Bifunctional Chloroplastic DJ-1B from Arabidopsis thaliana is an Oxidation-Robust Holdase and a Glyoxalase Sensitive to H2O2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cloning and Purification of Recombinant AtDJ-1B
2.2. Circular Dichroism (CD)
2.3. Thermal Unfolding
2.4. Glyoxalase Assay
2.5. Chaperone Assay
2.6. Oxidation of AtDJ-1B for Mass Spectrometric Analysis
2.7. Determination of Cysteine Oxidation States by Mass Spectrometry
2.8. Gene Expression Levels
2.9. Plant Material
2.10. RT-qPCR
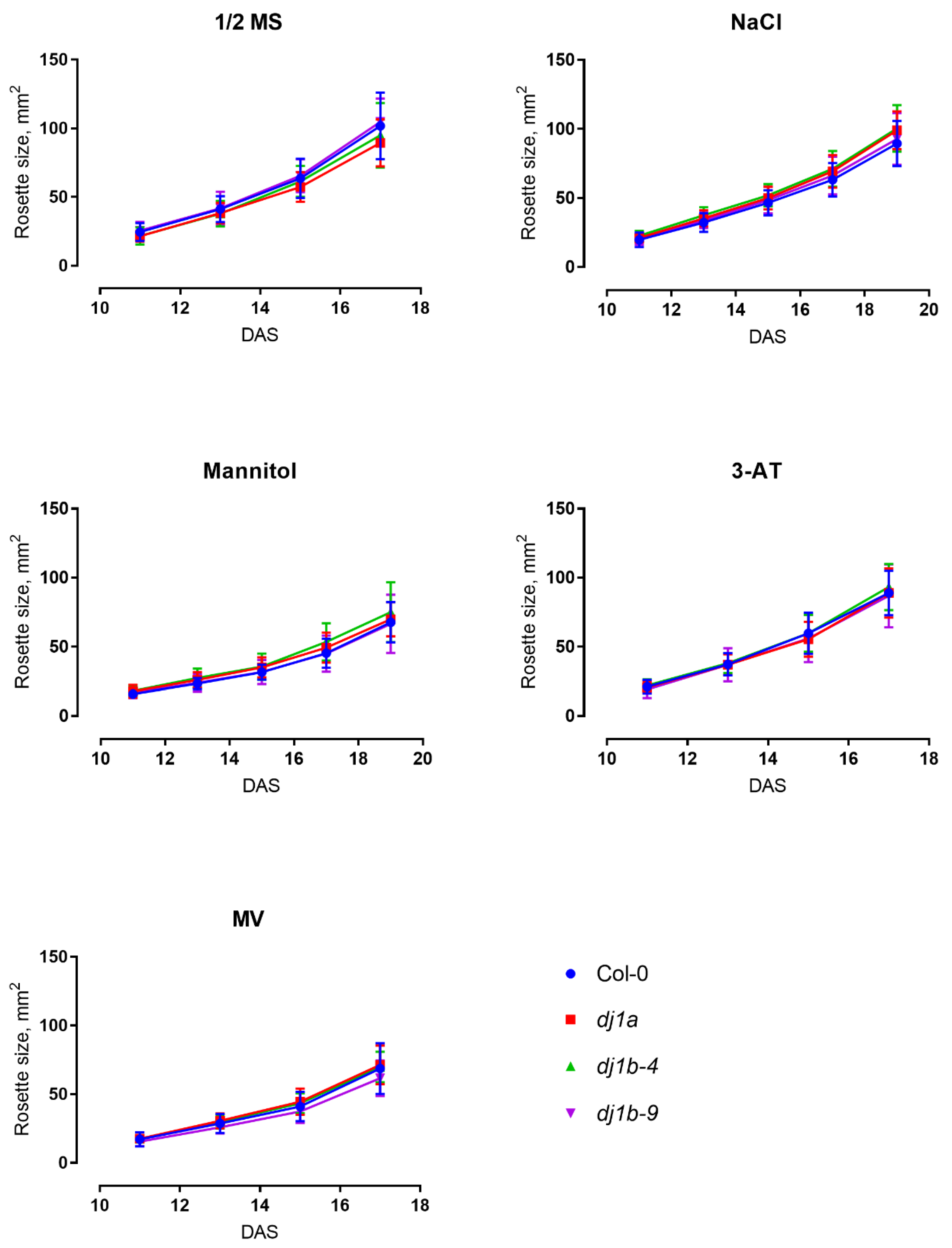
2.11. Growth Conditions and Plant Stress Assays
3. Results
3.1. AtDJ-1B Contains Multiple Oxidant-Sensitive Cysteines
3.2. Oxidized AtDJ-1B Becomes More Thermostable
3.3. Oxidation Inactivates the Glyoxalase Activity of AtDJ-1B
3.4. Oxidation does not Affect the Chaperone Activity of AtDJ-1B
3.5. DJ-1B-Deficient Plants are Phenotypically Identical to Wildtype
3.6. Transcript Levels of DJ-1B are Stress-Independent
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Vistoli, G.; De Maddis, D.; Cipak, A.; Zarkovic, N.; Carini, M.; Aldini, G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): An overview of their mechanisms of formation. Free Radic. Res. 2013, 47 (Suppl. 1), 3–27. [Google Scholar] [CrossRef] [PubMed]
- Wetzels, S.; Wouters, K.; Schalkwijk, C.G.; Vanmierlo, T.; Hendriks, J.J. Methylglyoxal-Derived Advanced Glycation Endproducts in Multiple Sclerosis. Int. J. Mol. Sci. 2017, 18, 421. [Google Scholar] [CrossRef] [PubMed]
- Chetyrkin, S.; Mathis, M.; Pedchenko, V.; Sanchez, O.A.; McDonald, W.H.; Hachey, D.L.; Madu, H.; Stec, D.; Hudson, B.; Voziyan, P. Glucose autoxidation induces functional damage to proteins via modification of critical arginine residues. Biochemistry 2011, 50, 6102–6112. [Google Scholar] [CrossRef] [PubMed]
- Takagi, D.; Inoue, H.; Odawara, M.; Shimakawa, G.; Miyake, C. The Calvin cycle inevitably produces sugar-derived reactive carbonyl methylglyoxal during photosynthesis: A potential cause of plant diabetes. Plant Cell Physiol. 2014, 55, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, J.; Dittmar, I.C.; Brockmann, J.D.; Schmidt, M.; Hudig, M.; Rossoni, A.W.; Maurino, V.G. Defense against Reactive Carbonyl Species Involves at Least Three Subcellular Compartments Where Individual Components of the System Respond to Cellular Sugar Status. Plant Cell 2017, 29, 3234–3254. [Google Scholar] [CrossRef] [PubMed]
- Misra, K.; Banerjee, A.B.; Ray, S.; Ray, M. Glyoxalase III from Escherichia coli: A single novel enzyme for the conversion of methylglyoxal into D-lactate without reduced glutathione. Biochem. J. 1995, 305 Pt 3, 999–1003. [Google Scholar] [CrossRef]
- Kahle, P.J.; Waak, J.; Gasser, T. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic. Biol. Med. 2009, 47, 1354–1361. [Google Scholar] [CrossRef]
