Strategies Used to Reduce Methane Emissions from Ruminants: Controversies and Issues
Abstract
:1. Introduction
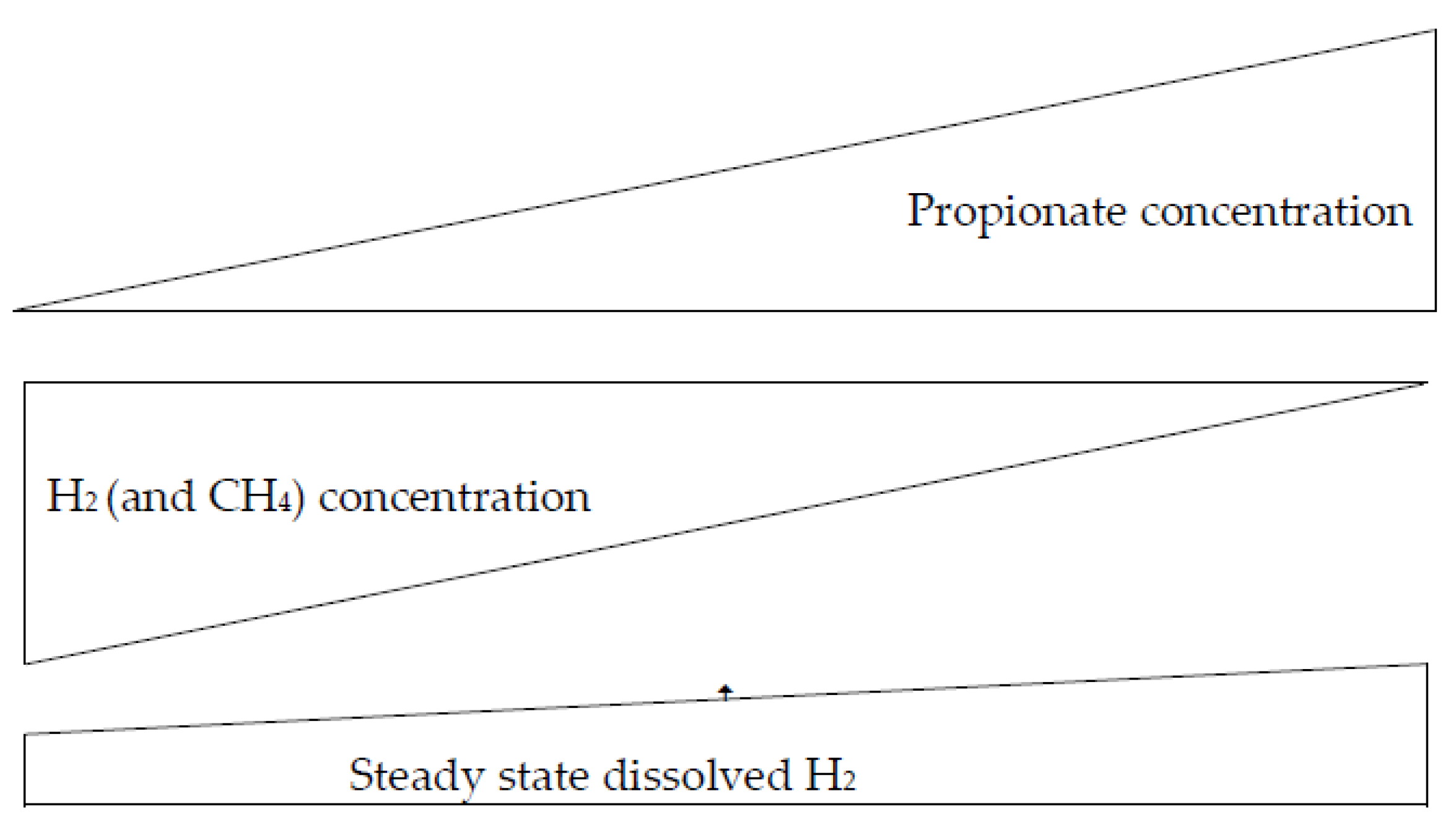
2. Methanogenesis
3. Mitigation Strategies
3.1. Mitigation through Feed Additives
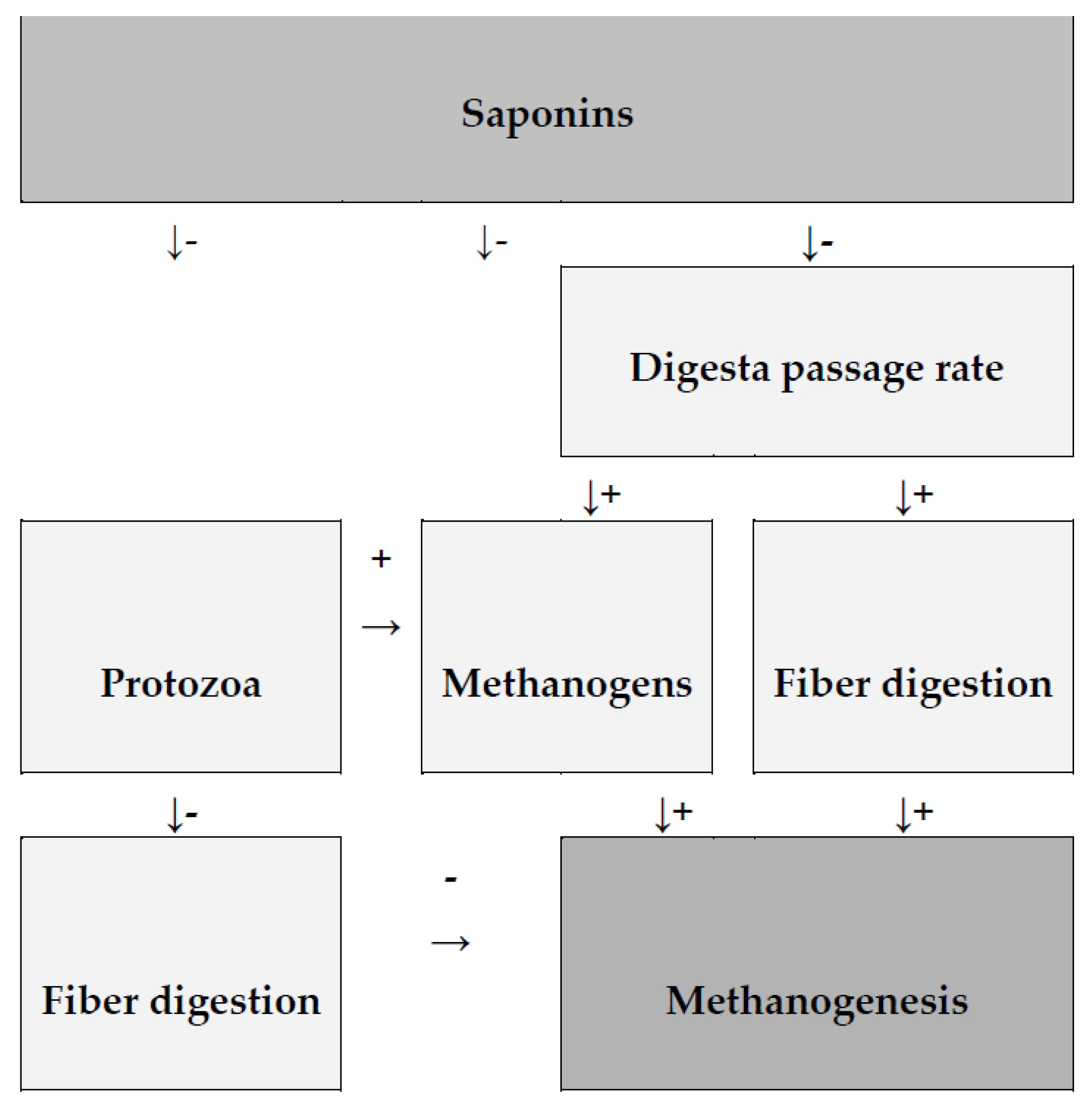
3.1.1. Saponins
3.1.2. Tannins
3.1.3. Flavonoids
3.1.4. Lipids
3.1.5. Plant Essential Oils
3.1.6. Algae
3.2. Mitigation through Microbiome Manipulation
3.2.1. Probiotics
3.2.2. Defaunation of the Rumen
3.2.3. Cellulolytic and Protozoan Activity Modification
3.2.4. Vaccination
3.3. Chemical Intervention
3.4. Reducing Methane Emissions through Genetic Selection
3.5. Forage Management
4. Perspectives and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Calabrò, P.S. Greenhouse gases emission from municipal waste management: The role of separate collection. Waste Manag. 2009, 29, 2178–2187. [Google Scholar] [CrossRef] [PubMed]
- Moss, A.R.; Jouany, J.-P.; Newbold, J. Methane production by ruminants: Its contribution to global warming. Ann. Zootech. 2000, 49, 231–253. [Google Scholar] [CrossRef] [Green Version]
- Kataria, R.P. Use of feed additives for reducing greenhouse gas emissions from dairy farms. Microbiol. Res. 2015, 6, 6120. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency. Global Mitigation of Non-CO2 Greenhouse Gases: 2010–2030, EPA Report 430R13011; United States Environmental Protection Agency: Washington, DC, USA, 2013. Available online: https://www.epa.gov/global-mitigation-non-co2-greenhouse-gases/global-mitigation-non-co2-ghgs-report-2010-2030 (accessed on 5 February 2023).
