Prevalence and Characterization of Salmonella Isolated from Chickens in Anhui, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Culture of Salmonella
2.2. Serotyping of Salmonella Isolates
2.3. Antimicrobial Susceptibility Testing
2.4. Prevalence of Drug Resistance and Virulence Genes in Salmonella Isolates
2.5. Biofilm Assay
2.6. Multilocus Sequence Typing (MLST)
2.7. Statistical Analysis
3. Results
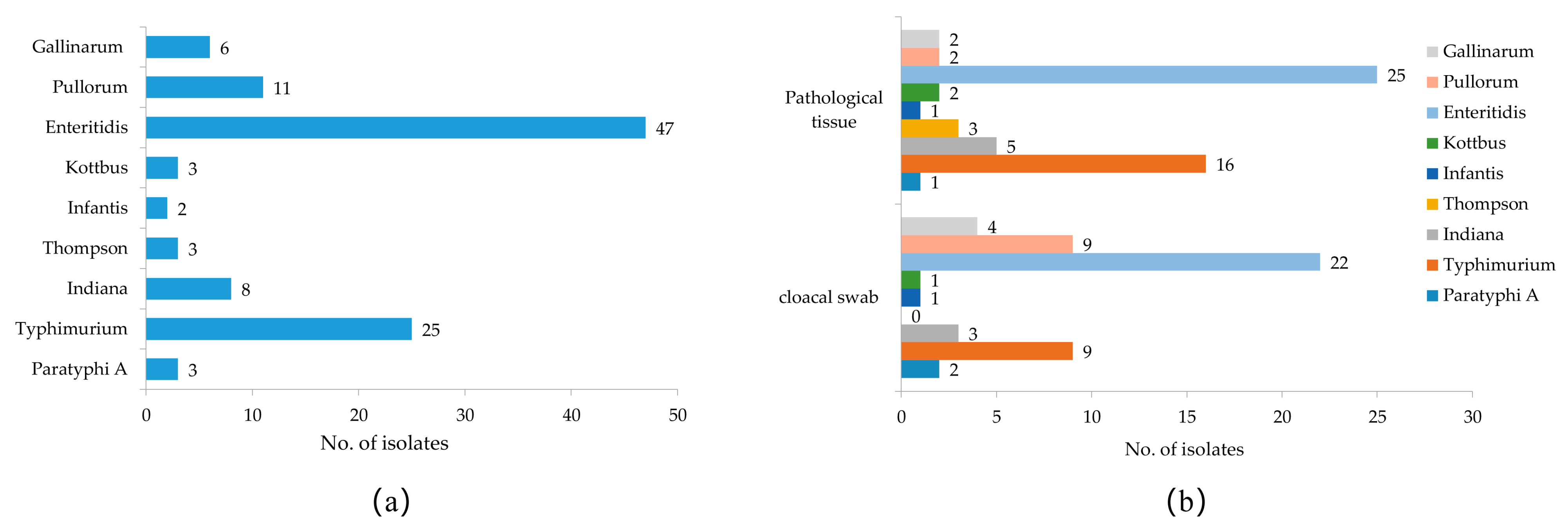
3.1. Isolation and Serotyping of Salmonella
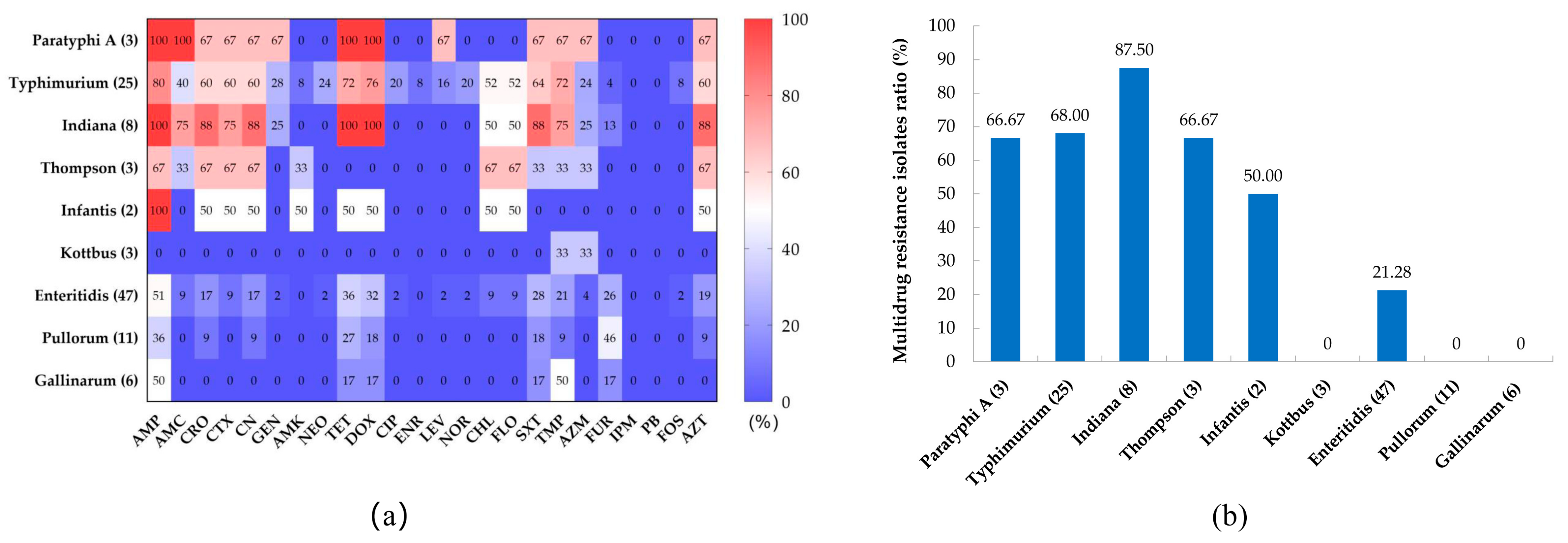
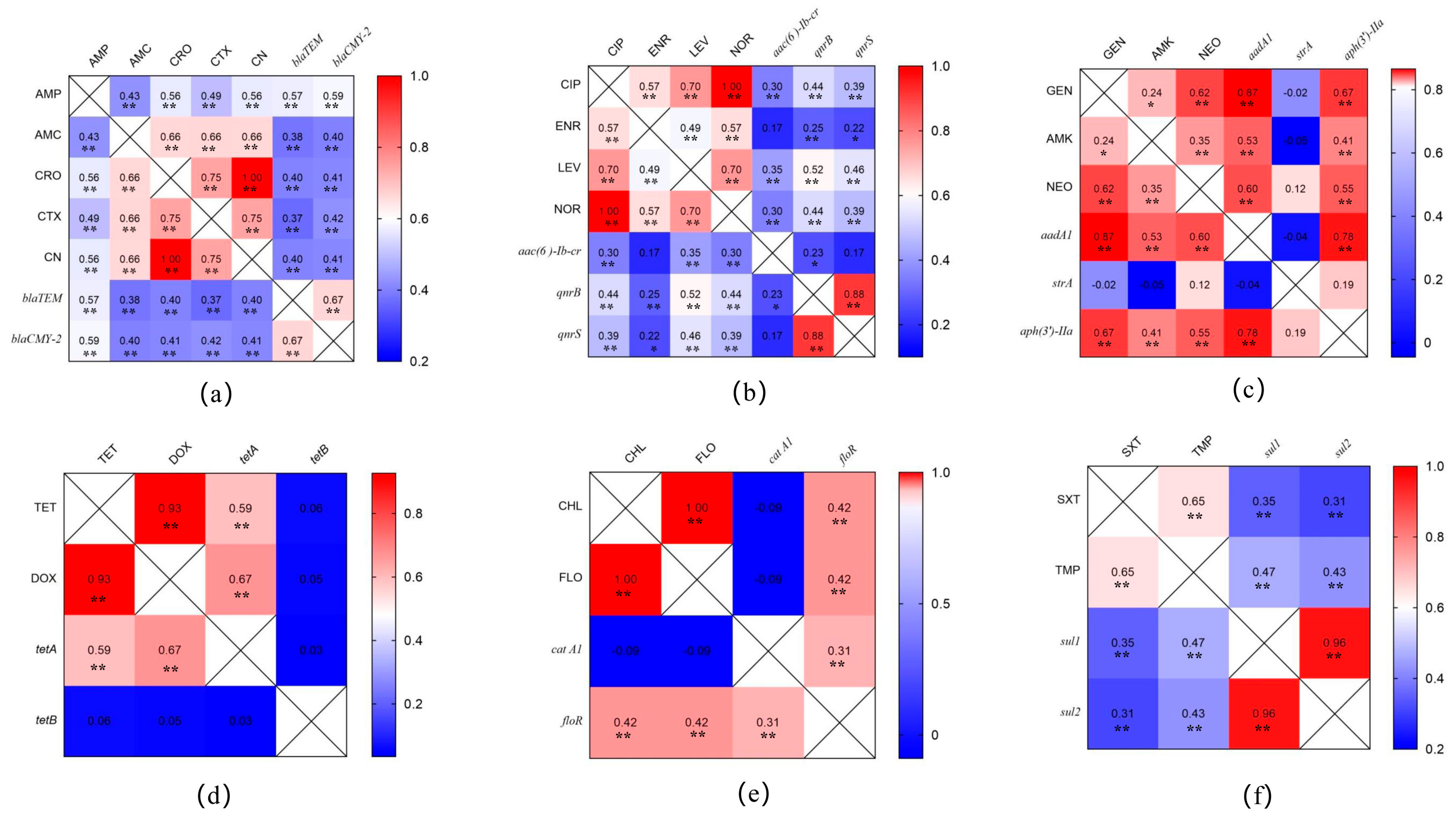
3.2. Antibiotic Susceptibility Testing
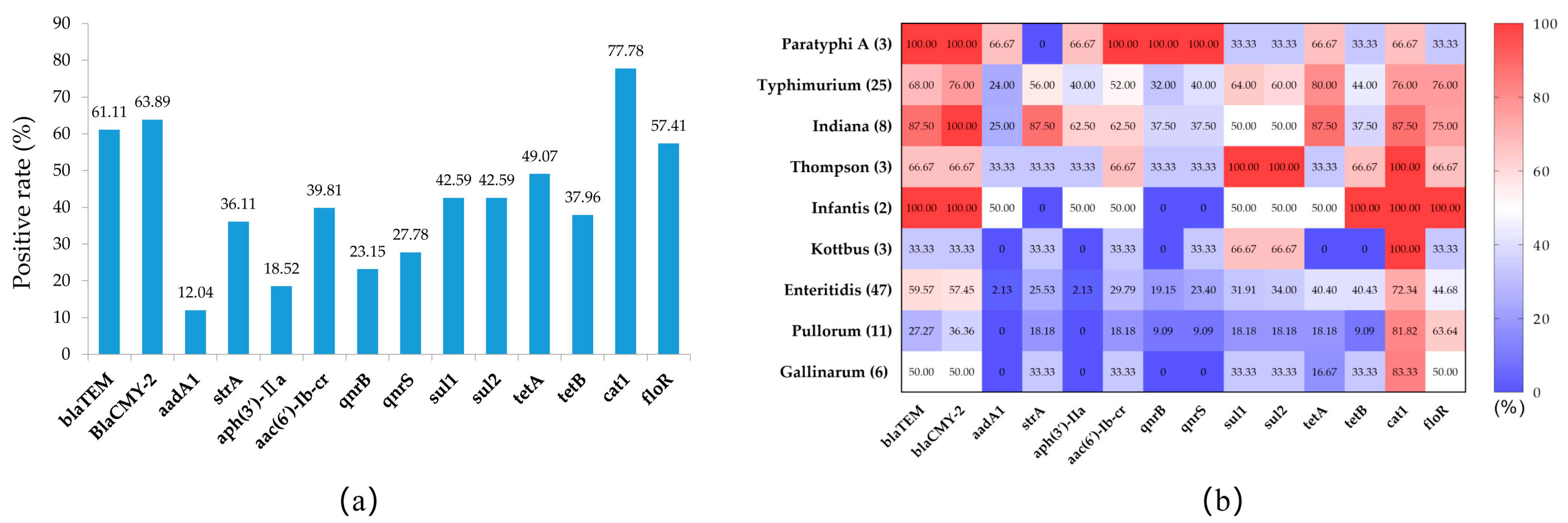
3.3. Prevalence of Antimicrobial-Resistance-Related Genes in Salmonella Isolates
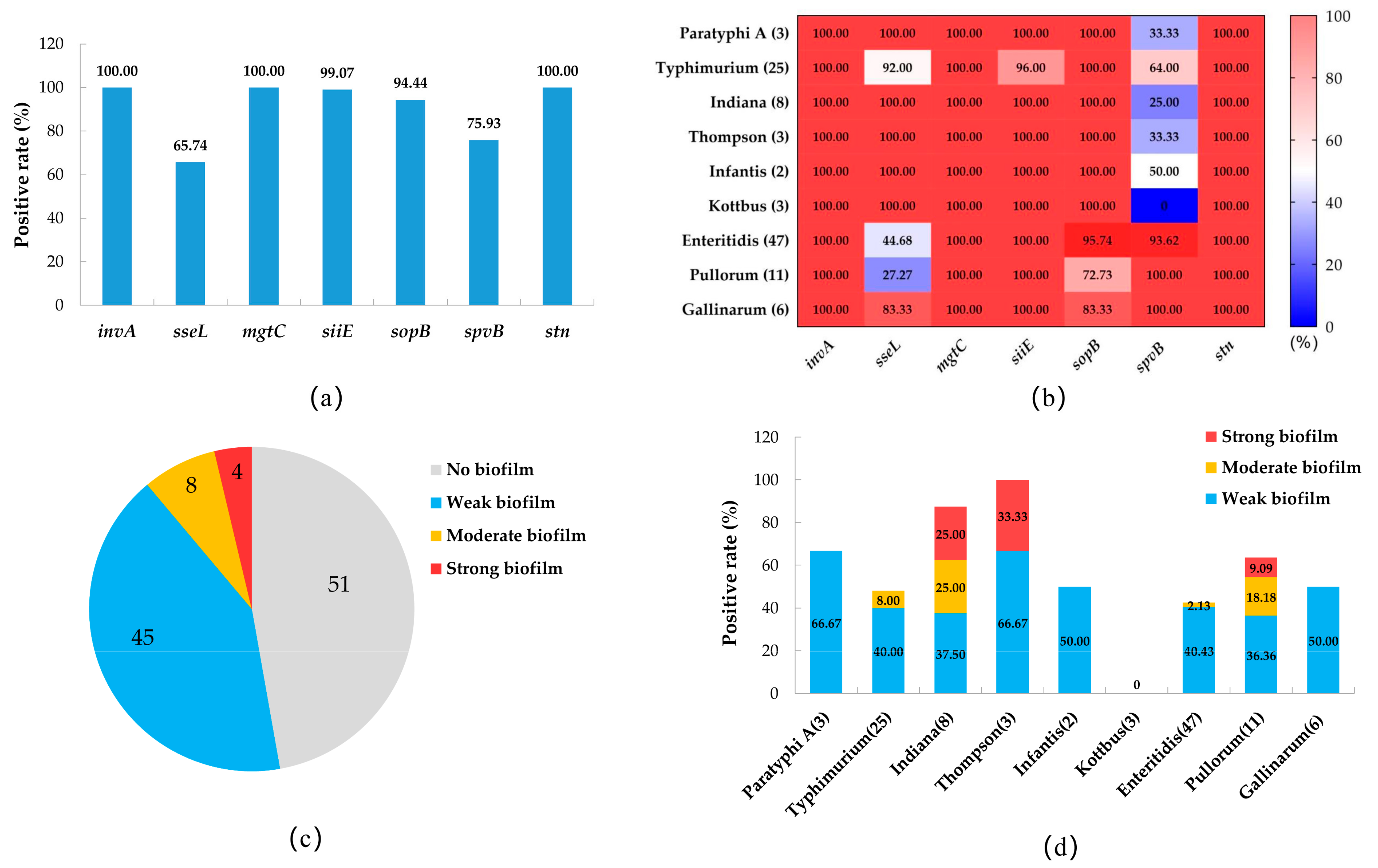
3.4. The Prevalence of Virulence Genes and the Biofilm-Producing Ability of Salmonella Isolates
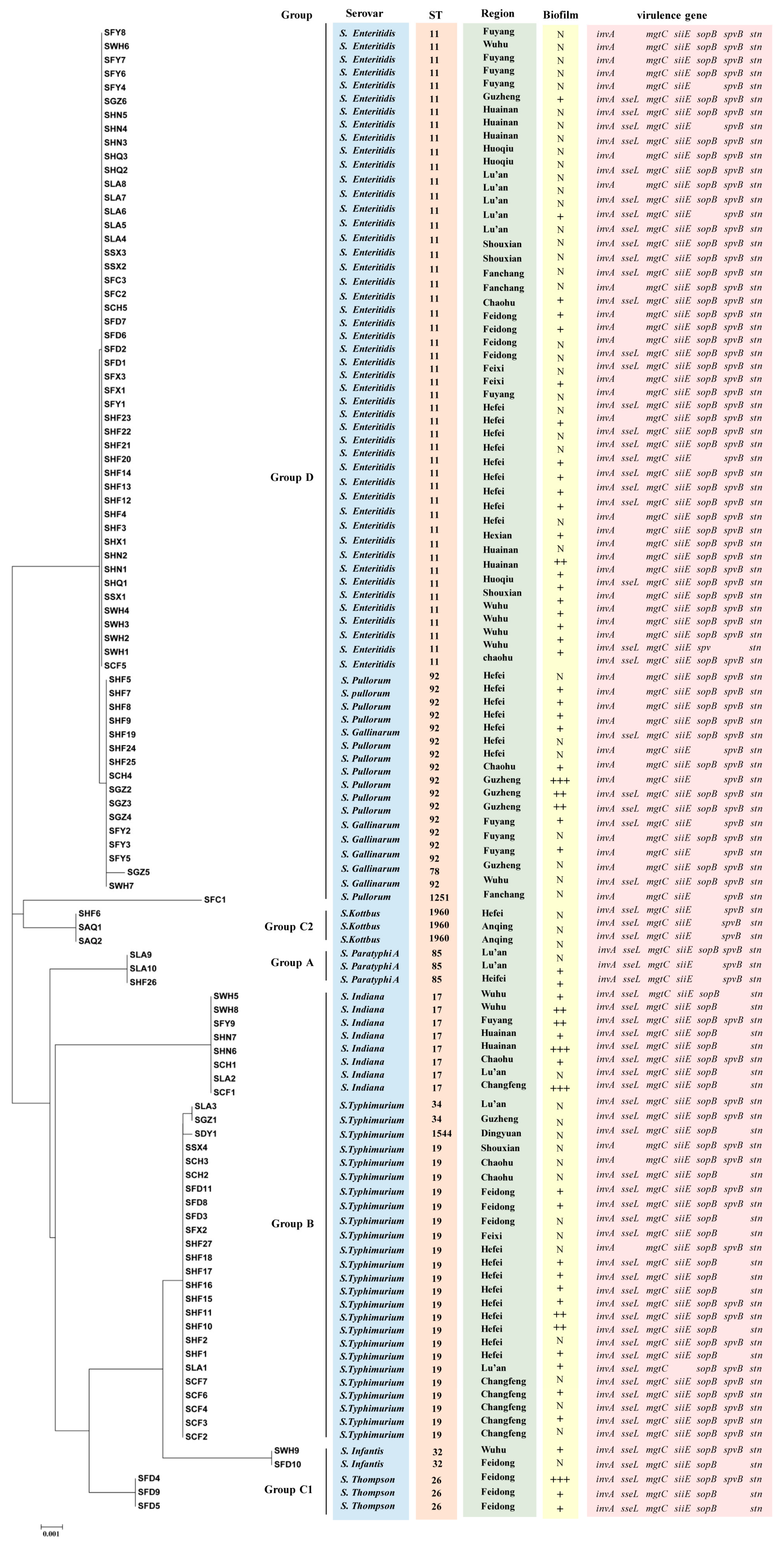
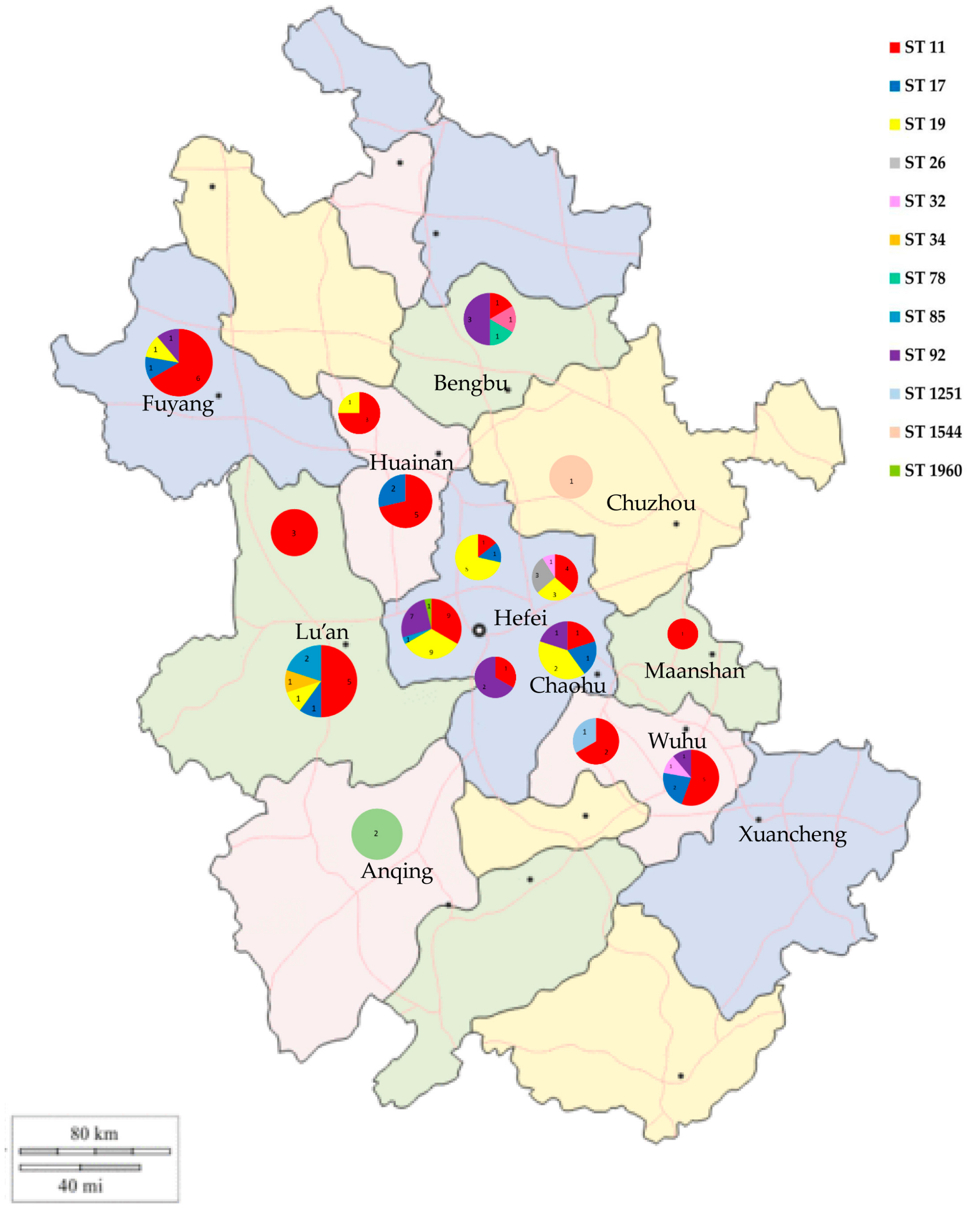
3.5. MLST Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takaya, A.; Yamamoto, T.; Tokoyoda, K. Humoral immunity vs. Salmonella. Front. Immunol. 2019, 10, 3155. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Moreno, L.Z.; Castellanos, L.R.; Chattaway, M.A.; McLauchlin, J.; Lodge, M.; O’Grady, J.; Zamudio, R.; Doughty, E.; Petrovska, L.; et al. Dynamics of Salmonella enterica and antimicrobial resistance in the Brazilian poultry industry and global impacts on public health. PLoS Genet. 2022, 18, e1010174. [Google Scholar] [CrossRef] [PubMed]
- Argüello, H.; Guerra, B.; Rodríguez, I.; Rubio, P.; Carvajal, A. Characterization of antimicrobial resistance determinants and class 1 and class 2 integrons in Salmonella enterica spp., multidrug-resistant isolates from pigs. Genes 2018, 9, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Q.; Dewey-Mattia, D.; Subramhanya, S.; Cui, Z.; Griffin, P.M.; Lance, S.; Lanier, W.; Wise, M.E.; Crowe, S.J. Food recalls associated with foodborne disease outbreaks, United States, 2006–2016. Epidemiol. Infect. 2021, 149, e190. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Efsa. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2011. EFSA J. 2011, 11, 19–73. [Google Scholar] [CrossRef]
- Zhao, X.; Hu, M.; Zhang, Q.; Zhao, C.; Zhang, Y.; Li, L.; Qi, J.; Luo, Y.; Zhou, D.; Liu, Y. Characterization of integrons and antimicrobial resistance in Salmonella from broilers in Shandong, China. Poult. Sci. 2020, 99, 7046–7054. [Google Scholar] [CrossRef]
- Alam, S.B.; Mahmud, M.; Akter, R.; Hasan, M.; Sobur, A.; Nazir, K.N.H.; Noreddin, A.; Rahman, T.; El Zowalaty, M.E.; Rahman, M. Molecular Detection of Multidrug Resistant Salmonella Species Isolated from Broiler Farm in Bangladesh. Pathogens 2020, 9, 201. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Li, Z.; Zhou, X.; Huang, C.; Hu, Y.; Geng, S.; Chen, X.; Li, Q.; Pan, Z.; Jiao, X. Induction of arthritis in chickens by infection with novel virulent Salmonella Pullorum strains. Vet. Microbiol. 2019, 228, 165–172. [Google Scholar] [CrossRef]
