A Polyclonal Antibody against a Burkholderia cenocepacia OmpA-like Protein Strongly Impairs Pseudomonas aeruginosa and B. multivorans Virulence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Protein Expression and Purification
2.3. Cell Line and Cell Culture
2.4. Polyclonal Goat Antibodies
2.5. Western Blot Analyses
2.6. Adhesion and Invasion of Epithelial Cells
2.7. Planktonic Cells Aggregation Assay
2.8. Biofilm Formation Assays
2.9. Galleria mellonella Killing Assays
2.10. Protein Bioinformatics Analyses
2.11. Statistical Analysis
3. Results
3.1. Identification of BCAL2645 Homolog Proteins in the B. multivorans and P. aeruginosa Genomes
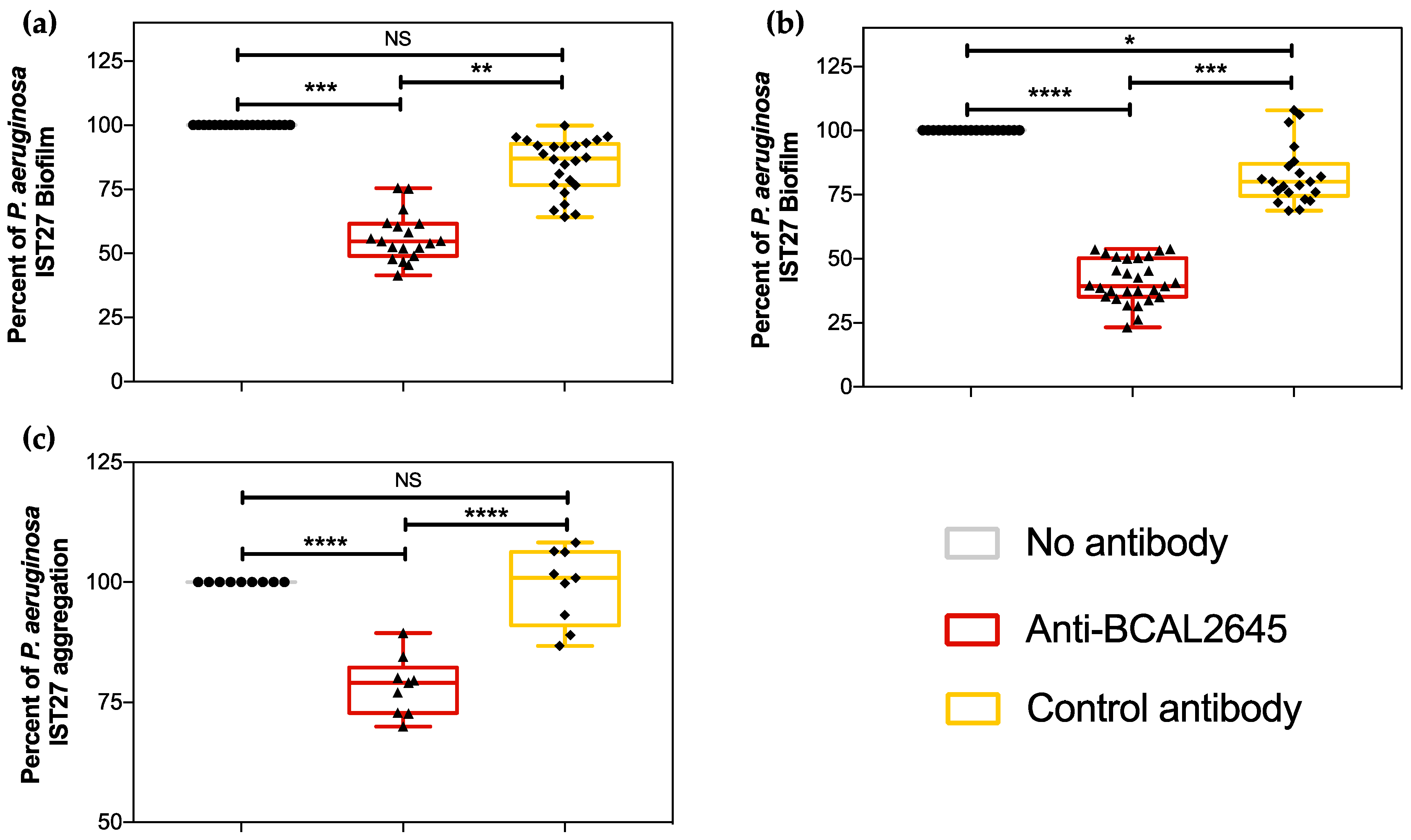
3.2. The Anti-BCAL2645 Polyclonal Antibody Interferes with P. aeruginosa Ability to Form Biofilms
3.3. The Anti-BCAL2645 Antibody Strongly Inhibits Adhesion and Invasion of Cultured Human Bronchial Epithelial Cells
3.4. The Anti-BCAL2645 Antibody Protects Galleria mellonella Larvae from B. cenocepacia, B. multivorans and P. aeruginosa Infections
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, J.; Garratt, A.; Hill, A. Worldwide rates of diagnosis and effective treatment for cystic fibrosis. J. Cyst. Fibros. 2022, 21, 456–462. [Google Scholar] [CrossRef]
- Bell, S.C.; De Boeck, K.; Amaral, M.D. New pharmacological approaches for cystic fibrosis: Promises, progress, pitfalls. Pharmacol. Ther. 2015, 145, 19–34. [Google Scholar] [CrossRef]
- Zolin, A.; Orenti, A.; Jung, A.; van Rens, J.; Prasad, V.; Fox, A.; Krasnyk, M.; Mayor, S.L.; Naehrlich, L.; Gkolia, P.; et al. CFSPR Annual Report 2021. Available online: https://www.ecfs.eu/sites/default/files/Annual%20Report_2021_09Jun2023_ECFSPR_final.pdf (accessed on 10 January 2024).
