TNFRSF1B Signaling Blockade Protects Airway Epithelial Cells from Oxidative Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
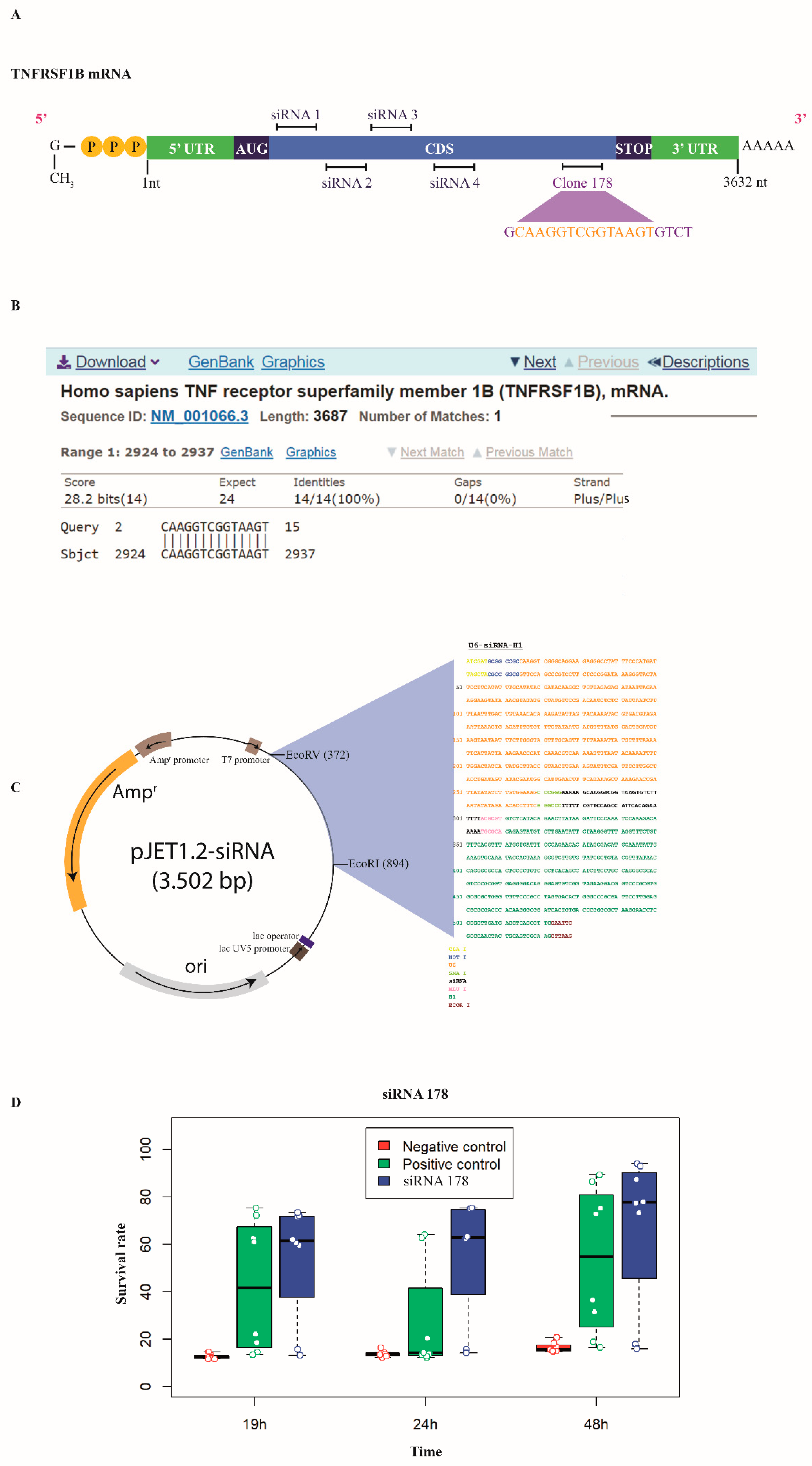
2.2. Cloning of Anti-TNFRSF1B siRNA
2.3. Reverse Cell Transfection and Short-Term Selection
2.4. Differential Gene Expression Analysis
2.5. Validation of siRNA-Mediated Knock-Down of TNFRSF1B Expression
2.6. Bioluminescent Reporter Assay
2.7. Cell Death Induction Trough Oxidative Stress
2.8. Identification of Putative siRNA Targets
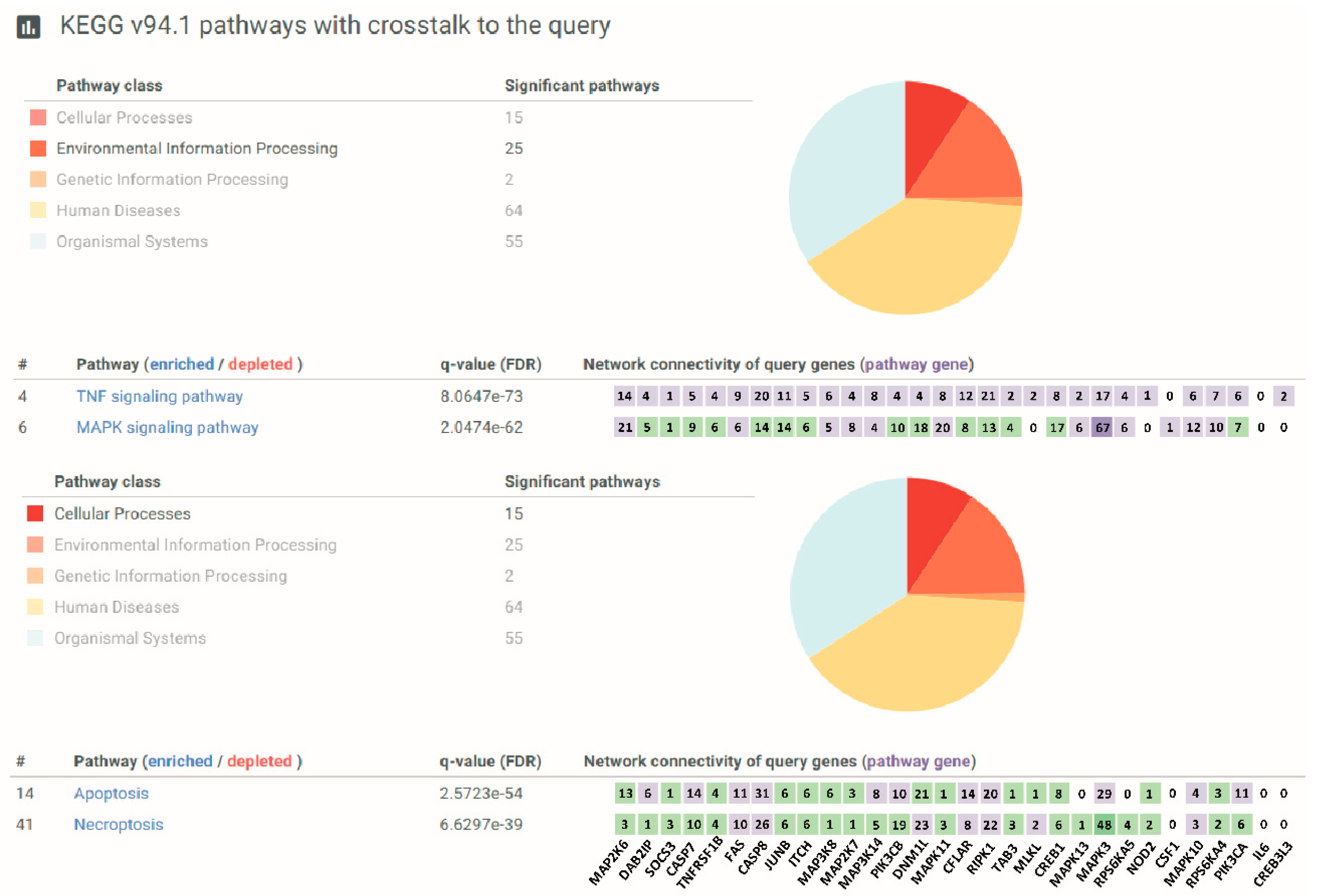
2.9. Pathways Overrepresentation Analysis (ORA)
2.10. Statistical Analysis
3. Results
3.1. The Involvement of the TNF Pathway and the TNFRSF1B Gene in Oxidative Stress in CF Airway Epithelial Cells
3.2. TNFRSF1B Transcript Knock-Down Induces Resistance to Oxidative Stress in Airway Epithelial Cells
3.3. The Levels of Lymphotoxins (LTs) Are Modulated by Oxidative Stress in Airway Epithelial Cells
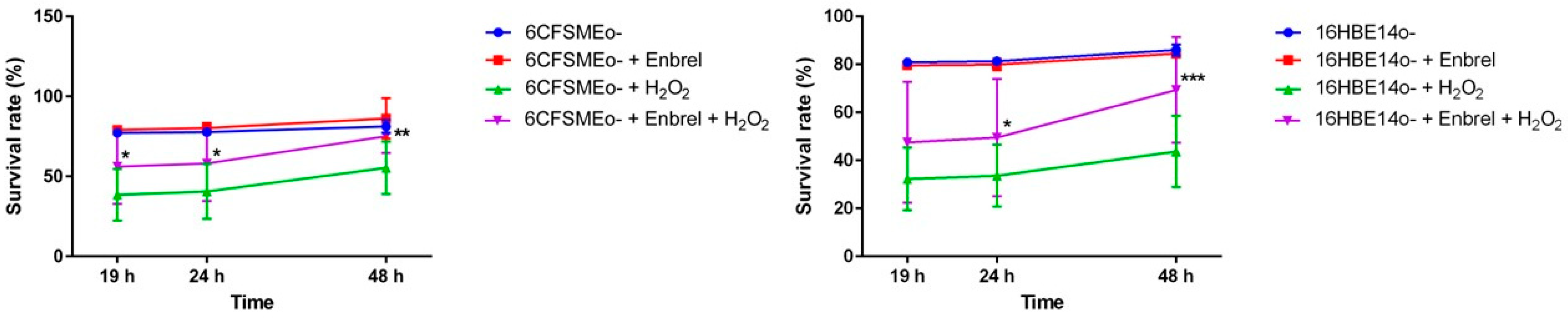
3.4. The Anti-TNF Biologic Etanercept Increases the Cell Survival of Airway Epithelial Cells under Oxidative Stress Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Csanády, L.; Vergani, P.; Gadsby, D.C. Structure, gating, and regulation of the CFTR anion channel. Physiol. Rev. 2019, 99, 707–738. [Google Scholar] [CrossRef]
- Shteinberg, M.; Haq, I.J.; Polineni, D.; Davies, J.C. Cystic fibrosis. Lancet 2021, 397, 2195–2211. [Google Scholar] [CrossRef]
- Haq, I.J.; Gray, M.A.; Garnett, J.P.; Ward, C.; Brodlie, M. Airway surface liquid homeostasis in cystic fibrosis: Pathophysiology and therapeutic targets. Thorax 2016, 71, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C.B.; Markovetz, M.R.; Ehre, C. Mucus, mucins, and cystic fibrosis. Pediatr. Pulmonol. 2019, 54 (Suppl. S3), S84–S96. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.T.; Yang, C.M. Role of NADPH oxidase/ROS in pro-inflammatory mediators-induced airway and pulmonary diseases. Biochem. Pharmacol. 2012, 84, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Checa, J.; Aran, J.M. Airway Redox Homeostasis and Inflammation Gone Awry: From Molecular Pathogenesis to Emerging Therapeutics in Respiratory Pathology. Int. J. Mol. Sci. 2020, 21, 9317. [Google Scholar] [CrossRef]