- Soria, M.C.; Soria, M.A.; Bueno, D.J. Comparison of 2 culture methods and PCR assays for Salmonella detection in poultry feces. Poult. Sci. 2012, 91, 616–626. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, X.; Jiang, Z.; Qi, Y.; Ed-Dra, A.; Yue, M. Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder chicken hatcheries in Henan, China. Front. Cell Infect. Microbiol. 2020, 10, 497. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ju, Z.; Wang, G.; Yang, J.; Wang, F.; Tang, H.; Zhao, X.; Sun, S. Prevalence and antimicrobial resistance of Salmonella isolated from dead-in-shell chicken embryos in Shandong, China. Front. Vet. Sci. 2021, 8, 581946. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, H.; Chen, J.; Chen, L.; Qi, X.; Zhang, R. Epidemiology of foodborne disease outbreaks caused by nontyphoidal Salmonella in Zhejiang Province, China, 2010-2019. Foodborne Pathog. Dis. 2021, 18, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Wang, Z.; Tian, Y.; Kang, X.; Meng, C.; Chen, X.; Pan, Z.; Jiao, X. Prevalence of Salmonella isolates and their distribution based on whole-genome sequence in a chicken slaughterhouse in Jiangsu, China. Front. Vet. Sci. 2020, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Gao, S.; Chang, Y.; Su, M.; Xie, Y.; Sun, S. Occurrence and characterization of Salmonella isolated from large-scale breeder farms in Shandong Province, China. Biomed Res. Int. 2019, 2019, 8159567. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Chen, J.; Shao, X.; Huang, P.; Zha, J.; Ye, Y. Occurrence and antimicrobial resistance of Salmonella isolated from retail meats in Anhui, China. Food Sci. Nutr. 2021, 9, 4701–4710. [Google Scholar] [CrossRef]
- Song, Y.; Wang, F.; Liu, Y.; Song, Y.; Zhang, L.; Zhang, F.; Gu, X.; Sun, S. Occurrence and characterization of Salmonella isolated from chicken breeder flocks in nine chinese provinces. Front. Vet. Sci. 2020, 7, 479. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef] [Green Version]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhao, S.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.; McDermott, P.F.; Ayers, S.; Meng, J. Characterization of multiple-antimicrobial-resistant salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Aarestrup, F.M.; Lertworapreecha, M.; Evans, M.C.; Bangtrakulnonth, A.; Chalermchaikit, T.; Hendriksen, R.S.; Wegener, H.C. Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J. Antimicrob. Chemother. 2003, 52, 715–718. [Google Scholar] [CrossRef]
- Hai, D.; Yin, X.; Lu, Z.; Lv, F.; Zhao, H.; Bie, X. Occurrence, drug resistance, and virulence genes of Salmonella isolated from chicken and eggs. Food Control. 2020, 113, 107109. [Google Scholar] [CrossRef]
- Skyberg, J.A.; Logue, C.M.; Nolan, L.K. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian Dis. 2006, 50, 77–81. [Google Scholar] [CrossRef] [Green Version]
- Yin, B.; Zhu, L.; Zhang, Y.; Dong, P.; Mao, Y.; Liang, R.; Niu, L.; Luo, X. The Characterization of biofilm formation and detection of biofilm-related genes in Salmonella isolated from beef processing plants. Foodborne Pathog. Dis. 2018, 15, 660–667. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Liu, F.; Cheng, Y.; Su, J. Characterization of Salmonella enterica isolates from diseased poultry in northern China between 2014 and 2018. Pathogens 2020, 9, 95. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, H.; Zheng, S.; Wang, Z.; Sheng, H.; Shi, C.; Shi, X.; Niu, Q.; Yang, B. Prevalence, serotype, antibiotic susceptibility, and genotype of Salmonella in eggs from poultry farms and marketplaces in Yangling, Shaanxi province, China. Front. Microbiol. 2020, 11, 1482. [Google Scholar] [CrossRef]
