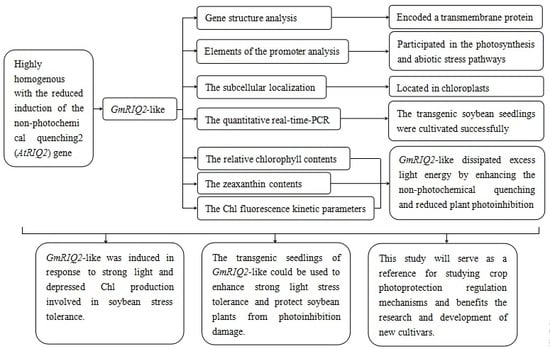
Overexpression of GmRIQ2-like (Glyma.04G174400) Enhances the Tolerance of Strong Light Stress and Reduces Photoinhibition in Soybean (Glycine max (L.) Merr.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Plant Materials, Growth Conditions, and RNA Isolation
2.3. Construction of Related Vectors
2.4. Subcellular Localization in Arabidopsis Protoplasts
2.5. Cultivation of Transgenic Soybeans
2.6. Detection of Transgenic Soybeans Using Quantitative Real-Time PCR
2.7. Measurements of Physiological Characteristics in Transgenic Agronomic Traits
2.8. Determination of Chl Fluorescence Kinetic Parameters
2.9. Statistical Analyses
3. Results
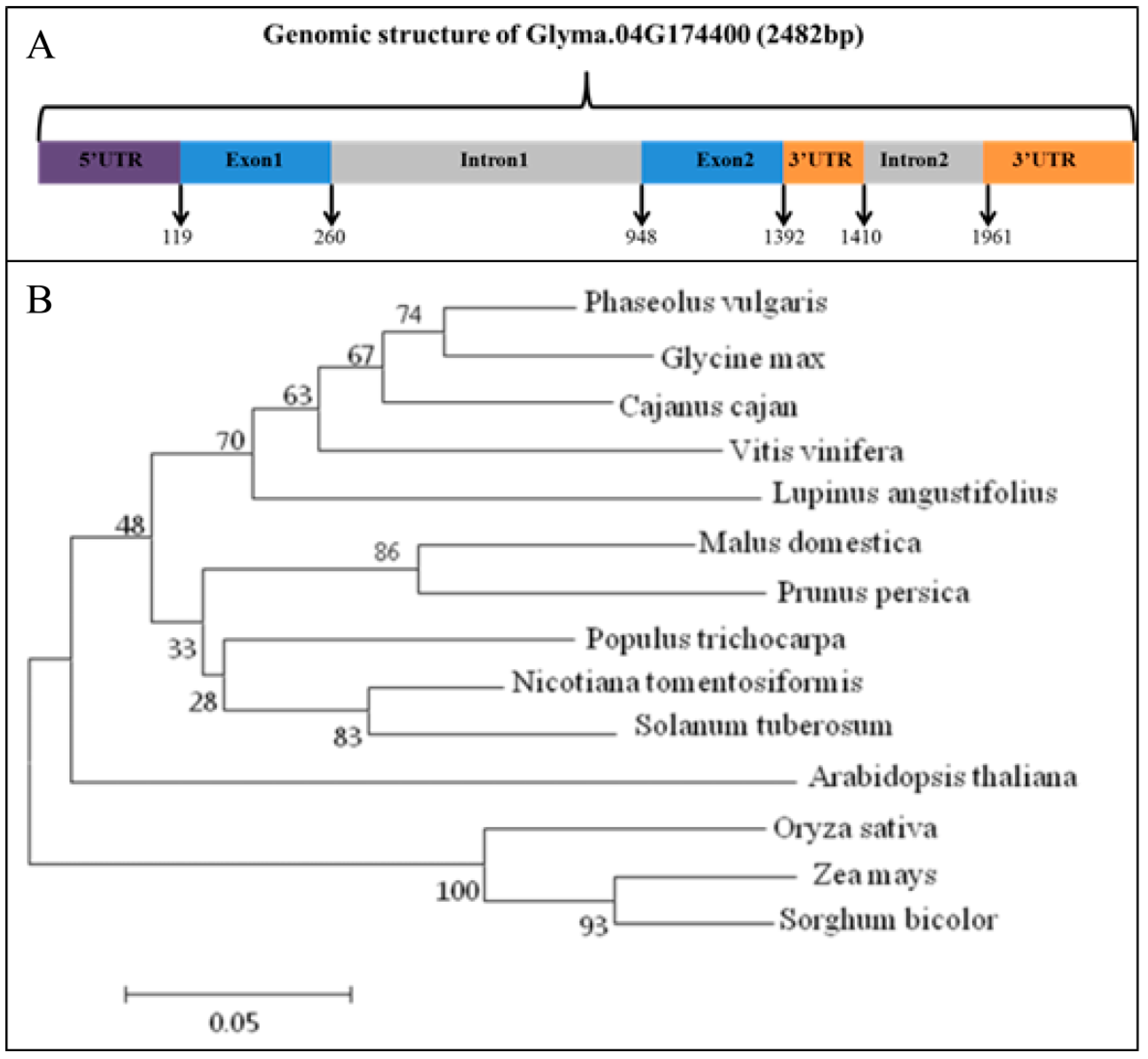
3.1. Gene Structure and Phylogenetic Analyses of GmRIQ2-like
3.2. Subcellular Localization of GmRIQ2-like
3.3. Identification of GmRIQ2-like Transgenic Soybeans
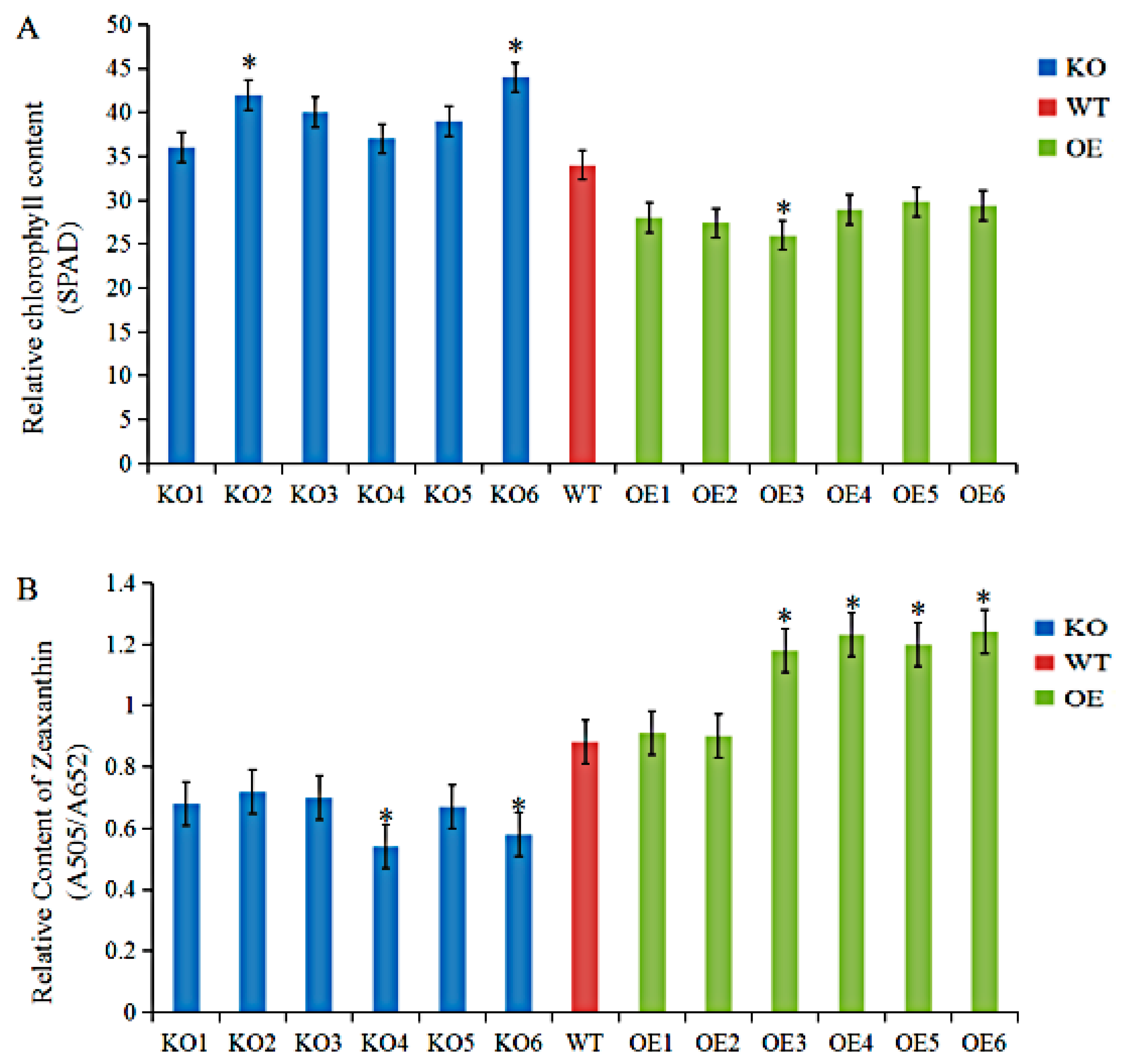
3.4. Agronomic Traits and Physiological Indices of GmRIQ2-like Transgenic Seedlings
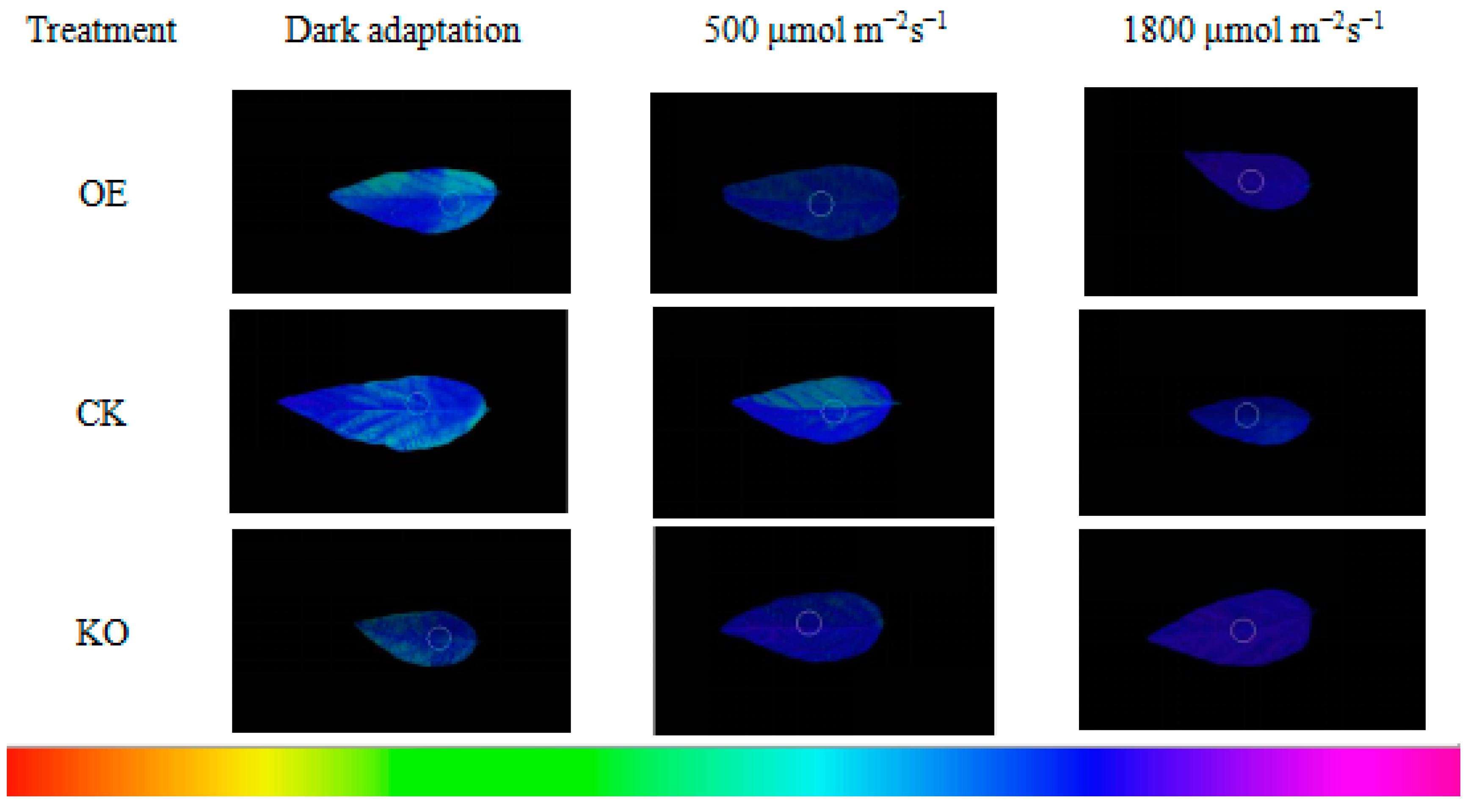
3.5. Chl fluorescence Kinetic Parameters of the T2 and T3 Generations of GmRIQ2-like Transgenic Seedlings
4. Discussion
4.1. Cloning and Functional Analysis of GmRIQ2-like
4.2. Yield Variation between GmRIQ2-like-OE, -KO, and WT Soybean Seedlings
4.3. Energy Dissipating Ability Comparison between GmRIQ2-like-OE and WT Seedlings
4.4. Xanthophyll Cycle Pool Analysis of GmRIQ2-like-OE, -KO, and WT Soybean Seedlings
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Duan, R.Y.; Wu, G.L.; Wang, Z.G. Effects of temperature stress on chlorophyll fluorescence parameters of strawberry. Chin. J. Spectrosc. Lab. 2011, 28, 2249–2252. [Google Scholar]
- Qi, Y.J.; Yang, S.P.; Gai, J.Y. Genetic transformation and functional analysis of soybean GmFtsH gene. In Proceedings of the Abstract of the 23rd National Soybean Research and Production Symposium, Daqing, China, 24–26 August 2012. [Google Scholar]
- Lou, Y.F. Study on Photoprotection and Related Gene Function of Phyllostachys Pubescens. Ph.D. Thesis, Chinese Academy of Forestry, Beijing, China, 2016. [Google Scholar]
- Jia, H.S.; Han, Y.Q. Progress in Photoinhibition of Photosynthesis in Higher Plants. Chin. J. Plant Sci. 2000, 17, 218–224. [Google Scholar] [CrossRef]