- Hoegger, M.J.; Fischer, A.J.; McMenimen, J.D.; Ostedgaard, L.S.; Tucker, A.J.; Awadalla, M.A.; Moninger, T.O.; Michalski, A.S.; Hoffman, E.A.; Zabner, J.; et al. Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science 2014, 345, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Drevinek, P.; Mahenthiralingam, E. Burkholderia cenocepacia in cystic fibrosis: Epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 2010, 16, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Nzula, S.; Vandamme, P.; Govan, J.R.W. Infuence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother. 2002, 50, 265–269. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J. Antimicrob. Chemother. 2009, 64, 801–809. [Google Scholar] [CrossRef]
- Scoffone, V.C.; Chiarelli, L.R.; Trespidi, G.; Mentasti, M.; Riccardi, G.; Buroni, S. Burkholderia cenocepacia infections in cystic fibrosis patients: Drug resistance and therapeutic approaches. Front. Microbiol. 2017, 8, 1592. [Google Scholar] [CrossRef]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung Infections Associated with Cystic Fibrosis Lung Infections Associated with Cystic Fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef] [PubMed]
- Regan, K.H.; Bhatt, J. Eradication therapy for burkholderia cepacia complex in people with cystic fibrosis. Cochrane Database Syst. Rev. 2019, 2019, CD009876. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Bjarnsholt, T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010, 5, 1663–1674. [Google Scholar] [CrossRef]
- Schwab, U.; Abdullah, L.H.; Perlmutt, O.S.; Albert, D.; William Davis, C.; Arnold, R.R.; Yankaskas, J.R.; Gilligan, P.; Neubauer, H.; Randell, S.H.; et al. Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infect. Immun. 2014, 82, 4729–4745. [Google Scholar] [CrossRef]
- Leitão, J.H.; Sousa, S.A.; Ferreira, A.S.; Ramos, C.G.; Silva, I.N.; Moreira, L.M. Pathogenicity, virulence factors, and strategies to fight against Burkholderia cepacia complex pathogens and related species. Appl. Microbiol. Biotechnol. 2010, 87, 31–40. [Google Scholar] [CrossRef]
- Isles, A.; Maclusky, I.; Corey, M.; Gold, R.; Prober, C.; Fleming, P.; Levison, H. Pseudomonas cepacia infection in cystic fibrosis: An emerging problem. J. Pediatr. 1984, 104, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.R.; Pressler, T.; Høiby, N. Early aggressive eradication therapy for intermittent Pseudomonas aeruginosa airway colonization in cystic fibrosis patients: 15 years experience. J. Cyst. Fibros. 2008, 7, 523–530. [Google Scholar] [CrossRef]
- Filkins, L.M.; O’Toole, G.A. Cystic Fibrosis Lung Infections: Polymicrobial, Complex, and Hard to Treat. PLoS Pathog. 2015, 11, e1005258. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Schloss, P.D.; Kalikin, L.M.; Carmody, L.A.; Foster, B.K.; Petrosino, J.F.; Cavalcoli, J.D.; VanDevanter, D.R.; Murray, S.; Li, J.Z.; et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl. Acad. Sci. USA 2012, 109, 5809–5814. [Google Scholar] [CrossRef]
- Seixas, A.M.M.; Sousa, S.A.; Leitão, J.H. Antibody-Based Immunotherapies as a Tool for Tackling Multidrug-Resistant Bacterial Infections. Vaccines 2022, 10, 1789. [Google Scholar] [CrossRef]
- Sousa, S.A.; Seixas, A.M.M.; Marques, J.M.M.; Leitão, J.H. Immunization and immunotherapy approaches against Pseudomonas aeruginosa and Burkholderia cepacia complex infections. Vaccines 2021, 9, 670. [Google Scholar] [CrossRef]
- Seixas, A.M.M.; Sousa, S.A.; Feliciano, J.R.; Gomes, S.C.; Ferreira, M.R.; Moreira, L.M.; Leitão, J.H. A polyclonal antibody raised against the Burkholderia cenocepacia ompa-like protein BCAL2645 impairs the bacterium adhesion and invasion of human epithelial cells in vitro. Biomedicines 2021, 9, 1788. [Google Scholar] [CrossRef]