- Cui, M.; Xie, M.; Qu, Z.; Zhao, S.; Wang, J.; Wang, Y.; He, T.; Wang, H.; Zuo, Z.; Wu, C. Prevalence and antimicrobial resistance of Salmonella isolated from an integrated broiler chicken supply chain in Qingdao, China. Food Control 2016, 62, 270–276. [Google Scholar] [CrossRef]
- Liu, C.; Yao, K.; Ren, D.; Xiao, Y. Prevalence and characterization of Salmonella from meat in slaughterhouses in Hangzhou, China. Int. J. Food Microbiol. 2022, 371, 109649. [Google Scholar] [CrossRef]
- Rasamsetti, S.; Berrang, M.E.; Cox, N.A.; Shariat, N.W. Assessing Salmonella prevalence and complexity through processing using different culture methods. Poult. Sci. 2022, 101, 101949. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, N.; Pan, H.; Elbediwi, M.; Zhou, X.; Peng, X.; Li, X.; Fang, W.; Yue, M. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. 2019, 19, 226. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Lai, J.; Wang, Y.; Liu, S.; Li, Y.; Liu, K.; Shen, J.; Wu, C. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int. J. Food Microbiol. 2013, 163, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.B.; Chen, J.; Huang, Y.Y.; Liu, B.; Shi, X.M. Serotype, genotype, and antimicrobial susceptibility profiles of Salmonella from chicken farms in Shanghai. J. Food Prot. 2010, 73, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Quino, W.; Caro-Castro, J.; Mestanza, O.; Hurtado, C.V.; Zamudio, M.L.; Gavilan, R.G. Phylogenetic structure of Salmonella Enteritidis provides context for a foodborne outbreak in Peru. Sci. Rep. 2020, 10, 22080. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.K.; Chen, S.Y.; Wong, M.Y.; Chiu, C.H.; Chu, C. Pathogenicity differences of Salmonella enterica serovars Typhimurium, Enteritidis, and Choleraesuis-specific virulence plasmids and clinical S. Choleraesuis strains with large plasmids to the human THP-1 cell death. Microb. Pathog. 2019, 128, 69–74. [Google Scholar] [CrossRef]
- Jørgensen, F.; McLauchlin, J.; Verlander, N.Q.; Aird, H.; Balasegaram, S.; Chattaway, M.A.; Dallman, T.; Herdman, M.T.; Hoban, A.; Lai, S.; et al. Levels and genotypes of Salmonella and levels of Escherichia coli in frozen ready-to-cook chicken and turkey products in England tested in 2020 in relation to an outbreak of S. Enteritidis. Int. J. Food Microbiol. 2022, 369, 109609. [Google Scholar] [CrossRef]
- Ansari-Lari, M.; Hosseinzadeh, S.; Manzari, M.; Khaledian, S. Survey of Salmonella in commercial broiler farms in Shiraz, southern Iran. Prev. Vet. Med. 2022, 198, 105550. [Google Scholar] [CrossRef]
- Ramtahal, M.A.; Amoako, D.G.; Akebe, A.L.K.; Somboro, A.M.; Bester, L.A.; Essack, S.Y. A public health insight into Salmonella in poultry in Africa: A review of the past decade: 2010–2020. Microb. Drug Resist. 2022, 28, 710–733. [Google Scholar] [CrossRef]