- Zheng, B.; Gao, Z.M. A Preliminary Study on the Characteristics of NPQ and ETR at the Seedling Stage of Phyllostachys Pubescens. World Newsl. 2011, 1, 18–20. [Google Scholar]
- Xu, D.Q. Photodamage Defense of Plant Photosynthetic Apparatus. Sci. Shanghai 2002, 54, 16–20. [Google Scholar]
- Kangasjarvi, E.; Naveh, L.; Adam, Z. The thylakoid lumen protease Deg l is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 2012, 19, 1039–1047. [Google Scholar]
- Salgado-Luarte, C.; Gianoli, E. Herbivory may modify functional responses to shade in seedlings of a light-demanding tree species. Funct. Ecol. 2011, 25, 492–499. [Google Scholar] [CrossRef]
- Zhou, L.Y.; Fu, Z.; Bu, Y. Research Progress of Plant Light Destruction Defense Mechanism. Subtrop. Plant Sci. 2016, 3. [Google Scholar] [CrossRef]
- Demmig-adams, B.; Adams, W.W., III. Photoprotection and Other Responses of Plants to High Light Stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 599–626. [Google Scholar] [CrossRef]
- Guo, L.W.; Xu, D.Q. Light suppression of light and scorpion of coral tree leaves under natural conditions. J. Plant Physiol. 1994, 20, 46–54. [Google Scholar]
- Lin, Z.F.; Peng, C.L.; Lin, Z.Q. Differences between 505nm and chlorophyll fluorescence in leaves induced by strong light. J. Trop. Subtrop. Bot. 1997, 3, 27–34. [Google Scholar]
- Li, M.; Zhou, J.; Peng, C.L. Response of Overexpression of GLDH Arabidopsis Plants to High Light Stress. J. South China Norm. Univ. Nat. Sci. Ed. 2016, 48, 91–95. [Google Scholar] [CrossRef]
- Tao, Z.Y.; Zou, Q. The Role of Mehler Reaction in Dissipating Excess Light Energy of Soybean Leaves. J. Plant Physiol. 2001, 1, 69–75. [Google Scholar]
- Cheng, M.; Li, Z.Q.; Jiang, C.D.; Shi, L.; Tang, Y.D.; Zhang, J.Z. Photosynthetic Characteristics and Photodamage Defense Mechanism of Highland Barley. Acta Agron. Sin. 2008, 34, 1805–1811. [Google Scholar] [CrossRef]
- Powles, S. Photoinhibition of photosynthesis induced by visible light. Annu. Rev. Plant Biol. 1984, 35, 14–44. [Google Scholar] [CrossRef]
- Guo, L.W.; Xu, D.Q.; Shen, Y.G. The reason for the decline in photosynthetic efficiency of cotton leaves in the field at noon. J. Plant Physiol. 1994, 4, 360–366. [Google Scholar]