- Patra, A.K. Enteric methane mitigation technologies for ruminant livestock: A synthesis of current research and future directions. Environ. Monit. Assess. 2012, 184, 1929–1952. [Google Scholar] [CrossRef] [PubMed]
- Gerber, P.; Steinfeld, H.; Henderson, B.; Mottet, A.; Opio, C.; Dijkman, J.; Falcucci, A.; Tempio, G. Tackling Climate Change Through Livestock: A Global Assessment of Emissions and Mitigation Opportunities; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2013. [Google Scholar]
- FAO. FAOSTAT Database Collections. Available online: http://faostat.fao.org/ (accessed on 10 December 2022).
- Haque, M.N. Dietary manipulation: A sustainable way to mitigate methane emissions from ruminants. J. Anim. Sci. Technol. 2018, 60, 15. [Google Scholar] [CrossRef] [Green Version]
- Opio, C.; Gerber, P.; Mottet, A.; Falcucci, A.; Tempio, G.; MacLeod, M.; Vellinga, T.; Henderson, B.; Steinfeld, H. Greenhouse Gas Emissions from Ruminant Supply Chains—A Global Life Cycle Assessment; Food and Agriculture Organization of the United Nations: Rome, Italy, 2013. [Google Scholar]
- Muller, R.A.; Muller, E.A. Fugitive methane and the role of atmospheric half-life. Geoinfor. Geostat. Overv. 2017, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Saunois, M.; Stavert, A.R.; Poulter, B.; Bousquet, P.; Canadell, J.G.; Jackson, R.B.; Raymond, P.A.; Dlugokencky, E.J.; Houweling, S.; Patra, P.K.; et al. The global methane budget 2000–2017. Earth Syst. Sci. Data 2020, 12, 1561–1623. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Ungerfeld, E.M.; Eckard, R.J.; Wang, M. Review: Fifty years of research on rumen methanogenesis: Lessons learned and future challenges for mitigation. Animal 2020, 14, s2–s16. [Google Scholar] [CrossRef] [Green Version]
- Guzman, C.E.; Bereza-Malcolm, L.T.; De Groef, B.; Franks, A.E. Presence of selected methanogens, fibrolytic bacteria, and proteobacteria in the gastrointestinal tract of neonatal dairy calves from birth to 72 hours. PLoS ONE 2015, 10, e0133048. [Google Scholar] [CrossRef] [Green Version]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Abecia, L.; Angarita, E.; Aravena, P.; Nora Arenas, G.; Ariza, C.; et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [Green Version]
- Hungate, R.E. The Rumen and Its Microbes; Academic Press: New York, NY, USA, 1966. [Google Scholar]
- Farah Naz, F.; Saba, K. Methanogenic diversity and taxonomy in the gastro intestinal tract of ruminants. In Extremophilic Microbes and Metabolites; Afef, N., Ameur, C., Haïtham, S., Hadda Imene, O., Eds.; IntechOpen: Rijeka, Croatia, 2019; p. 7. [Google Scholar]
- Kumar, S.; Puniya, A.K.; Puniya, M.; Dagar, S.S.; Sirohi, S.K.; Singh, K.; Griffith, G.W. Factors affecting rumen methanogens and methane mitigation strategies. World J. Microbiol. Biotechnol. 2009, 25, 1557–1566. [Google Scholar] [CrossRef]
- Mauerhofer, L.-M.; Zwirtmayr, S.; Pappenreiter, P.; Bernacchi, S.; Seifert, A.H.; Reischl, B.; Schmider, T.; Taubner, R.-S.; Paulik, C.; Rittmann, S.K.M.R. Hyperthermophilic methanogenic archaea act as high-pressure CH4 cell factories. Commun. Biol. 2021, 4, 289. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, G.; Yañez-Ruiz, D.R.; Seradj, A.R.; Balcells, J.; Belanche, A. Methanogenesis in animals with foregut and hindgut fermentation: A review. Anim. Prod. Sci. 2019, 59, 2109–2122. [Google Scholar] [CrossRef]