- Cui, M.; Zhang, P.; Li, J.; Sun, C.; Song, L.; Zhang, C.; Zhao, Q.; Wu, C. Prevalence and characterization of fluoroquinolone resistant Salmonella isolated from an integrated broiler chicken supply chain. Front. Microbiol. 2019, 10, 1865. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wang, Y.; Gao, Y.; Li, C.; Ma, B.; Wang, H. Antimicrobial resistance and CRISPR typing among Salmonella isolates from poultry farms in China. Front. Microbiol. 2021, 12, 730046. [Google Scholar] [CrossRef] [PubMed]
- Lyu, N.; Feng, Y.; Pan, Y.; Huang, H.; Liu, Y.; Xue, C.; Zhu, B.; Hu, Y. Genomic characterization of Salmonella enterica isolates from retail meat in Beijing, China. Front. Microbiol. 2021, 12, 636332. [Google Scholar] [CrossRef] [PubMed]
- Wigley, P.; Jones, M.A.; Barrow, P.A. Salmonella enterica serovar Pullorum requires the Salmonella pathogenicity island 2 type III secretion system for virulence and carriage in the chicken. Avian Pathol. 2002, 31, 501–506. [Google Scholar] [CrossRef]
- Florez-Cuadrado, D.; Moreno, M.A.; Ugarte-Ruíz, M.; Domínguez, L. Antimicrobial resistance in the food chain in the European Union. Adv. Food Nutr. Res. 2018, 86, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Dodds, D.R. Antibiotic resistance: A current epilogue. Biochem. Pharmacol. 2017, 134, 139–146. [Google Scholar] [CrossRef]
- Xie, T.; Wu, G.; He, X.; Lai, Z.; Zhang, H.; Zhao, J. Antimicrobial resistance and genetic diversity of Salmonella enterica from eggs. Food Sci. Nutr. 2019, 7, 2847–2853. [Google Scholar] [CrossRef] [Green Version]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef] [Green Version]
- Willmann, M.; El-Hadidi, M.; Huson, D.H.; Schütz, M.; Weidenmaier, C.; Autenrieth, I.B.; Peter, S. Antibiotic selection pressure determination through sequence-based metagenomics. Antimicrob. Agents Chemother. 2015, 59, 7335–7345. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Z.; Zhou, X.; Cui, Y.; Shi, C.; Shi, X. Prevalence and characterization of antimicrobial resistance in Salmonella enterica isolates from retail foods in Shanghai, China. Foodborne Pathog. Dis. 2020, 17, 35–43. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, J.; Xu, X.; Zhou, X.; Shi, C.; Zhao, X.; Liu, Y.; Shi, X. Co-existence of mphA, oqxAB and blaCTX-M-65 on the IncHI2 Plasmid in highly drug-resistant Salmonella enterica serovar Indiana ST17 isolated from retail foods and humans in China. Food Control 2020, 118, 107269. [Google Scholar] [CrossRef]
- Abd El-Aziz, N.K.; Tartor, Y.H.; Gharieb, R.M.A.; Erfan, A.M.; Khalifa, E.; Said, M.A.; Ammar, A.M.; Samir, M. Extensive drug-resistant Salmonella enterica isolated from poultry and humans: Prevalence and molecular determinants behind the co-resistance to ciprofloxacin and tigecycline. Front. Microbiol. 2021, 12, 738784. [Google Scholar] [CrossRef]
- Bourély, C.; Cazeau, G.; Jarrige, N.; Jouy, E.; Haenni, M.; Lupo, A.; Madec, J.Y.; Leblond, A.; Gay, E. Co-resistance to amoxicillin and tetracycline as an indicator of multidrug resistance in Escherichia coli isolates from animals. Front. Microbiol. 2019, 10, 2288. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, M.G.; de Melo, R.T.; Dumont, C.F.; Peixoto, J.L.M.; Ferreira, G.R.A.; Chueiri, M.C.; Iasbeck, J.R.; Timóteo, M.F.; de Araújo Brum, B.; Rossi, D.A. Agents of campylobacteriosis in different meat matrices in Brazil. Int. J. Environ. Res. Public Health 2022, 19, 87. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Duarte, D.J.; Oldenkamp, R.; Ragas, A.M.J. Modelling environmental antibiotic-resistance gene abundance: A meta-analysis. Sci. Total Environ. 2019, 659, 335–341. [Google Scholar] [CrossRef] [Green Version]