- Kogan, I.; Ramjeesingh, M.; Li, C.; Kidd, J.F.; Wang, Y.; Leslie, E.M.; Cole, S.P.C.; Bear, C.E. CFTR directly mediates nucleotide-regulated glutathione flux. EMBO J. 2003, 22, 1981–1989. [Google Scholar] [CrossRef]
- Checa, J.; Martínez-González, I.; Maqueda, M.; Mosquera, J.L.; Aran, J.M. Genome-wide RNAi screening identifies novel pathways/genes involved in oxidative stress and repurposable drugs to preserve cystic fibrosis airway epithelial cell integrity. Antioxidants 2021, 10, 1936. [Google Scholar] [CrossRef]
- Medler, J.; Wajant, H. Tumor necrosis factor receptor-2 (TNFR2): An overview of an emerging drug target. Expert Opin. Ther. Targets 2019, 23, 295–307. [Google Scholar] [CrossRef]
- MacEwan, D.J. TNF ligands and receptors—A matter of life and death. Br. J. Pharmacol. 2002, 135, 855–875. [Google Scholar] [CrossRef]
- Sedger, L.M.; McDermott, M.F. TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants-past, present and future. Cytokine Growth Factor Rev. 2014, 25, 453–472. [Google Scholar] [CrossRef]
- Gruenert, D.C.; Willems, M.; Cassiman, J.J.; Frizzell, R.A. Established cell lines used in cystic fibrosis research. J. Cyst. Fibros. 2004, 3, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.; Cairns, M.J.; Dawes, I.W.; Arndt, G.M. Expressing functional siRNAs in mammalian cells using convergent transcription. BMC Biotechnol. 2003, 3, 21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Ogris, C.; Helleday, T.; Sonnhammer, E.L.L. PathwAX: A web server for network crosstalk based pathway annotation. Nucleic Acids Res. 2016, 44, W105–W109. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Aguade-Gorgorio, J.; McComb, S.; Eckert, C.; Guinot, A.; Marovca, B.; Mezzatesta, C.; Jenni, S.; Abduli, L.; Schrappe, M.; Dobay, M.P.; et al. TNFR2 is required for RIP1-dependent cell death in human leukemia. Blood Adv. 2020, 4, 4823–4833. [Google Scholar] [CrossRef]