- Janssen, P.H. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 2010, 160, 1–22. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef]
- Van Zijderveld, S.M.; Gerrits, W.J.J.; Apajalahti, J.A.; Newbold, J.R.; Dijkstra, J.; Leng, R.A.; Perdok, H.B. Nitrate and sulfate: Effective alternative hydrogen sinks for mitigation of ruminal methane production in sheep. J. Dairy Sci. 2010, 93, 5856–5866. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, P.K.; Jena, R.; Tomar, S.K.; Puniya, A.K. Reducing enteric methanogenesis through alternate hydrogen sinks in the rumen. Methane 2022, 1, 320–341. [Google Scholar] [CrossRef]
- Buddle, B.M.; Denis, M.; Attwood, G.T.; Altermann, E.; Janssen, P.H.; Ronimus, R.S.; Pinares-Patiño, C.S.; Muetzel, S.; Neil Wedlock, D. Strategies to reduce methane emissions from farmed ruminants grazing on pasture. Vet. J. 2011, 188, 11–17. [Google Scholar] [CrossRef]
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Diverse hydrogen production and consumption pathways influence methane production in ruminants. ISME J. 2019, 13, 2617–2632. [Google Scholar] [CrossRef]
- Malik, P.K.; Bhatta, R.; Gagen, E.J.; Sejian, V.; Soren, N.M.; Prasad, C.S. Alternate H2 sinks for reducing rumen methanogenesis. In Climate Change Impact on Livestock: Adaptation and Mitigation; Sejian, V., Gaughan, J., Baumgard, L., Prasad, C., Eds.; Springer: New Delhi, India, 2015; pp. 303–320. [Google Scholar]
- Holmes, D.E.; Smith, J.A. Biologically produced methane as a renewable energy source. Adv. Appl. Microbiol. 2016, 97, 1–61. [Google Scholar] [CrossRef]
- Belanche, A.; de la Fuente, G.; Newbold, C.J. Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol. Ecol. 2014, 90, 663–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, R.A. Interactions between microbial consortia in biofilms: A paradigm shift in rumen microbial ecology and enteric methane mitigation. Anim. Prod. Sci. 2014, 54, 519–543. [Google Scholar] [CrossRef] [Green Version]
- Valle, E.R.; Henderson, G.; Janssen, P.H.; Cox, F.; Alexander, T.W.; McAllister, T.A. Considerations in the use of fluorescence in situ hybridization (FISH) and confocal laser scanning microscopy to characterize rumen methanogens and define their spatial distributions. Can. J. Microbiol. 2015, 61, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, R.; Dicksved, J.; Sun, L.; Gonda, H.; Müller, B.; Schnürer, A.; Bertilsson, J. Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front. Microbiol. 2017, 8, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, P.H.; Kirs, M. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 2008, 74, 3619–3625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leahy, S.C.; Kelly, W.J.; Altermann, E.; Ronimus, R.S.; Yeoman, C.J.; Pacheco, D.M.; Li, D.; Kong, Z.; McTavish, S.; Sang, C.; et al. The genome sequence of the rumen methanogen methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS ONE 2010, 5, e8926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patra, A.; Park, T.; Kim, M.; Yu, Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechnol. 2017, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Teklebrhan, T.; Tan, Z.; Wang, M.; Wang, R. Rumen methanogens community as drivers of methane emission. J. Vet. Sci. Ani. Husb. 2018, 6, 406. [Google Scholar]
- Hungate, R.E. Hydrogen as an intermediate in the rumen fermentation. Arch. Mikrobiol. 1967, 59, 158–164. [Google Scholar] [CrossRef]