- Xu, D.Q. Several Problems in the Study of Plant Light Stress. Plant Physiol. Commun. 2003, 5, 493–495. [Google Scholar]
- Zeng, J.Q.; Liu, H.X.; Wang, Y.R. Photoinhibition and restoration of cotyledons of cucumber seedlings at low temperature. Chin. J. Plant Physiol. 1997, 1, 15–20. [Google Scholar]
- Bertamini, M.; Muthuchelian, K.; Rubinigg, M.M. Low-night temperature increased the photoinhibition of photosynthesis in grapevine (Vitis vinifera L. cv. Riesling) leaves. Environ. Exp. Bot. 2006, 57, 25–31. [Google Scholar] [CrossRef]
- Murata, N.; Takahashi, S.; Nishiyama, Y. Photoinhibition of PhotosystemⅡ under environmental stress. Biochim. Biophys. Acta 2007, 1767, 414–421. [Google Scholar] [CrossRef]
- Donald, R.O. When there is too much light. Plant Physiol. 2001, 125, 29–32. [Google Scholar]
- Jiang, C.D.; Gao, H.Y.; Zou, Q. Iron deficiency increases the dissipation of excitation energy of soybean leaves. J. Plant Physiol. Mol. Biol. 2002, 28, 127–132. [Google Scholar]
- Zhang, Y.R. Using Chlorophyll Fluorescence Technique to Reveal the Response of Artificially Cultivated Cucumber Seedlings to Stress Temperature, Light and Salinity; Ocean University of China: Qingdao, China, 2009. [Google Scholar]
- Shi, S.B.; Li, T.C.; Li, M. Interactive effects of soil drought and glare on non-photochemical quenching of PSII reaction center in Kobresia humilis. Acta Physiol. Sin. Sin. 2015, 332, 155–163. [Google Scholar]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 2, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.E.; Liu, W.J.; Su, Y.Q.; Cui, J.M.; Zhang, Z.W.; Yuan, M.; Zhang, H.Y.; Yuan, S. Different response of photosystem II to short and long term drought stress in Arabidopsis thaliana. Physiol Plantarum. 2016, 158, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Baroli, I.; Do, A.D.; Yamane, T.; Niyogi, K.K. Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell 2003, 15, 992–1008. [Google Scholar] [CrossRef]
- Yamamoto, H.Y.; Nakayama, T.O.M.; Chichester, C.O. Studies on the light and dark interconversions of leaf xanthophylls. Arch. Biochem. Biophys. 1962, 97, 168–173. [Google Scholar] [CrossRef]