- Gough, P.; Myles, I.A. Tumor Necrosis Factor Receptors: Pleiotropic Signaling Complexes and Their Differential Effects. Front. Immunol. 2020, 11, 585880. [Google Scholar] [CrossRef] [PubMed]
- Checa, J.; Aran, J.M. Reactive Oxygen Species: Drivers of Physiological and Pathological Processes. J. Inflamm. Res. 2020, 13, 1057–1073. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Y.; Mei, K.; Zhang, S.; Sun, X.; Ren, F.; Liu, S.; Yang, Z.; Wang, X.; Qin, Z.; et al. Tumor necrosis factor receptor 2 (TNFR2)•interleukin-17 receptor d (IL-17RD) heteromerization reveals a novel mechanism for NF-κB activation. J. Biol. Chem. 2015, 290, 861–871. [Google Scholar] [CrossRef]
- Wajant, H.; Pfizenmaier, K.; Scheurich, P. Tumor necrosis factor signaling. Cell Death Differ. 2003, 10, 45–65. [Google Scholar] [CrossRef] [PubMed]
- Joardar, S.; Dewanjee, S.; Bhowmick, S.; Dua, T.K.; Das, S.; Saha, A.; De Feo, V. Rosmarinic acid attenuates cadmium-induced nephrotoxicity via inhibition of oxidative stress, apoptosis, inflammation and fibrosis. Int. J. Mol. Sci. 2019, 20, 2027. [Google Scholar] [CrossRef] [PubMed]
- Ware, C.F.; VanArsdale, S.; VanArsdale, T.L. Apoptosis mediated by the TNF-related cytokine and receptor families. J. Cell. Biochem. 1996, 60, 47–55. [Google Scholar] [CrossRef]
- Ware, C.F. Network communications: Lymphotoxins, LIGHT, and TNF. Annu. Rev. Immunol. 2005, 23, 787–819. [Google Scholar] [CrossRef]
- Idriss, H.T.; Naismith, J.H. TNFα and the TNF receptor superfamily: Structure-function relationship(s). Microsc. Res. Tech. 2000, 50, 184–195. [Google Scholar] [CrossRef]
- Bodmer, J.L.; Schneider, P.; Tschopp, J. The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 2002, 27, 19–26. [Google Scholar] [CrossRef]
- Etemadi, N.; Holien, J.K.; Chau, D.; Dewson, G.; Murphy, J.M.; Alexander, W.S.; Parker, M.W.; Silke, J.; Nachbur, U. Lymphotoxin α induces apoptosis, necroptosis and inflammatory signals with the same potency as tumour necrosis factor. FEBS J. 2013, 280, 5283–5297. [Google Scholar] [CrossRef]