- Ghafourian, S.; Sadeghifard, N.; Soheili, S.; Sekawi, Z. Extended spectrum beta-lactamases: Definition, classification and epidemiology. Curr. Issues Mol. Biol. 2015, 17, 11–21. Available online: https://pubmed.ncbi.nlm.nih.gov/24821872/ (accessed on 20 February 2023).
- Mei, X.; Ma, B.; Zhai, X.; Zhang, A.; Lei, C.; Zuo, L.; Yang, X.; Zhou, C.; Wang, H. Florfenicol enhances colonization of a Salmonella enterica Serovar Enteritidis floR mutant with major alterations to the intestinal microbiota and metabolome in neonatal chickens. Appl. Environ. Microbiol. 2021, 87, e0168121. [Google Scholar] [CrossRef]
- Gupta, A.; Sinha, P.; Nema, V.; Gupta, P.K.; Chakraborty, P.; Kulkarni, S.; Rastogi, N.; Anupurba, S. Detection of Beijing strains of MDR M. tuberculosis and their association with drug resistance mutations in katG, rpoB, and embB genes. BMC Infect. Dis. 2020, 20, 752. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial resistance in bacteria: Mechanisms, evolution, and persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Shen, X.; Yin, L.; Ma, H.; Pan, X.; Zhang, D.; Zhao, R.; Dai, Y.; Hou, H.; Hu, X. Comprehensive genomic analysis and characterization of a new ST 174 type Klebsiella variicola strain isolated from chicken embryos. Infect. Genet. Evol. 2021, 90, 104768. [Google Scholar] [CrossRef]
- Ijaz, A.; Veldhuizen, E.J.A.; Broere, F.; Rutten, V.; Jansen, C.A. The interplay between Salmonella and intestinal innate immune cells in chickens. Pathogens 2021, 10, 1512. [Google Scholar] [CrossRef] [PubMed]
- Andesfha, E.; Indrawati, A.; Mayasari, N.; Rahayuningtyas, I.; Jusa, I. Detection of Salmonella pathogenicity island and Salmonella plasmid virulence genes in Salmonella Enteritidis originated from layer and broiler farms in Java Island. J. Adv. Vet. Anim. Res. 2019, 6, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhuang, L.; Wang, C.; Zhang, P.; Zhang, T.; Shao, H.; Han, X.; Gong, J. Virulence gene distribution of Salmonella Pullorum isolates recovered from chickens in China (1953–2015). Avian Dis. 2018, 62, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ju, Z.; Yang, Y.; Zhao, X.; Jiang, Z.; Sun, S. Serotype, antimicrobial susceptibility and genotype profiles of Salmonella isolated from duck farms and a slaughterhouse in Shandong province, China. BMC Microbiol. 2019, 19, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, S.; Wang, Y.; Xue, Y.; Wang, H.; Cai, Y.; Zhang, J.; Barrow, P.; Pan, Z.; Jiao, X. The SseL protein inhibits the intracellular NF-κB pathway to enhance the virulence of Salmonella Pullorum in a chicken model. Microb. Pathog. 2019, 129, 1–6. [Google Scholar] [CrossRef]
- Sun, L.; Yang, S.; Deng, Q.; Dong, K.; Li, Y.; Wu, S.; Huang, R. Salmonella effector SpvB disrupts intestinal epithelial barrier integrity for bacterial translocation. Front. Cell Infect. Microbiol. 2020, 10, 606541. [Google Scholar] [CrossRef]