- Stransky, H.; Hager, A. Das Carotenoidmuster und dieVerbreitung des lichtinduzierten Xanthophyllcyclus in versc-hiedenen Algenklassen. Arch. Mikrobiol. 1970, 73, 315–323. [Google Scholar] [CrossRef]
- Niyogi, K.K. Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 333–359. [Google Scholar] [CrossRef]
- Endo, T.; Uebayashi, N.; Ishida, S. Light energy allocation at PSⅡ under field light conditions: How much energy is lost in NPQ-associated dissipation? Plant Physiol. Biochem. 2014, 81, 115–220. [Google Scholar] [CrossRef]
- Walters, R.G. Towards an understanding of photosynthetic acclimation. J. Exp. Bot. 2005, 56, 435–447. [Google Scholar] [CrossRef]
- Pinheiro, C.; Chaves, M.M. Photosynthesis and drought: Can we make metabolic connections from available data? J. Exp. Bot. 2011, 62, 869–882. [Google Scholar] [CrossRef]
- Jia, H.; Oguchi, R.; Hope, A.; Barber, J.; Chow, W. Differential effects of severe water stress on linear and cyclic electron fluxes through Photosystem I in spinach leaf discs in CO2-enriched air. Planta 2008, 228, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Meng, L.Z.; Cao, K.F. Sustained diurnal photosynthetic depression in uppermost-canopy leaves of four dipterocarp species in the rainy and dry seasons: Does photorespiration play a role in photoprotection? Tree Physiol. 2008, 29, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol. 2010, 125, 27–58. [Google Scholar] [CrossRef]
- Wang, Y.Q. Response Mechanism of Photosynthetic Apparatus of Apple Peel under High Temperature and Strong Light Inhibition; Northwest A & F University: Xi’an, China, 2015. [Google Scholar]
- Xu, Z.F.; Luo, G.H. Effects of strong light and reactive oxygen species on photosynthesis of soybean. Chin. J. Bot. Engl. Ed. 1999, 8, 862–866. [Google Scholar]
- Dietz, K.J.; Thomas, P. Novel regulations in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol. 2011, 155, 1477–1485. [Google Scholar] [CrossRef]
- Escalona, J.M.F.J.; Medrano, H. Water stress induces different levels of photosynthesis and electron transport rate regulation in grapevines. Plant Cell Environ. 1999, 22, 39–48. [Google Scholar]