- Shendelman, S.; Jonason, A.; Martinat, C.; Leete, T.; Abeliovich, A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004, 2, e362. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Song, J.; Kwon, K.; Jang, S.; Kim, C.; Baek, K.; Kim, J.; Park, C. Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 2012, 21, 3215–3225. [Google Scholar] [CrossRef] [Green Version]
- Richarme, G.; Liu, C.; Mihoub, M.; Abdallah, J.; Leger, T.; Joly, N.; Liebart, J.-C.; Jurkunas, U.V.; Nadal, M.; Bouloc, P.; et al. Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science 2017, 357, 208–211. [Google Scholar] [CrossRef]
- Hod, Y.; Pentyala, S.N.; Whyard, T.C.; El-Maghrabi, M.R. Identification and characterization of a novel protein that regulates RNA-protein interaction. J. Cell Biochem. 1999, 72, 435–444. [Google Scholar] [CrossRef]
- Kwon, K.; Choi, D.; Hyun, J.K.; Jung, H.S.; Baek, K.; Park, C. Novel glyoxalases from Arabidopsis thaliana. FEBS J. 2013, 280, 3328–3339. [Google Scholar] [CrossRef] [PubMed]
- Kinumi, T.; Kimata, J.; Taira, T.; Ariga, H.; Niki, E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2004, 317, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Canet-Aviles, R.M.; Wilson, M.A.; Miller, D.W.; Ahmad, R.; McLendon, C.; Bandyopadhyay, S.; Baptista, M.J.; Ringe, D.; Petsko, G.A.; Cookson, M.R. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 2004, 101, 9103–9108. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Lin, H.; Maple, J.; Björkblom, B.; Alves, G.; Larsen, J.P.; Møller, S.G. The Arabidopsis DJ-1a protein confers stress protection through cytosolic SOD activation. J Cell Sci 2010, 123, 1644–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, L.; Luchinat, E.; Banci, L. Intracellular metal binding and redox behavior of human DJ-1. J. Biol. Inorg. Chem. 2018, 23, 61–69. [Google Scholar] [CrossRef]
- Lin, J.; Nazarenus, T.J.; Frey, J.L.; Liang, X.; Wilson, M.A.; Stone, J.M. A plant DJ-1 homolog is essential for Arabidopsis thaliana chloroplast development. PLoS ONE 2011, 6, e23731. [Google Scholar] [CrossRef]
- Akter, S.; Huang, J.; Bodra, N.; De Smet, B.; Wahni, K.; Rombaut, D.; Pauwels, J.; Gevaert, K.; Carroll, K.; Van Breusegem, F.; et al. DYn-2 based identification of Arabidopsis sulfenomes. Mol. Cell. Proteom. 2015, 14, 1183–1200. [Google Scholar] [CrossRef]
- De Smet, B.; Willems, P.; Fernandez-Fernandez, A.D.; Alseekh, S.; Fernie, A.R.; Messens, J.; Van Breusegem, F. In vivo detection of protein cysteine sulfenylation in plastids. Plant J. 2018. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Provencher, S.W.; Glöckner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981, 20, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Lobley, A.; Whitmore, L.; Wallace, B.A. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 2002, 18, 211–212. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, W668–W673. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, C.; Ponces Freire, A. Methylglyoxal assay in cells as 2-methylquinoxaline using 1,2-diaminobenzene as derivatizing reagent. Anal. Biochem. 1996, 234, 221–224. [Google Scholar] [CrossRef]