- De Soyza, A.; Hall, A.J.; Mahenthiralingam, E.; Drevinek, P.; Kaca, W.; Drulis-Kawa, Z.; Stoitsova, S.R.; Toth, V.; Coenye, T.; Zlosnik, J.E.A.; et al. Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen 2013, 2, 1010–1023. [Google Scholar] [CrossRef]
- Varga, J.J.; Losada, L.; Zelazny, A.M.; Kim, M.; McCorrison, J.; Brinkac, L.; Sampaio, E.P.; Greenberg, D.E.; Singh, I.; Heiner, C.; et al. Draft Genome Sequences of Burkholderia cenocepacia ET12 Lineage Strains K56-2 and BC7. Genome Announc. 2013, 1, e00841-13. [Google Scholar] [CrossRef]
- Silva, I.N.; Santos, P.M.; Santos, M.R.; Zlosnik, J.E.A.; Speert, D.P.; Buskirk, S.W.; Bruger, E.L.; Waters, C.M.; Cooper, V.S.; Moreira, L.M. Long-Term Evolution of Burkholderia multivorans during a Chronic Cystic Fibrosis Infection Reveals Shifting Forces of Selection. mSystems 2016, 1, e00029-16. [Google Scholar] [CrossRef]
- Bruscia, E.; Sangiuolo, F.; Sinibaldi, P.; Goncz, K.K.; Gruenert, D.C. Isolation of CF cell lines corrected at ΔF508-CFTR locus by SFHR-mediated targeting. Gene Ther. 2002, 9, 683–685. [Google Scholar] [CrossRef]
- Mil-Homens, D.; Leç, M.I.; Fernandes, F.; Pinto, S.N.; Fialho, A.M. Characterization of BCAM0224, a multifunctional trimeric autotransporter from the human pathogen Burkholderia cenocepacia. J. Bacteriol. 2014, 196, 1968–1979. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.S.; Silva, I.N.; Fernandes, F.; Pilkington, R.; Callaghan, M.; McClean, S.; Moreira, L.M. The tyrosine kinase BceF and the phosphotyrosine phosphatase BceD of Burkholderia contaminans are required for efficient invasion and epithelial disruption of a cystic fibrosis lung epithelial cell line. Infect. Immun. 2015, 83, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Ray, V.A.; Hill, P.J.; Stover, K.C.; Roy, S.; Sen, C.K.; Yu, L.; Wozniak, D.J.; Digiandomenico, A. Anti-Psl Targeting of Pseudomonas aeruginosa Biofilms for Neutrophil-Mediated Disruption. Sci. Rep. 2017, 7, 16065. [Google Scholar] [CrossRef] [PubMed]
- Cunha, M.V.; Sousa, S.A.; Leitão, J.H.; Moreira, L.M.; Videira, P.A.; Sá-Correia, I. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J. Clin. Microbiol. 2004, 42, 3052–3058. [Google Scholar] [CrossRef] [PubMed]
- Seed, K.D.; Dennis, J.J. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect. Immun. 2008, 76, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, A.I.; Kilcoyne, M.; Bernardes, N.; Mil-Homens, D.; Joshi, L.; Fialho, A.M. Burkholderia cenocepacia BCAM2418-induced antibody inhibits bacterial adhesion, confers protection to infection and enables identification of host glycans as adhesin targets. Cell. Microbiol. 2021, 23, e13340. [Google Scholar] [CrossRef] [PubMed]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef]
- Hulo, N.; Bairoch, A.; Bulliard, V.; Cerutti, L.; De Castro, E.; Langendijk-Genevaux, P.S.; Pagni, M.; Sigrist, C.J.A. The PROSITE database. Nucleic Acids Res. 2006, 34, 227–230. [Google Scholar] [CrossRef]
- Winsor, G.L.; Khaira, B.; Van Rossum, T.; Lo, R.; Whiteside, M.D.; Brinkman, F.S.L. The Burkholderia Genome Database: Facilitating flexible queries and comparative analyses. Bioinformatics 2008, 24, 2803–2804. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonasgenomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.A.; Seixas, A.M.M.; Mandal, M.; Rodríguez-Ortega, M.J.; Leitão, J.H. Characterization of the Burkholderia cenocepacia J2315 surface-exposed immunoproteome. Vaccines 2020, 8, 509. [Google Scholar] [CrossRef]
- Confer, A.W.; Ayalew, S. The OmpA family of proteins: Roles in bacterial pathogenesis and immunity. Vet. Microbiol. 2013, 163, 207–222. [Google Scholar] [CrossRef]