- Yokoyama, R.; Yamamoto, H.; Kondo, M.; Takeda, S.; Ifuku, K.; Fukao, Y.; Kamei, Y.; Nishimura, M.; Shikanai, T. Grana-localized Proteins, RIQ1 and RIQ2, Affect the Dynamics of Light-harvesting Complex II and Grana Stacking in Arabidopsis. Plant Cell 2016, 28, 2261–2275. [Google Scholar] [CrossRef]
- Labate, M.T.; Ko, K.; Ko, Z.W.; Pinto, L.S.; Real, M.J.; Romano, M.R.; Barja, P.R.; Granell, A.; Friso, G.; Wijk, K. Constitutive expression of pea Lhcb 1-2 in tobacco affects plant development, morphology and photosynthetic capacity. Plant Mol. Biol. 2004, 55, 701–714. [Google Scholar] [CrossRef]
- Kim, E.H.; Li, X.P.; Razeghifard, R.; Anderson, J.M.; Niyogi, K.K.; Pogson, B.J.; Chow, W.S. The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: A study using two chlorophyll b-less mutants. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 973–984. [Google Scholar] [CrossRef]
- Cui, Y.L.; Jia, Q.S.; Yin, Q.Q.; Lin, G.N.; Kong, M.M.; Yang, Z.N. The GDC1 Gene Encodes a Novel Ankyrin Domain-Containing Protein That Is Essential for Grana Formation in Arabidopsis. Plant Physiol. 2011, 155, 130–141. [Google Scholar] [CrossRef][Green Version]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef] [PubMed]
- Caffarri, S.; Kouřil, R.; Kereïche, S.; Boekema, E.J.; Croce, R. Unctional architecture of higher plant photosystem II supercomplexes. EMBO J. 2009, 28, 3052–3063. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyeva, Y.; Suorsa, M.; Rossi, F.; Pavesi, A.; Kater, M.M.; Antonacci, A.; Tadini, L.; Pribil, M.; Schneider, A.; Wanner, G. Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. Cell Mol. Biol. 2013, 75, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, H.W.; Liu, R.F. Responses of Heat Dissipation in Rice to Stress. Chin. J. Rice Sci. 2016, 4, 30. [Google Scholar]
- Zhu, C.G.; Chen, Y.N.; Li, W.H.; Fu, A.H.; Yang, Y.H. Effect of Drought Stress on Photochemical Efficiency and Dissipation of Excited Energy in Photosystem Ⅱ of Populus euphratica. Chin. Bull. Bot. 2011, 46, 413–424. [Google Scholar]
- Zhao, X. Photosynthetic Response of Environmental Factors to Non-Photochemical Quenching of Rice Leaves; Huazhong Agricultural University: Wuhan, China, 2017. [Google Scholar]
- TAIR. Available online: https://www.arabidopsis.org/ (accessed on 3 April 2016).