- Choi, S.; Jeong, J.; Na, S.; Lee, H.S.; Kim, H.Y.; Lee, K.J.; Paek, E. New algorithm for the identification of intact disulfide linkages based on fragmentation characteristics in tandem mass spectra. J. Proteome Res. 2010, 9, 626–635. [Google Scholar] [CrossRef]
- Kerchev, P.; Waszczak, C.; Lewandowska, A.; Willems, P.; Shapiguzov, A.; Li, Z.; Alseekh, S.; Mühlenbock, P.; Hoeberichts, F.A.; Huang, J.; et al. Lack of GLYCOLATE OXIDASE1, but not GLYCOLATE OXIDASE2, attenuates the photorespiratory phenotype of catalase2-deficient arabidopsis. Plant Physiol. 2016, 171, 1704–1719. [Google Scholar] [CrossRef]
- Zhang, X.; Ivanova, A.; Vandepoele, K.; Radomiljac, J.; Van de Velde, J.; Berkowitz, O.; Willems, P.; Xu, Y.; Ng, S.; Van Aken, O.; et al. The Transcription Factor MYB29 Is a Regulator of ALTERNATIVE OXIDASE1a. Plant Physiol. 2017, 173, 1824–1843. [Google Scholar] [CrossRef]
- Kerchev, P.I.; De Clercq, I.; Denecker, J.; Muhlenbock, P.; Kumpf, R.; Nguyen, L.; Audenaert, D.; Dejonghe, W.; Van Breusegem, F. Mitochondrial perturbation negatively affects auxin signaling. Mol. Plant 2014, 7, 1138–1150. [Google Scholar] [CrossRef]
- Waszczak, C.; Kerchev, P.I.; Muhlenbock, P.; Hoeberichts, F.A.; Van Der Kelen, K.; Mhamdi, A.; Willems, P.; Denecker, J.; Kumpf, R.P.; Noctor, G.; et al. SHORT-ROOT Deficiency Alleviates the Cell Death Phenotype of the Arabidopsis catalase2 Mutant under Photorespiration-Promoting Conditions. Plant Cell 2016, 28, 1844–1859. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerchev, P.; Muhlenbock, P.; Denecker, J.; Morreel, K.; Hoeberichts, F.A.; Van Der Kelen, K.; Vandorpe, M.; Nguyen, L.; Audenaert, D.; Van Breusegem, F. Activation of auxin signalling counteracts photorespiratory H2O2-dependent cell death. Plant Cell Environ. 2015, 38, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid. Redox Signal. 2011, 15, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Lakshminarasimhan, M.; Maldonado, M.T.; Zhou, W.; Fink, A.L.; Wilson, M.A. Structural impact of three Parkinsonism-associated missense mutations on human DJ-1. Biochemistry 2008, 47, 1381–1392. [Google Scholar] [CrossRef]
- Hasim, S.; Hussin, N.A.; Alomar, F.; Bidasee, K.R.; Nickerson, K.W.; Wilson, M.A. A glutathione-independent glyoxalase of the DJ-1 superfamily plays an important role in managing metabolically generated methylglyoxal in Candida albicans. J. Biol. Chem. 2014, 289, 1662–1674. [Google Scholar] [CrossRef]