- Elemfareji, O.I.; Thong, K.L. Comparative virulotyping of Salmonella typhi and Salmonella enteritidis. Indian J. Microbiol. 2013, 53, 410–417. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Sanguiné, A.Y.; D’Alessandro, B.; Langleib, M.; Traglia, G.M.; Mónaco, A.; Durán, R.; Chabalgoity, J.A.; Betancor, L.; Yim, L. Salmonella enterica Serovars Dublin and Enteritidis comparative proteomics reveals differential expression of proteins involved in stress resistance, virulence, and anaerobic Metabolism. Infect. Immun. 2021, 89, e00606-20. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [Green Version]
- Biswas, R.; Agarwal, R.K.; Bhilegaonkar, K.N.; Kumar, A.; Nambiar, P.; Rawat, S.; Singh, M. Cloning and sequencing of biofilm-associated protein (bapA) gene and its occurrence in different serotypes of Salmonella. Lett. Appl. Microbiol. 2011, 52, 138–143. [Google Scholar] [CrossRef]
- Wang, R.; King, D.A.; Kalchayanand, N. Evaluation of Salmonella biofilm cell transfer from common food contact surfaces to beef products. J. Food Prot. 2022, 85, 632–638. [Google Scholar] [CrossRef]
- Borges, K.A.; Furian, T.Q.; de Souza, S.N.; Menezes, R.; de Lima, D.A.; Fortes, F.B.B.; Salle, C.T.P.; Moraes, H.L.S.; Nascimento, V.P. Biofilm formation by Salmonella Enteritidis and Salmonella Typhimurium isolated from avian sources is partially related with their in vivo pathogenicity. Microb. Pathog. 2018, 118, 238–241. [Google Scholar] [CrossRef]
- Akinola, S.A.; Tshimpamba, M.E.; Mwanza, M.; Ateba, C.N. Biofilm production potential of Salmonella Serovars isolated from chickens in north west province, South Africa. Pol. J. Microbiol. 2020, 69, 427–439. [Google Scholar] [CrossRef]
- Manafi, L.; Aliakbarlu, J.; Dastmalchi Saei, H. Antibiotic resistance and biofilm formation ability of Salmonella serotypes isolated from beef, mutton, and meat contact surfaces at retail. J. Food Sci. 2020, 85, 2516–2522. [Google Scholar] [CrossRef]
- Silva, P.; Goulart, L.R.; Reis, T.F.M.; Mendonça, E.P.; Melo, R.T.; Penha, V.A.S.; Peres, P.; Hoepers, P.G.; Beletti, M.E.; Fonseca, B.B. Biofilm formation in different Salmonella serotypes isolated from poultry. Curr. Microbiol. 2019, 76, 124–129. [Google Scholar] [CrossRef]
- Dougue, A.N.; El-Kholy, M.A.; Giuffrè, L.; Galeano, G.; D′Aleo, F.; Kountchou, C.L.; Nangwat, C.; Dzoyem, J.P.; Giosa, D.; Pernice, I.; et al. Multilocus sequence typing (MLST) analysis reveals many novel genotypes and a high level of genetic diversity in Candida tropicalis isolates from Italy and Africa. Mycoses 2022, 65, 989–1000. [Google Scholar] [CrossRef]
- Tedersoo, T.; Roasto, M.; Mäesaar, M.; Kisand, V.; Ivanova, M.; Meremäe, K. The prevalence, counts, and MLST genotypes of Campylobacter in poultry meat and genomic comparison with clinical isolates. Poult. Sci. 2022, 101, 101703. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, H.; Cheng, Y.; Zhang, W.; Luo, Q.; Wen, G.; Wang, G.; Shao, H.; Zhang, T. Quinolone resistance phenotype and genetic characterization of Salmonella enterica serovar Pullorum isolates in China, during 2011 to 2016. BMC Microbiol. 2018, 18, 225. