al. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J. Clin. Microbiol. 2010, 48, 4404–4409. [Google Scholar] [CrossRef] [PubMed]
- Abarca, K.; Martínez-Valdebenito, C.; Angulo, J.; Jiang, J.; Farris, C.; Richards, A.; Acosta-Jamett, G.; Weitzel, T. Molecular description of a novel Orientia species causing scrub typhus in South America. Emerg. Infect. Dis. 2020, 26, 2148–2156. [Google Scholar] [CrossRef]
| Strain | Country | Year Isolated | Size (bp) | GC Content | Genes | RNA | CDSs | Pseudogenes | GenBank Accession | RefSeq Accession | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Boryong | South Korea | late 1980s † | 2,127,051 | 30.5% | 2264 | 40 | 2224 | 533 | AM494475 | NC_009488 | [29] |
| Ikeda | Japan | 1979 † | 2,008,987 | 30.5% | 2131 | 40 | 2091 | 423 | AP008981 | NC_010793 § | [30] |
| Gilliam * | Burma | 1943 † | 2,465,012 | 30.5% | 2600 | 40 | 2560 | 566 | LS398551 | NZ_LS398551 | [31] |
| Karp * | New Guinea | 1943 † | 2,469,803 | 30.8% | 2525 | 40 | 2485 | 488 | LS398548 | NZ_LS398548 | [31] |
| Kato * | Japan | 1955 † | 2,319,449 | 30.8% | 2339 | 41 | 2298 | 431 | LS398550 | NZ_LS398550 | [31] |
| TA686 | Thailand | 1963 ‡ | 2,254,553 | 30.6% | 2537 | 40 | 2497 | 1025 | LS398549 | NZ_LS398549 ¶ | [31] |
| UT76 | Thailand | 2003 † | 2,078,193 | 30.5% | 2203 | 40 | 2163 | 451 | LS398552 | NZ_LS398552 | [31] |
| UT176 | Thailand | 2004 † | 1,932,116 | 30.2% | 1990 | 41 | 1949 | 416 | LS398547 | NZ_LS398547 | [31] |
| Wuj/2014 | China | 2014 † | 1,972,387 | 30.5% | 2054 | 40 | 2014 | 421 | CP044031 | NZ_CP044031 | unpublished |
| TW-1 | Taiwan | 2007 † | 2,008,429 | 30.5% | 2067 | 40 | 2027 | 400 | CP142421 | pending | this study |
| TW-22 | Taiwan | 2007 † | 2,044,475 | 30.5% | 2192 | 40 | 2152 | 414 | CP142420 | pending | this study |
| Maximum Likelihood | Neighbor-Joining | |
|---|---|---|
| core | core | |
| core | 1.0000 | 1.0000 |
| TSA56 | 0.5910 | 0.5910 |
| ScaA | 0.0818 | 0.0818 |
| ScaC | 0.3180 | 0.4000 |
| ScaD | 0.1680 | 0.2450 |
| ScaE | 0.2640 | 0.2270 |
| ScaA + TSA56 | 0.9270 | 0.9640 |
| ScaC + TSA56 | 0.5910 | 0.5640 |
| ScaD + TSA56 | 0.6730 | 0.6450 |
| ScaE + TSA56 | 0.6360 | 0.6360 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minahan, N.T.; Yen, T.-Y.; Guo, Y.-L.L.; Shu, P.-Y.; Tsai, K.-H. Concatenated ScaA and TSA56 Surface Antigen Sequences Reflect Genome-Scale Phylogeny of Orientia tsutsugamushi: An Analysis Including Two Genomes from Taiwan. Pathogens 2024, 13, 299. https://doi.org/10.3390/pathogens13040299
Minahan NT, Yen T-Y, Guo Y-LL, Shu P-Y, Tsai K-H. Concatenated ScaA and TSA56 Surface Antigen Sequences Reflect Genome-Scale Phylogeny of Orientia tsutsugamushi: An Analysis Including Two Genomes from Taiwan. Pathogens. 2024; 13(4):299. https://doi.org/10.3390/pathogens13040299
Chicago/Turabian StyleMinahan, Nicholas T., Tsai-Ying Yen, Yue-Liang Leon Guo, Pei-Yun Shu, and Kun-Hsien Tsai. 2024. "Concatenated ScaA and TSA56 Surface Antigen Sequences Reflect Genome-Scale Phylogeny of Orientia tsutsugamushi: An Analysis Including Two Genomes from Taiwan" Pathogens 13, no. 4: 299. https://doi.org/10.3390/pathogens13040299