- Bankapalli, K.; Saladi, S.; Awadia, S.S.; Goswami, A.V.; Samaddar, M.; D’Silva, P. Robust glyoxalase activity of Hsp31, a ThiJ/DJ-1/PfpI family member protein, is critical for oxidative stress resistance in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 26491–26507. [Google Scholar] [CrossRef]
- Tsai, C.-J.; Aslam, K.; Drendel, H.M.; Asiago, J.M.; Goode, K.M.; Paul, L.N.; Rochet, J.-C.; Hazbun, T.R. Hsp31 is a stress response chaperone that intervenes in the protein misfolding process. J. Biol. Chem. 2015, 290, 24816–24834. [Google Scholar] [CrossRef]
- Amm, I.; Norell, D.; Wolf, D.H. Absence of the Yeast Hsp31 chaperones of the DJ-1 superfamily perturbs cytoplasmic protein quality control in late growth phase. PLoS ONE 2015, 10, e0140363. [Google Scholar] [CrossRef]
- Aslam, K.; Hazbun, T.R. Hsp31, a member of the DJ-1 superfamily, is a multitasking stress responder with chaperone activity. Prion 2016, 10, 103–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willems, P.; Mhamdi, A.; Stael, S.; Storme, V.; Kerchev, P.; Noctor, G.; Gevaert, K.; Van Breusegem, F. The ROS wheel: Refining ROS transcriptional footprints. Plant Physiol. 2016, 171, 1720–1733. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A. Genome-Wide Identification of Glyoxalase Genes in Medicago truncatula and Their Expression Profiling in Response to Various Developmental and Environmental Stimuli. Front. Plant Sci. 2017, 8, 836. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kim, J.; Ha, S.; Kwon, K.; Kim, E.-H.; Lee, H.-Y.; Ryu, K.-S.; Park, C. Stereospecific mechanism of DJ-1 glyoxalases inferred from their hemithioacetal-containing crystal structures. FEBS J. 2014, 281, 5447–5462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 2005, 337, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Melvin, P.; Bankapalli, K.; D’Silva, P.; Shivaprasad, P.V. Methylglyoxal detoxification by a DJ-1 family protein provides dual abiotic and biotic stress tolerance in transgenic plants. Plant Mol. Biol. 2017, 94, 381–397. [Google Scholar] [CrossRef]
- Saito, R.; Yamamoto, H.; Makino, A.; Sugimoto, T.; Miyake, C. Methylglyoxal functions as Hill oxidant and stimulates the photoreduction of O(2) at photosystem I: A symptom of plant diabetes. Plant Cell Environ. 2011, 34, 1454–1464. [Google Scholar] [CrossRef]
- Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J. 2014, 80, 93–105. [Google Scholar] [CrossRef] [Green Version]
- Haslbeck, M.; Buchner, J. Assays to characterize molecular chaperone function in vitro. Methods Mol. Biol. 2015, 1292, 39–51. [Google Scholar] [CrossRef]
- Lin, J.; Prahlad, J.; Wilson, M.A. Conservation of oxidative protein stabilization in an insect homologue of parkinsonism-associated protein DJ-1. Biochemistry 2012, 51, 3799–3807. [Google Scholar] [CrossRef] [PubMed]