- Phytozome. Available online: https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 3 April 2016).
- ExPasy. Available online: http://web.expasy.org/translate/ (accessed on 3 April 2016).
- TMHMM. Available online: http://www.cbs.dtu.dk/services/TMHMM/ (accessed on 3 April 2016).
- MEGA5.0. Available online: http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 3 April 2016).
- NCBI Database. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome (accessed on 3 April 2016).
- CRISPRdirect. Available online: http://crispr.dbcls.jp/ (accessed on 5 April 2016).
- Li, M.T.; Tan, W.Y.; Sun, M.Y.; Li, Y.G.; Li, W.B. Construction of TA Cloning Plant Expression Vector with Herbicide Resistance. Genom. Appl. Biol. 2015, 34, 2436–2440. [Google Scholar]
- Yu, Y.Z.; Du, J.; Ji, J. Study on Freezing-thawing Method of Introducing Recombinant Plasmid into Agrobacterium tumefaciens. J. Jilin Agric. Univ. 2003, 3, 257–259. [Google Scholar]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Jia, Y.; Yao, X.; Zhao, M.; Zhao, Q.; Du, Y.; Yu, C.; Xie, F. Comparison of Soybean Transformation Efficiency and Plant Factors Affecting Transformation during the Agrobacterium Infection Process. Int. J. Mol. Sci. 2015, 16, 18522–18543. [Google Scholar] [CrossRef]
- Li, J.; Song, Y.; Wu, K. Effects of Cr2O3 nanoparticles on the chlorophyll fluorescence and chloroplast ultrastructure of soybean (Glycine max). Environ. Sci. Pollut. Res. 2018, 3, 1–12. [Google Scholar] [CrossRef]
- Hong, T.; Xu, D.Q. Non-photochemical quenching of chlorophyll fluorescence in coral and soybean leaves. Chin. J. Plant Physiol. 1999, 1, 15–21. [Google Scholar]
- Dekker, J.P.; Boekema, E.J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta (BBA) Bioenerg. 2005, 1706, 12–39. [Google Scholar] [CrossRef] [PubMed]
- Mullineaux, C.W. Function and evolution of grana. Trends Plant Sci. 2005, 10, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Ruban, A.V.; Johnson, M.P.; Duffy, C.D. The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta. 2012, 1817, 167–181. [Google Scholar] [CrossRef]
- Takahashi, S.; Milward, S.E.; Fan, D.Y.B.; Chow, W.S.; Badger, M.R. How does cyclic electron flow alleviate photoinhibition in Arabidopsis? Plant Physiol. 2009, 149, 1560–1567. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Aminaka, R.; Yoshioka, M.; Khatoon, M.; Komayama, K.; Takenaka, D.; Yamashita, A.; Nijo, N.; Inagawa, K.; Morit, N.; et al. Quality control of photosystem II: Impact of light and heat stresses. Photosynth Res. 2008, 98, 589–608. [Google Scholar] [CrossRef]
- Flexas, J.; Diaz-Espejo, A.; Gago, J.; Gallé, A.; Galmés, J.; Gulías, J.; Medrano, H. Photosynthetic limitations in Mediterranean plants: A review. Environ. Exp. Bot. 2014, 103, 12–23. [Google Scholar] [CrossRef]
- Hu, M.J.; Guo, Y.P.; Shen, Y.G. Environmental Regulation of Photosynthesis in Citrus. Chin. J. Appl. Ecol. 2006, 3, 181–186. [Google Scholar]
- Moustakas, M.; Sperdouli, I.; Kouna, T.; Antonopoulou, C.I.; Therios, I. Exogenous proline induces soluble sugar accumulation and alleviates drought stress effects on photosystem II functioning of Arabidopsis thaliana leaves. Plant Growth Regul. 2011, 65, 315–325. [Google Scholar] [CrossRef]
- Zhang, B.; Xiao, M.; Li, H.Y. The changes of hydrogen peroxide metabolism and related enzyme activities during the senescence of tobacco leaves caused by high light intensity. J. Anhui Agric. Univ. 2007, 34, 477–480. [Google Scholar]