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Gao, Y.; Ye, C.; Yang, L.; Wang, T.; Chang, W. Prevalence and characteristics of Salmonella isolated from free-range chickens in Shandong Province, China. Biomed Res. Int. 2016, 2016, 8183931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakaria, Z.; Hassan, L.; Sharif, Z.; Ahmad, N.; Ali, R.M.; Husin, S.A.; Hazis, N.; Sohaimi, N.F.M.; Bakar, S.A.; Garba, B. Analysis of Salmonella enterica serovar Enteritidis isolates from chickens and chicken meat products in Malaysia using PFGE, and MLST. BMC Vet. Res. 2020, 16, 393. [Google Scholar] [CrossRef] [PubMed]
- Ktari, S.; Ksibi, B.; Gharsallah, H.; Mnif, B.; Maalej, S.; Rhimi, F.; Hammami, A. Molecular epidemiological characteristics of Salmonella enterica serovars Enteritidis, Typhimurium and Livingstone strains isolated in a Tunisian university hospital. Apmis 2016, 124, 194–200. [Google Scholar] [CrossRef]
- Zhang, K.; Ge, H.; He, J.; Hu, M.; Xu, Z.; Jiao, X.; Chen, X. Salmonella Typhimurium ST34 isolate was more resistant than the ST19 isolate in China, 2007– 2019. Foodborne Pathog. Dis. 2022, 19, 62–69. [Google Scholar] [CrossRef]
- Sun, J.; Ke, B.; Huang, Y.; He, D.; Li, X.; Liang, Z.; Ke, C. The molecular epidemiological characteristics and genetic diversity of Salmonella typhimurium in Guangdong, China, 2007–2011. PLoS ONE 2014, 9, e113145. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Huang, J.; Zhang, Y.; Liu, S.; Chen, L.; Xiao, C.; Zeng, H.; Wei, X.; Gu, Q.; Li, Y.; et al. Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 2020, 713, 136385. [Google Scholar] [CrossRef]
- Hu, Y.; Fanning, S.; Nguyen, S.V.; Wang, W.; Liu, C.; Cui, X.; Dong, Y.; Gan, X.; Xu, J.; Li, F. Emergence of a Salmonella enterica serovar Typhimurium ST34 isolate, CFSA629, carrying a novel mcr-1.19 variant cultured from egg in China. J. Antimicrob. Chemother. 2021, 76, 1776–1785. [Google Scholar] [CrossRef]
- Vaid, R.K.; Thakur, Z.; Anand, T.; Kumar, S.; Tripathi, B.N. Comparative genome analysis of Salmonella enterica serovar Gallinarum biovars Pullorum and Gallinarum decodes strain specific genes. PLoS ONE 2021, 16, e0255612. [Google Scholar] [CrossRef]
| Location/Function | Gene | Primer Sequence (5′-3′) | Reference |
|---|---|---|---|
| ß-Lactams | blaTEM | F: CAGCGGTAAGATCCTTGAGA R: ACTCCCCGTCGTGTAGATAA | [20] |
| blaCMY-2 | F: TGGCCGTTGCCGTTATCTAC R: CCCGTTTTATGCACCCAT GA | [20] | |
| Aminoglycoside resistance genes | aadA | F: ATCCTTCGGCGCGATTTTG R: GCAGCGCAATGACATTCTTG | [21] |
| strA | F: CCAATCGCAGATAGAAGGC R: CTTGGTGATAACGGCAATTC | [21] | |
| aph(3′)-IIa | F: TCCGGTGCCCTGAATGAACT R: ACG GGT AGC CAA CGC TAT GT | [20] | |
| Quinolone resistance genes | qnrB | F: GATCGTGAAAGCCAGAAAGG R: ACGATGCCTGGTAGTTGTCC | [17] |
| qnrS | F: ACGACATTCGTCAACTGCAA R: TAAATTGGCACCCTGTAGGC | [17] | |
| aac(6′)-Ib-cr | F: TTGCGATGCTCTATGAGTGGCTA R: CTCGAATGCCTGGCGTGTTT | [17] | |
| Tetracycline resistance genes | tetA | F: GCGCCTTTCCTTTGGGTTCT R: CCACCCGTTCCACGTTGTTA | [17] |
| tetB | F: CATTAATAGGCGCATCGCTG R: TGAAGGTCATCGATAGCAGG | [17] | |
| Sulfonamide resistance genes | sul1 | F: CTTCGATGAGAGCCGGCGGC R: GCAAGGCGGAAACCCGCGCC | [17] |
| sul2 | F: GCGCTCAAGGCAGATGGCATT R: GCGTTTGATACCGGCACCCGT | [17] | |
| Chloramphenicol resistance genes | cat1 | F: CTTGTCGCCTTGCGTATAAT R: ATCCCAATGGCATCGTAAAG | [21] |
