Genomic Recombination Leading to Decreased Virulence of Group B Streptococcus in a Mouse Model of Adult Invasive Disease
Abstract
:1. Introduction
2. Results
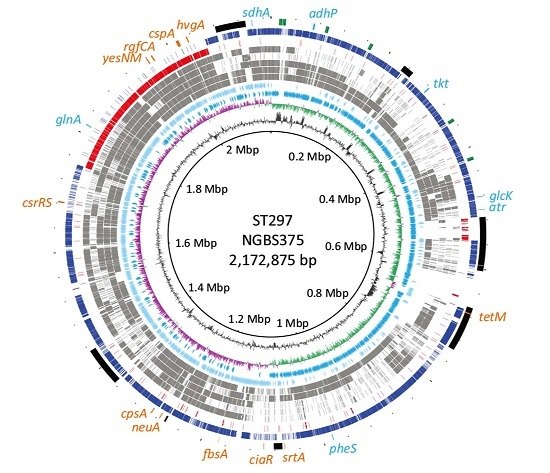
2.1. Genome Comparisons Show that Strain NGBS375 Has an ST1 Genome Backbone but Has Acquired an Area of 308,916 bp by Homologous Recombination from an ST17 Donor
2.2. Genetic Content in the Recombined Region of ST297Strain NGBS375
2.3. Ortholog Analysis of GBS Strains
2.4. Altered Virulence of the Mosaic GBS Strain in An Adult Mouse Model of Systemic Infection
2.5. In vivo Systemic Inflammatory Response induced by the ST297 GBS Strain
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Genome Sequencing and Analysis
4.3. Mouse Experimental Infections
4.4. Cytokine Quantification by ELISA
4.5. Statistical Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Farley, M.M. Group b streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 2001, 33, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Berti, F.; Campisi, E.; Toniolo, C.; Morelli, L.; Crotti, S.; Rosini, R.; Romano, M.R.; Pinto, V.; Brogioni, B.; Torricelli, G.; et al. Structure of the type IX group B Streptococcus capsular polysaccharide and its evolutionary relationship with types V and VII. J. Biol. Chem. 2014, 289, 23437–23448. [Google Scholar] [CrossRef] [PubMed]
- Cieslewicz, M.J.; Chaffin, D.; Glusman, G.; Kasper, D.; Madan, A.; Rodrigues, S.; Fahey, J.; Wessels, M.R.; Rubens, C.E. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 2005, 73, 3096–3103. [Google Scholar] [CrossRef] [PubMed]
- Lamagni, T.L.; Keshishian, C.; Efstratiou, A.; Guy, R.; Henderson, K.L.; Broughton, K.; Sheridan, E. Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991–2010. Clin. Infect. Dis. 2013, 57, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Joubrel, C.; Tazi, A.; Six, A.; Dmytruk, N.; Touak, G.; Bidet, P.; Raymond, J.; Trieu Cuot, P.; Fouet, A.; Kerneis, S.; et al. Group B Streptococcus neonatal invasive infections, France 2007–2012. Clin. Microbiol. Infect. 2015, 21, 910–916. [Google Scholar] [CrossRef] [PubMed]
- Phares, C.R.; Lynfield, R.; Farley, M.M.; Mohle-Boetani, J.; Harrison, L.H.; Petit, S.; Craig, A.S.; Schaffner, W.; Zansky, S.M.; Gershman, K.; et al. Epidemiology of invasive group b streptococcal disease in the United States, 1999–2005. JAMA J. Am. Med. Assoc. 2008, 299, 2056–2065. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.; Bohnsack, J.F.; Takahashi, S.; Oliver, K.A.; Chan, M.S.; Kunst, F.; Glaser, P.; Rusniok, C.; Crook, D.W.; Harding, R.M.; et al. Multilocus sequence typing system for group B Streptococcus. J. Clin. Microbiol. 2003, 41, 2530–2536. [Google Scholar] [CrossRef] [PubMed]
- Honsa, E.; Fricke, T.; Stephens, A.J.; Ko, D.; Kong, F.; Gilbert, G.L.; Huygens, F.; Giffard, P.M. Assignment of Streptococcus agalactiae isolates to clonal complexes using a small set of single nucleotide polymorphisms. BMC Microbiol. 2008, 8, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Cunha, V.; Davies, M.R.; Douarre, P.E.; Rosinski-Chupin, I.; Margarit, I.; Spinali, S.; Perkins, T.; Lechat, P.; Dmytruk, N.; Sauvage, E.; et al. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat. Commun. 2014, 5, 4544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, S.L.; Granlund, M.; Sellin, M.; Lagergard, T.; Spratt, B.G.; Norgren, M. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 2005, 43, 3727–3733. [Google Scholar] [CrossRef] [PubMed]
- Lefebure, T.; Stanhope, M.J. Evolution of the core and pan-genome of Streptococcus: Positive selection, recombination, and genome composition. Genome Biol. 2007, 8, R71. [Google Scholar] [CrossRef] [PubMed]