- Alhede, M.; Kragh, K.N.; Qvortrup, K.; Allesen-Holm, M.; van Gennip, M.; Christensen, L.D.; Jensen, P.Ø.; Nielsen, A.K.; Parsek, M.; Wozniak, D.; et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS ONE 2011, 6, e27943. [Google Scholar] [CrossRef]
- Valvano, M.A. Intracellular survival of Burkholderia cepacia complex in phagocytic cells. Can. J. Microbiol. 2015, 61, 607–615. [Google Scholar] [CrossRef]
- Steere, A.C.; Sikand, V.K.; Meurice, F.; Parenti, D.L.; Fikrig, E.; Schoen, R.T.; Nowakowski, J.; Schmid, C.H.; Laukamp, S.; Buscarino, C.; et al. Vaccination against Lyme Disease with Recombinant Borrelia burgdorferi Outer-Surface Lipoprotein A with Adjuvant. N. Engl. J. Med. 1998, 339, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.G.J.; Mahon, V.; Lambert, M.A.; Fagan, R.P. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 2007, 273, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Szabo, G.; Tamm, L.K. Electrostatic couplings in OmpA ion-channel gating suggest a mechanism for pore opening. Nat. Chem. Biol. 2006, 2, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Li, Y.; Tutt, C.B.; Xin, L.; Eaves-Pyles, T.; Soong, L. Outer membrane protein A of Escherichia coli O157:H7 stimulates dendritic cell activation. Infect. Immun. 2006, 74, 2676–2685. [Google Scholar] [CrossRef] [PubMed]
- Vila-Farrés, X.; Parra-Millán, R.; Sánchez-Encinales, V.; Varese, M.; Ayerbe-Algaba, R.; Bayó, N.; Guardiola, S.; Pachón-Ibáñez, M.E.; Kotev, M.; García, J.; et al. Combating virulence of Gram-negative bacilli by OmpA inhibition. Sci. Rep. 2017, 7, 14683. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Comolli, J.C.; Waite, L.L.; Mostov, K.E.; Engel, J.N. Pili Binding to Asialo-GM1 on Epithelial Cells Can Mediate Cytotoxicity or Bacterial Internalization by Pseudomonas aeruginosa. Infect. Immun. 1999, 67, 3207–3214. [Google Scholar] [CrossRef]
- Fleiszig, S.M.; Zaidi, T.S.; Pier, G.B. Pseudomonas aeruginosa invasion of and multiplication within corneal epithelial cells in vitro. Infect. Immun. 1995, 63, 4072–4077. [Google Scholar] [CrossRef]
- Del Mar Cendra, M.; Torrents, E. Differential adaptability between reference strains and clinical isolates of Pseudomonas aeruginosa into the lung epithelium intracellular lifestyle. Virulence 2020, 11, 862–876. [Google Scholar] [CrossRef]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef]
- Mullen, T.; Callaghan, M.; McClean, S. Invasion of Burkholderia cepacia complex isolates into lung epithelial cells involves glycolipid receptors. Microb. Pathog. 2010, 49, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.W.; Mohr, C.D. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 2000, 68, 24–29. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seixas, A.M.M.; Gomes, S.C.; Silva, C.; Moreira, L.M.; Leitão, J.H.; Sousa, S.A. A Polyclonal Antibody against a Burkholderia cenocepacia OmpA-like Protein Strongly Impairs Pseudomonas aeruginosa and B. multivorans Virulence. Vaccines 2024, 12, 207. https://doi.org/10.3390/vaccines12020207
Seixas AMM, Gomes SC, Silva C, Moreira LM, Leitão JH, Sousa SA. A Polyclonal Antibody against a Burkholderia cenocepacia OmpA-like Protein Strongly Impairs Pseudomonas aeruginosa and B. multivorans Virulence. Vaccines. 2024; 12(2):207. https://doi.org/10.3390/vaccines12020207
Chicago/Turabian StyleSeixas, António M. M., Sara C. Gomes, Carolina Silva, Leonilde M. Moreira, Jorge H. Leitão, and Sílvia A. Sousa. 2024. "A Polyclonal Antibody against a Burkholderia cenocepacia OmpA-like Protein Strongly Impairs Pseudomonas aeruginosa and B. multivorans Virulence" Vaccines 12, no. 2: 207. https://doi.org/10.3390/vaccines12020207