- Scott, L.J. Etanercept: A review of its use in autoimmune inflammatory diseases. Drugs 2014, 74, 1379–1410. [Google Scholar] [CrossRef] [PubMed]
- Visser, S.; Martin, M.; Serisier, D.J. Improvements in cystic fibrosis lung disease and airway inflammation associated with etanercept therapy for rheumatoid arthritis: A case report. Lung 2012, 190, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Adelsten, T.; Rasmussen, N.; Katzenstein, T.L.; Nielsen, C.T. Safe and effective tumour necrosis factor-α inhibitor (etanercept) treatment of chronic episodic arthritis in a patient with cystic fibrosis. Scand. J. Rheumatol. 2016, 45, 330–331. [Google Scholar] [CrossRef] [PubMed]
- Dudchenko, O.; Ordovas-Montanes, J.; Bingle, C.D. Respiratory epithelial cell types tates and fates in the era of single-cell RNA-sequencing. Biochem. J. 2023, 480, 921–939. [Google Scholar] [CrossRef]


| Primer Sequences | |||
|---|---|---|---|
| Name | Sequence (5′ → 3′) | Number of nt | |
| ClaI/NotI F | CCGCGCATCGATGCGGCCGCCAAGGTCGGGCAGGAAGAGGGCCTA | 45 | |
| EcoRI-H1 R | CGCGAATTCGAACGCTGACGTCATCAACCCGCTCCAAGGAATCGC | 45 | |
| pJET 1.2 F | CGACTCACTATAGGGAGAGCGGC | 23 | |
| pJET 1.2 R | AAGAACATCGATTTTCCATGGCAG | 24 | |
| TNFR2 F | GCGCGCTAGCTAGTTCGGGAACAGAACCGCATC | 33 | |
| TNFR2 R | GCGCGTCGACTAGTGGCCTTATCGGCAGGCAAGT | 34 | |
| Pre-Designed siRNAs (TNFRSF1B) | |||
| siRNA Name | Sequence (5′ → 3′) | ||
| ON-TARGET plus Non-targeting siRNA (C-) | UGGUUUACAUGUCGACUAA | ||
| ON-TARGET plus SMART pool siRNA J-003934-05 (siRNA1) | CGACUUCGCUCUUCCAGUU | ||
| ON-TARGET plus SMART pool siRNA J-003934-06 (siRNA2) | GGAAUGUGCCUUUCGGUCA | ||
| ON-TARGET plus SMART pool siRNA J-003934-07 (siRNA3) | CAUCAGACGUGGUGUGCAA | ||
| ON-TARGET plus SMART pool siRNA J-003934-08 (siRNA4) | AGCCUUGGGUCUACUAAUA | ||
| Target Symbol (Accession #) | Target Entrez | siRNA id * | Homology (%) * |
|---|---|---|---|
| DAB2IP (NM_138709) | 153090 | 39/66/132 | 63/63/73 |
| RPS6KA4 (NM_003942) | 8986 | 146/172 | 63/73 |
| TNFRSF1B (NM_001066) | 7133 | 178 | 73 |