- Brochet, M.; Rusniok, C.; Couve, E.; Dramsi, S.; Poyart, C.; Trieu-Cuot, P.; Kunst, F.; Glaser, P. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 2008, 105, 15961–15966. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, U.B.; Poulsen, K.; Ghezzo, C.; Margarit, I.; Kilian, M. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 2010, 1, e00178-10. [Google Scholar] [CrossRef] [PubMed]
- Bellais, S.; Six, A.; Fouet, A.; Longo, M.; Dmytruk, N.; Glaser, P.; Trieu-Cuot, P.; Poyart, C. Capsular switching in group B Streptococcus CC17 hypervirulent clone: A future challenge for polysaccharide vaccine development. J. Infect. Dis. 2012, 206, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Teatero, S.; McGeer, A.; Low, D.E.; Li, A.; Demczuk, W.; Martin, I.; Fittipaldi, N. Characterization of invasive group B Streptococcus strains from the greater Toronto area, Canada. J. Clin. Microbiol. 2014, 52, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Tazi, A.; Disson, O.; Bellais, S.; Bouaboud, A.; Dmytruk, N.; Dramsi, S.; Mistou, M.Y.; Khun, H.; Mechler, C.; Tardieux, I.; et al. The surface protein HvgA mediates group B Streptococcus hypervirulence and meningeal tropism in neonates. J. Exp. Med. 2010, 207, 2313–2322. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.R.; Galloway-Pena, J.; Sahasrabhojane, P.; Saldana, M.; Yao, H.; Su, X.; Ajami, N.J.; Holder, M.E.; Petrosino, J.F.; Thompson, E.; et al. Sequence type 1 group B Streptococcus, an emerging cause of invasive disease in adults, evolves by small genetic changes. Proc. Natl. Acad. Sci. USA 2015, 112, 6431–6436. [Google Scholar] [CrossRef] [PubMed]
- Marttinen, P.; Hanage, W.P.; Croucher, N.J.; Connor, T.R.; Harris, S.R.; Bentley, S.D.; Corander, J. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012, 40, e6. [Google Scholar] [CrossRef] [PubMed]
- Seifert, K.N.; McArthur, W.P.; Bleiweis, A.S.; Brady, L.J. Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: Surface localization, enzymatic activity, and protein-protein interactions. Can. J. Microbiol. 2003, 49, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Madureira, P.; Baptista, M.; Vieira, M.; Magalhaes, V.; Camelo, A.; Oliveira, L.; Ribeiro, A.; Tavares, D.; Trieu-Cuot, P.; Vilanova, M.; et al. Streptococcus agalactiae GAPDH is a virulence-associated immunomodulatory protein. J. Immunol. 2007, 178, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Bryan, J.D.; Shelver, D.W. Streptococcus agalactiae CspA is a serine protease that inactivates chemokines. J. Bacteriol. 2009, 191, 1847–1854. [Google Scholar] [CrossRef] [PubMed]
- Al Safadi, R.; Mereghetti, L.; Salloum, M.; Lartigue, M.F.; Virlogeux-Payant, I.; Quentin, R.; Rosenau, A. Two-component system rgfA/C activates the fbsb gene encoding major fibrinogen-binding protein in highly virulent CC17 clone group B Streptococcus. PLoS ONE 2011, 6, e14658. [Google Scholar] [CrossRef] [PubMed]
- Teatero, S.; Ramoutar, E.; McGeer, A.; Li, A.; Melano, R.G.; Wasserscheid, J.; Dewar, K.; Fittipaldi, N. Clonal complex 17 group B Streptococcus strains causing invasive disease in neonates and adults originate from the same genetic pool. Sci. Rep. 2016, 6, 20047. [Google Scholar] [CrossRef] [PubMed]
- Puymege, A.; Bertin, S.; Guedon, G.; Payot, S. Analysis of Streptococcus agalactiae pan-genome for prevalence, diversity and functionality of integrative and conjugative or mobilizable elements integrated in the trna(lys ctt) gene. Mol. Genet. Genom. MGG 2015, 290, 1727–1740. [Google Scholar] [CrossRef] [PubMed]
- Puliti, M.; Uematsu, S.; Akira, S.; Bistoni, F.; Tissi, L. Toll-like receptor 2 deficiency is associated with enhanced severity of group B streptococcal disease. Infect. Immun. 2009, 77, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Wennekamp, J.; Henneke, P. Induction and termination of inflammatory signaling in group B streptococcal sepsis. Immunol. Rev. 2008, 225, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Landwehr-Kenzel, S.; Henneke, P. Interaction of Streptococcus agalactiae and cellular innate immunity in colonization and disease. Front. Immunol. 2014, 5, 519. [Google Scholar] [CrossRef] [PubMed]