- Gong, D.H.; Wang, Z.Z.; Ji, X. Effects of low temperature and strong light stress on photosynthetic rate and photosynthetic pigment content of Algae sinensis. J. Inn. Mong. Agric. Univ. (Nat. Sci. Ed.) 2016, 6, 71–75. [Google Scholar]
- Woo, N.S.; Badger, M.R.; Pogson, B.J. A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods. 2008, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, Y.Z.; Chen, L.S.; Peng, S.F.; Wang, X.N.; Yang, X.H.; Ma, L. Correlation analysis of SPAD value and chlorophyⅡ content in leaves of Camellia oleifera. J. Cent. South Univ. For. Technol. 2013, 33, 77–80. [Google Scholar]
- German, T.C. The role of ΔpH-dependent dissipation of excitation energy in protecting photosystem II against light-induced damage in Arabidopsis thaliana. Plant Physiol. Biochem. 2002, 40, 41–49. [Google Scholar]
- Yamamoto, H.Y.; Bugos, R.C.; Hieber, A.D. Biochemistry and Molecular Biology of the Xanthophyll Cycle; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Govindjee, G.; Seufferheld, M. Non-photochemical quenching of chlorophyll a fluorescence: Early history and characterization of two xanthophyll-cycle mutants of Chlamydomonas reinhardtii. Funct. Plant Biol. 2002, 29, 1141–1155. [Google Scholar] [CrossRef]
- Björkman, O. High-Irradiance Stress in Higher Plants and Interaction with other Stress Factors. In Progress in Photosynthesis Research; Springer: New York, NY, USA, 1987; pp. 11–18. [Google Scholar]
- Bilger, W.; Bjorkman, O. Temperature dependence of violaxanthin de-epoxidation and non-photochemical fluorescence quenching in intact leaves of Gossypium hirsutum L. and Malva parvijora L. Planta 1991, 184, 226. [Google Scholar] [CrossRef]
- Adams, W.W.; Demmig-Adams, B.; Winter, K. Relative contributions of zeaxanthin-related and zeaxanthin-unrelated types of ‘high-energy-state’ quenching of chlorophyII fluorescence in spinach leaves exposed to various environmental conditions. Plant Physiol. 1990, 92, 302–309. [Google Scholar] [CrossRef]
- Oberhuber, W.; Bauer, H. Photoinhibition of photosynthesis under natural conditions in ivy (Hedera helix L.) growing in an understory of deciduous trees. Planta 1991, 185, 545–553. [Google Scholar] [CrossRef]
- Schöner, S.; Krause, G.H. Protective systems against active oxygen species in spinach: Response to cold acclimation in excess light. Planta 1990, 180, 383–389. [Google Scholar] [CrossRef]







| Primers | Primer Sequences (5′–3′) |
|---|---|
| GmRIQ2-like-OE-S | 5′-GCAACCTCGTCCCAACAAC-3′ |
| GmRIQ2-like-OE-A | 5′-CACCACCGAACCACCCATT-3′ |
| GmRIQ2-like-qRT-S | 5′-CCATTGTGCTCATTCCCG-3′ |
| GmRIQ2-like-qRT-A | 5′-ACCACTTCGCAACCATCT-3′ |
| Actin4-S | 5′-GTGTCAGCCATACTGTCCCCATT-3′ |
| Actin4-A | 5′-GTTTCAAGCTCTTGCTCGTAATCA-3′ |
| GmRIQ2-like-GFP-S | 5′-ATGGCGGTCCCTGCTACATCCTCA-3′ |
| GmRIQ2-like-GFP-A | 5′-GCAAGCCGCCGAGCACGAC-3′ |
| T7-gRNA-FPg | 5′-TAATACGACTCACTATAGGAGAAGGAGTGATTGGTGGTTTTAGTACTCTGGAAACAG-3′ |
| gRNA-RP | 5′-ATCTCGCCAACAAGTTGACGAG-3′ |
| CRISPR-GmRIQ2-like-S | 5′-TTGGAGAAGGAGTGATTGGTG-3′ |
| CRISPR-GmRIQ2-like-A | 5′-AACCACCAATCACTCCTTCTC-3′ |
| CRISPR-seq-A | 5′-CTCCTTCCTTCCGTCCACTTCATC-3′ |
| Solution | pH | Composition (g/L) |
|---|---|---|
| GM | 5.8 | B5: 3.21 g; Sucrose: 20 g; Agar: 7 g; 6-BA: 1 mg/L |
| LCCM | 5.4 | B5: 0.321 g; Sucrose: 30 g; Mes: 3.9 g; Acetosyringone (As): 160 mg/L;Gibberellic acid (GA): 10 mg/L; 6-BA: 1 mg/L |