| SOCS3 (NM_003955) | 9021 | 139 | 63 |
| NOD2 (NM_022162) | 64127 | 161 | 68 |
| RPS6KA5 (NM_182398) | 9252 | 6 | 68 |
| MAP2K7 (NM_145185) | 5609 | 29 | 68 |
| CASP8 (NM_001228) | 841 | 122 | 78 |
| IL6 (NM_000600) | 3569 | 32 | 89 (+1 mismatch) |
| CSF1 (NM_000757) | 1435 | 132 | 73 |
| MAP3K14 (NM_003954) | 9020 | 8/136 | 73/63 |
| PIK3CA (NM_006218) | 5290 | 176 | 68 |
| MAPK11 (NM_002751) | 5600 | 141 | 73 |
| MAPK3 (NM_002746) | 5595 | 54 | 68 |
| MAPK10 (NM_002753) | 5602 | 20/26/147 | 73/63/63 |
| MAP2K6 (NM_002758) | 5608 | 75/112 | 63/63 |
| CFLAR (NM_003879) | 8837 | 113 | 63 |
| FAS (NM_000043) | 355 | 119 | 63 |
| CASP7 (NM_001227) | 840 | 124 | 63 |
| RIPK1 (NM_003804) | 8737 | 125 | 63 |
| MLKL (NM_152649) | 197259 | 146 | 63 |
| JUNB (NM_002229) | 3726 | 148 | 63 |
| CREB3L3 (NM_032607) | 84699 | 149 | 63 |
| ITCH (NM_031483) | 83737 | 153 | 63 |
| PIK3CB (NM_006219) | 5291 | 27/56 | 63/68 |
| MAP3K8 (NM_005204) | 1326 | 52 | 63 |
| DNM1L (NM_012062) | 10059 | 73 | 63 |
| LIF (NM_002309) | 3976 | 8 | 73 |
| MAPK13 (NM_002754) | 5603 | 83 | 63 |
| TAB3 (NM_152787) | 257397 | 89 | 63 |
| CREB1 (NM_004379) | 1385 | 96 | 63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Checa, J.; Fiol, P.; Guevara, M.; Aran, J.M. TNFRSF1B Signaling Blockade Protects Airway Epithelial Cells from Oxidative Stress. Antioxidants 2024, 13, 368. https://doi.org/10.3390/antiox13030368
Checa J, Fiol P, Guevara M, Aran JM. TNFRSF1B Signaling Blockade Protects Airway Epithelial Cells from Oxidative Stress. Antioxidants. 2024; 13(3):368. https://doi.org/10.3390/antiox13030368
Chicago/Turabian StyleCheca, Javier, Pau Fiol, Marta Guevara, and Josep M. Aran. 2024. "TNFRSF1B Signaling Blockade Protects Airway Epithelial Cells from Oxidative Stress" Antioxidants 13, no. 3: 368. https://doi.org/10.3390/antiox13030368
APA StyleCheca, J., Fiol, P., Guevara, M., & Aran, J. M. (2024). TNFRSF1B Signaling Blockade Protects Airway Epithelial Cells from Oxidative Stress. Antioxidants, 13(3), 368. https://doi.org/10.3390/antiox13030368