- Biondo, C.; Mancuso, G.; Midiri, A.; Signorino, G.; Domina, M.; Lanza Cariccio, V.; Venza, M.; Venza, I.; Teti, G.; Beninati, C. Essential role of interleukin-1 signaling in host defenses against group B Streptococcus. mBio 2014, 5, e01428-14. [Google Scholar] [CrossRef] [PubMed]
- Puliti, M.; Bistoni, F.; Orefici, G.; Tissi, L. Exacerbation of group B streptococcal sepsis and arthritis in diabetic mice. Microbes Infect. 2006, 8, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Fahey, T.J., 3rd; Tracey, K.J.; Tekamp-Olson, P.; Cousens, L.S.; Jones, W.G.; Shires, G.T.; Cerami, A.; Sherry, B. Macrophage inflammatory protein 1 modulates macrophage function. J. Immunol. 1992, 148, 2764–2769. [Google Scholar] [PubMed]
- Didier, P.J.; Paradis, T.J.; Gladue, R.P. The CC chemokine MIP-1alpha induces a selective monocyte infiltration following intradermal injection into nonhuman primates. Inflammation 1999, 23, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Lindell, D.M.; Standiford, T.J.; Mancuso, P.; Leshen, Z.J.; Huffnagle, G.B. Macrophage inflammatory protein 1alpha/CCL3 is required for clearance of an acute Klebsiella pneumoniae pulmonary infection. Infect. Immun. 2001, 69, 6364–6369. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, M.A.; Gargiulo, F.; Negrini, R.; Gelmi, M.; Manca, N. Different sequence strains of Streptococcus agalactiae elicit various levels of cytokine production. Immunol. Investig. 2008, 37, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Lemire, P.; Calzas, C.; Segura, M. The NOD2 receptor does not play a major role in the pathogenesis of group B Streptococcus in mice. Microb. Pathog. 2013, 65, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Teatero, S.; McGeer, A.; Li, A.; Gomes, J.; Seah, C.; Demczuk, W.; Martin, I.; Wasserscheid, J.; Dewar, K.; Melano, R.G.; et al. Population structure and antimicrobial resistance of invasive serotype IV group B Streptococcus, Toronto, Ontario, Canada. Emerg. Infect. Dis. 2015, 21, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, J.; Yang, J.; Sun, S.; Xiao, J.; Yu, J. Pgap: Pan-genomes analysis pipeline. Bioinformatics 2012, 28, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Nusbaum, C.; Ohsumi, T.K.; Gomez, J.; Aquadro, J.; Victor, T.C.; Warren, R.M.; Hung, D.T.; Birren, B.W.; Lander, E.S.; Jaffe, D.B. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat. Methods 2009, 6, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. Blast ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]




| Strain | ST a | No. of SNPs b |
|---|---|---|
| NGBS572 | 452 | 15,244 |
| NEM316 | 23 | 13,951 |
| A909 | 7 | 13,800 |
| NGBS061 | 459 | 11,142 |
| 2603V/R | 110 | 10,987 |
| NGBS128 | 17 | 21,022 |
| NGBS357 | 1 | 2645 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teatero, S.; Lemire, P.; Dewar, K.; Wasserscheid, J.; Calzas, C.; Mallo, G.V.; Li, A.; Athey, T.B.T.; Segura, M.; Fittipaldi, N. Genomic Recombination Leading to Decreased Virulence of Group B Streptococcus in a Mouse Model of Adult Invasive Disease. Pathogens 2016, 5, 54. https://doi.org/10.3390/pathogens5030054
Teatero S, Lemire P, Dewar K, Wasserscheid J, Calzas C, Mallo GV, Li A, Athey TBT, Segura M, Fittipaldi N. Genomic Recombination Leading to Decreased Virulence of Group B Streptococcus in a Mouse Model of Adult Invasive Disease. Pathogens. 2016; 5(3):54. https://doi.org/10.3390/pathogens5030054
Chicago/Turabian StyleTeatero, Sarah, Paul Lemire, Ken Dewar, Jessica Wasserscheid, Cynthia Calzas, Gustavo V. Mallo, Aimin Li, Taryn B.T. Athey, Mariela Segura, and Nahuel Fittipaldi. 2016. "Genomic Recombination Leading to Decreased Virulence of Group B Streptococcus in a Mouse Model of Adult Invasive Disease" Pathogens 5, no. 3: 54. https://doi.org/10.3390/pathogens5030054