| SCCM | 5.4 | B5: 0.321 g; Sucrose: 30 g; Mes: 3.9 g; Agar: 5 g; As: 160 mg/L; GA: 10 mg/L;BA: 1 mg/L; DTT: 0.154 g; L-cysteine: 1 g; Na2S2O3: 0.248 g |
| SSIM− | 5.6 | B5: 3.21 g; Sucrose: 30 g; Mes: 0.59 g; Agar: 7.5 g; 6-BA: 1 mg/L; Cefotaxime (cef): 250 mg/L |
| SSIM+ | 5.6 | B5: 3.21 g; Sucrose: 30 g; Mes: 0.59 g; Agar: 7.5 g; 6-BA: 1 mg/L;Phosphinothricin (PPT): 5 mg/L; cef: 250 mg/L |
| SEM | 5.6 | Murashige and Skoog (MS): 4.43 g; Sucrose: 30 g; Mes: 0.59 g; Agar: 7.5 g;Aspirin (Asp): 50 mg/L; N,6-isopentenyladenine (ZR): 1 mg/L; Gibberellin (GA3): 1 mg/LIndole-3-acetic acid (IAA): 1 mg/L; cef: 250 mg/L |
| RM | 5.6 | B5: 3.21 g; Sucrose: 20 g; Mes: 0.59 g; Agar: 7.0 g; Indole-3-butytric acid (IBA): 1 mg/mL |
| Name | Number | Function |
|---|---|---|
| G-Box | 8 | involved in light responsiveness |
| box-4 | 5 | |
| box II | 2 | |
| ATCT-motif | 1 | |
| CATT-motif | 1 | |
| CG-motif | 1 | |
| ACE | 1 | |
| GATA-motif | 1 | |
| I-box | 1 | |
| ABRE | 5 | involved in abscisic acid responsiveness |
| HSE | 3 | involved in heat stress responsiveness |
| TCA-element | 2 | involved in salicylic acid responsiveness |
| AuxRR-core | 1 | involved in auxin responsiveness |
| TC-rich repeats | 1 | involved in defense and stress responsiveness |
| circadian | 1 | involved in circadian control |
| ARE | 1 | essential for anaerobic induction |
| (A) | |||
| Agronomic Traits | WT | GmRIQ2-like-OE | GmRIQ2-like-KO |
| Plant height (cm) | 68.5 ± 3.2 | 64.8 ± 2.4 | 71.0 ± 4.8 |
| Number of main nodes (no.) | 15 ± 2 | 15 ± 1 | 16 ± 2 |
| Number of pods (no.) | 27 ± 3 | 26 ± 2 | 28 ± 3 |
| Number of seeds (no.) | 71 ± 4 | 65 ± 3 | 74 ± 5 |
| Weight of seed per plant (g) | 6.3 ± 0.5 | 5.3 ± 0.4 | 6.9 ± 0.7 |
| Hundred-grain weight (g) | 8.9 ± 0.2 | 8.1 ± 0.2 | 9.3 ± 0.3 |
| (B) | |||
| Agronomic Traits | WT | GmRIQ2-like-OE | GmRIQ2-like-KO |
| Plant height (cm) | 67.8 ± 2.8 | 62.7 ± 2.5 | 69.3 ± 3.2 |
| Number of main nodes (no.) | 15 ± 2 | 14 ± 2 | 16 ± 1 |
| Number of pods (no.) | 26 ± 1 | 25 ± 2 | 29 ± 2 |
| Number of seeds (no.) | 68 ± 3 | 66 ± 2 | 73 ± 4 |
| Weight of seed per plant (g) | 5.9 ± 0.5 | 5.5 ± 0.3 | 6.7 ± 0.7 |
| Hundred-grain weight (g) | 8.7 ± 0.3 | 8.4 ± 0.2 | 9.2 ± 0.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, J.; Li, D.; Yin, H.; Ma, L.; Zhang, J.; Zhang, B. Overexpression of GmRIQ2-like (Glyma.04G174400) Enhances the Tolerance of Strong Light Stress and Reduces Photoinhibition in Soybean (Glycine max (L.) Merr.). Agriculture 2020, 10, 157. https://doi.org/10.3390/agriculture10050157
Deng J, Li D, Yin H, Ma L, Zhang J, Zhang B. Overexpression of GmRIQ2-like (Glyma.04G174400) Enhances the Tolerance of Strong Light Stress and Reduces Photoinhibition in Soybean (Glycine max (L.) Merr.). Agriculture. 2020; 10(5):157. https://doi.org/10.3390/agriculture10050157
Chicago/Turabian StyleDeng, Jing, Dongmei Li, Huayi Yin, Li Ma, Jiukun Zhang, and Binbin Zhang. 2020. "Overexpression of GmRIQ2-like (Glyma.04G174400) Enhances the Tolerance of Strong Light Stress and Reduces Photoinhibition in Soybean (Glycine max (L.) Merr.)" Agriculture 10, no. 5: 157. https://doi.org/10.3390/agriculture10050157
APA StyleDeng, J., Li, D., Yin, H., Ma, L., Zhang, J., & Zhang, B. (2020). Overexpression of GmRIQ2-like (Glyma.04G174400) Enhances the Tolerance of Strong Light Stress and Reduces Photoinhibition in Soybean (Glycine max (L.) Merr.). Agriculture, 10(5), 157. https://doi.org/10.3390/agriculture10050157