- King, E.E.; Smith, R.P.; St-Pierre, B.; Wright, A.D. Differences in the rumen methanogen populations of lactating Jersey and Holstein dairy cows under the same diet regimen. Appl. Environ. Microbiol. 2011, 77, 5682–5687. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kumar, S.; Lee, G.H.; Chang, D.H.; Rhee, M.S.; Yoon, M.H.; Kim, B.C. Methanobrevibacter boviskoreani sp. nov., isolated from the rumen of Korean native cattle. Int. J. Syst. Evol. Microbiol. 2013, 63, 4196–4201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Thauer, R.K. The Wolfe cycle comes full circle. Proc. Natl. Acad. Sci. USA 2012, 109, 15084–15085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Nan, X.; Chu, K.; Tong, J.; Yang, L.; Zheng, S.; Zhao, G.; Jiang, L.; Xiong, B. Shifts of hydrogen metabolism from methanogenesis to propionate production in response to replacement of forage fiber with non-forage fiber sources in diets in vitro. Front. Microbiol. 2018, 9, 2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Giller, K.; Kreuzer, M.; Ulbrich, S.E.; Braun, U.; Schwarm, A. Contribution of ruminal fungi, archaea, protozoa, and bacteria to the methane suppression caused by oilseed supplemented diets. Front. Microbiol. 2017, 8, 1864. [Google Scholar] [CrossRef]
- Yang, C.; Rooke, J.A.; Cabeza, I.; Wallace, R.J. Nitrate and inhibition of ruminal methanogenesis: Microbial ecology, obstacles, and opportunities for lowering methane emissions from ruminant livestock. Front. Microbiol. 2016, 7, 132. [Google Scholar] [CrossRef]
- Lan, W.; Yang, C. Ruminal methane production: Associated microorganisms and the potential of applying hydrogen-utilizing bacteria for mitigation. Sci. Total Environ. 2019, 654, 1270–1283. [Google Scholar] [CrossRef]
- Costa, K.C.; Wong, P.M.; Wang, T.; Lie, T.J.; Dodsworth, J.A.; Swanson, I.; Burn, J.A.; Hackett, M.; Leigh, J.A. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. USA 2010, 107, 11050–11055. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.; Lee, S.S. Advanced estimation and mitigation strategies: A cumulative approach to enteric methane abatement from ruminants. J. Anim. Sci. Technol. 2019, 61, 122–137. [Google Scholar] [CrossRef] [Green Version]
- Morgavi, D.P.; Forano, E.; Martin, C.; Newbold, C.J. Microbial ecosystem and methanogenesis in ruminants. Animal 2010, 4, 1024–1036. [Google Scholar] [CrossRef] [Green Version]
- Morgavi, D.P.; Forano, E.; Martin, C.; Newbold, C.J. Microbial ecosystem and methanogenesis in ruminants—CORRIGENDUM. Animal 2012, 6, 871. [Google Scholar] [CrossRef] [Green Version]
- Ragsdale, S.W.; Pierce, E. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 2008, 1784, 1873–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldwin, R.L.; Allison, M.J. Rumen metabolism. J. Anim. Sci. 1983, 57 (Suppl. 2), 461–477. [Google Scholar] [PubMed]
- Kobayashi, Y. Abatement of methane production from ruminants: Trends in the manipulation of rumen fermentation. Asian-Australas. J. Anim. Sci. 2010, 23, 410–416. [Google Scholar] [CrossRef]
- Martinez-Fernandez, G.; Denman, S.E.; Cheung, J.; McSweeney, C.S. Phloroglucinol degradation in the rumen promotes the capture of excess hydrogen generated from methanogenesis inhibition. Front. Microbiol. 2017, 8, 1871. [Google Scholar] [CrossRef]
- Latham, E.A.; Anderson, R.C.; Pinchak, W.E.; Nisbet, D.J. Insights on alterations to the rumen ecosystem by nitrate and nitrocompounds. Front. Microbiol. 2016, 7, 228. [Google Scholar] [CrossRef] [Green Version]