- Culleton, B.A.; Lall, P.; Kinsella, G.K.; Doyle, S.; McCaffrey, J.; Fitzpatrick, D.A.; Burnell, A.M. A role for the Parkinson’s disease protein DJ-1 as a chaperone and antioxidant in the anhydrobiotic nematode Panagrolaimus superbus. Cell Stress Chaperones 2015, 20, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Richarme, G.; Marguet, E.; Forterre, P.; Ishino, S.; Ishino, Y. DJ-1 family Maillard deglycases prevent acrylamide formation. Biochem. Biophys. Res. Commun. 2016, 478, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Mihoub, M.; Abdallah, J.; Gontero, B.; Dairou, J.; Richarme, G. The DJ-1 superfamily member Hsp31 repairs proteins from glycation by methylglyoxal and glyoxal. Biochem. Biophys. Res. Commun. 2015, 463, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, J.; Mihoub, M.; Gautier, V.; Richarme, G. The DJ-1 superfamily members YhbO and YajL from Escherichia coli repair proteins from glycation by methylglyoxal and glyoxal. Biochem. Biophys. Res. Commun. 2016, 470, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Richarme, G.; Dairou, J. Parkinsonism-associated protein DJ-1 is a bona fide deglycase. Biochem. Biophys. Res. Commun. 2017, 483, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Arsovski, A.A.; Pradinuk, J.; Guo, X.Q.; Wang, S.; Adams, K.L. Evolution of Cis-Regulatory Elements and Regulatory Networks in Duplicated Genes of Arabidopsis. Plant Physiol. 2015, 169, 2982–2991. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef]
- Rensing, S.A. Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 2014, 17, 43–48. [Google Scholar] [CrossRef]
- Islam, T.; Ghosh, A. Genome-wide dissection and expression profiling of unique glyoxalase III genes in soybean reveal the differential pattern of transcriptional regulation. Sci. Rep. 2018, 8, 4848. [Google Scholar] [CrossRef]
- Ghosh, A.; Kushwaha, H.R.; Hasan, M.R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 2016, 6, 18358. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewandowska, A.; Vo, T.N.; Nguyen, T.-D.H.; Wahni, K.; Vertommen, D.; Van Breusegem, F.; Young, D.; Messens, J. Bifunctional Chloroplastic DJ-1B from Arabidopsis thaliana is an Oxidation-Robust Holdase and a Glyoxalase Sensitive to H2O2. Antioxidants 2019, 8, 8. https://doi.org/10.3390/antiox8010008
Lewandowska A, Vo TN, Nguyen T-DH, Wahni K, Vertommen D, Van Breusegem F, Young D, Messens J. Bifunctional Chloroplastic DJ-1B from Arabidopsis thaliana is an Oxidation-Robust Holdase and a Glyoxalase Sensitive to H2O2. Antioxidants. 2019; 8(1):8. https://doi.org/10.3390/antiox8010008
Chicago/Turabian StyleLewandowska, Aleksandra, Trung Nghia Vo, Thuy-Dung Ho Nguyen, Khadija Wahni, Didier Vertommen, Frank Van Breusegem, David Young, and Joris Messens. 2019. "Bifunctional Chloroplastic DJ-1B from Arabidopsis thaliana is an Oxidation-Robust Holdase and a Glyoxalase Sensitive to H2O2" Antioxidants 8, no. 1: 8. https://doi.org/10.3390/antiox8010008