| floR | F: AACCCGCCCTCTGGATCAAGTCAA R: CAAATCACGGGCCACGCTGTATC | [22] | |
| SPI-1 | invA | F:CTGGCGGTGGGTTTTGTTGTCTTCTCTATT R:AGTTTCTCCCCCTCTTCATGCGTTACCC | [23] |
| SPI-2 | sseL | F: GCCCCTTCCAGATTACTTTATATG R: TGCTTAATATATTTTCTTTGGTGG | [22] |
| SPI-3 | mgtC | F: AAAGACAATGGCGTCAACGTATGG R: TTCTTTATAGCCCTGTTCCTGAGC | [22] |
| SPI-4 | siiE | F:GGAGTATCGATAAAGATGTT R: GCGCGTAACGTCAGAATCAA | [23] |
| SPI-5 | sopB | F:CGGACCGGCCAGCAACAAAACAAGAAG R: TAGTGATGCCCGTTATGCGTGAGTGTATT | [23] |
| Salmonella plasmid virulence | spvB | F:CTATCAGCCCCGCACGGAGAGCAGTTTT R: GGAGGAGGCGGTGGCGGTGGCATCATA | [23] |
| Salmonella enterotoxin | stn | F: AGCGTTCAGGTACAGATTCAACA R: AAATTCGTAACCCGCTCTCGT | [22] |
| Sample Type | Number of Samples | Number of Isolates | Isolation Rate (%) |
|---|---|---|---|
| Cloacal swab | 1500 | 51 | 3.40 |
| Pathological tissue | 408 | 57 | 13.97 |
| Total | 1908 | 108 | 5.66 |
| Classes | Antimicrobials | Concentrations(µg) | Number of Isolates | Resistance (%) |
|---|---|---|---|---|
| Penicillin | AMP | 10 | 66 | 61.11 |
| β-lactams | AMC | 20/10 | 24 | 22.22 |
| Cephems | CRO | 30 | 36 | 33.33 |
| CTX | 30 | 30 | 27.78 | |
| CN | 30 | 36 | 33.33 | |
| Aminoglycosides | GEN | 10 | 12 | 11.11 |
| AMK | 30 | 4 | 3.70 | |
| NEO | 30 | 7 | 6.48 | |
| Tetracyclines | TET | 30 | 51 | 47.22 |
| DOX | 30 | 49 | 45.37 | |
| Quinolones | CIP | 5 | 6 | 5.56 |
| ENR | 10 | 2 | 1.85 | |
| LEV | 5 | 8 | 7.41 | |
| NOR | 10 | 6 | 5.56 | |
| Chloramphenicols | CHL | 30 | 24 | 22.22 |
| FLO | 30 | 24 | 22.22 | |
| Sulfonamides | SXT | 23.75/1.25 | 42 | 48.89 |
| TMP | 5 | 42 | 48.89 | |
| Macrolides | AZM | 15 | 14 | 12.96 |
| Nitrofurans | FUR | 100 | 20 | 18.52 |
| Carbapenems | IPM | 10 | 0 | 0 |
| Polypeptide | PB | 300 | 0 | 0 |
| Fosfomycin | FOS | 200 | 4 | 3.70 |
| Monobactams | AZT | 30 | 37 | 34.26 |
| Antimicrobials | Gene | No. (%) |
|---|---|---|
| ß-Lactamase (n = 66) | blaTEM | 55 (83.33) |
| blaCMY-2 | 59 (89.39) | |
| Quinolones (n = 9) | aac(6′)-Ib-cr | 9 (100.00) |
| qnrB | 9 (100.00) | |
| qnrS | 9 (100.00) | |
| Aminoglycoside (n = 15) | aadA1 | 14 (93.33) |
| strA | 5 (33.33) | |
| aph(3′)-IIa | 14 (93.33) | |
| Tetracycline (n = 52) | tetA | 42 (80.77) |
| tetB | 22 (42.31) | |
| Sulfonamide (n = 51) | sul1 | 32 (62.75) |
| sul2 | 31 (60.78) | |
| Chloramphenicol (n = 24) | catA1 | 17 (70.83) |
| floR | 23 (95.83) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Yin, L.; Zhang, A.; Zhao, R.; Yin, D.; Wang, J.; Dai, Y.; Hou, H.; Pan, X.; Hu, X.; et al. Prevalence and Characterization of Salmonella Isolated from Chickens in Anhui, China. Pathogens 2023, 12, 465. https://doi.org/10.3390/pathogens12030465
Shen X, Yin L, Zhang A, Zhao R, Yin D, Wang J, Dai Y, Hou H, Pan X, Hu X, et al. Prevalence and Characterization of Salmonella Isolated from Chickens in Anhui, China. Pathogens. 2023; 12(3):465. https://doi.org/10.3390/pathogens12030465
Chicago/Turabian StyleShen, Xuehuai, Lei Yin, Anyun Zhang, Ruihong Zhao, Dongdong Yin, Jieru Wang, Yin Dai, Hongyan Hou, Xiaocheng Pan, Xiaomiao Hu, and et al. 2023. "Prevalence and Characterization of Salmonella Isolated from Chickens in Anhui, China" Pathogens 12, no. 3: 465. https://doi.org/10.3390/pathogens12030465