- Olijhoek, D.W.; Hellwing, A.L.F.; Brask, M.; Weisbjerg, M.R.; Højberg, O.; Larsen, M.K.; Dijkstra, J.; Erlandsen, E.J.; Lund, P. Effect of dietary nitrate level on enteric methane production, hydrogen emission, rumen fermentation, and nutrient digestibility in dairy cows. J. Dairy Sci. 2016, 99, 6191–6205. [Google Scholar] [CrossRef] [Green Version]
- Ungerfeld, E.M.; Kohn, R.A. The role of thermodynamics in the control of ruminal fermentation. In Ruminant Physiology: Digestion, Metabolism and Impact of Nutrition on Gene Expression, Immunology and Stress; Sejrsen, K., Hvelplund, T., Nielsen, M.O., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2006; pp. 55–85. [Google Scholar]
- Raleng, A. The Potential of Feeding Nitrate to Reduce Enteric Methane Production in Ruminants; The Department of Climate Change Commonwealth Government of Australia: Canberra, Australia, 2008.
- Simon, J. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 2002, 26, 285–309. [Google Scholar] [CrossRef]
- Uniyal, S.; Mishra, A.K.; Aswin, K.; Sahoo, J.K.; Munde, V.K.; Mishra, G.K. Use of nitrates and sulphates as hydrogen sink in reducing enteric methane production. Research & reviews. J. Vet. Sci. Technol. 2016, 5, 5–8. [Google Scholar]
- Huisingh, J.; McNeill, J.J.; Matrone, G. Sulfate reduction by a Desulfovibrio species isolated from sheep rumen. Appl. Microbiol. 1974, 28, 489–497. [Google Scholar] [CrossRef]
- Negussie, E.; de Haas, Y.; Dehareng, F.; Dewhurst, R.J.; Dijkstra, J.; Gengler, N.; Morgavi, D.P.; Soyeurt, H.; van Gastelen, S.; Yan, T.; et al. Invited review: Large-scale indirect measurements for enteric methane emissions in dairy cattle: A review of proxies and their potential for use in management and breeding decisions. J. Dairy Sci. 2017, 100, 2433–2453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Choudhury, P.K.; Carro, M.D.; Griffith, G.W.; Dagar, S.S.; Puniya, M.; Calabro, S.; Ravella, S.R.; Dhewa, T.; Upadhyay, R.C.; et al. New aspects and strategies for methane mitigation from ruminants. Appl. Microbiol. Biotechnol. 2014, 98, 31–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patra, A.K.; Saxena, J. The effect and mode of action of saponins on the microbial populations and fermentation in the rumen and ruminant production. Nutr. Res. Rev. 2009, 22, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Romero-Pérez, G.A.; Ominski, K.H.; McAllister, T.A.; Krause, D.O. Effect of environmental factors and influence of rumen and hindgut biogeography on bacterial communities in steers. Appl. Environ. Microbiol. 2011, 77, 258–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos-Morales, E.; de la Fuente, G.; Duval, S.; Wehrli, C.; Bouillon, M.; Lahmann, M.; Preskett, D.; Braganca, R.; Newbold, C.J. Antiprotozoal effect of saponins in the rumen can be enhanced by chemical modifications in their structure. Front. Microbiol. 2017, 8, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayanegara, A.; Wina, E.; Takahashi, J. Meta-analysis on methane mitigating properties of saponin-rich sources in the rumen: Influence of addition levels and plant sources. Asian-Australas. J. Anim. Sci. 2014, 27, 1426–1435. [Google Scholar] [CrossRef]
- Hartinger, T.; Gresner, N.; Südekum, K.H. Does intra-ruminal nitrogen recycling waste valuable resources? A review of major players and their manipulation. J. Anim. Sci. Biotechnol. 2018, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Newbold, C.J.; de la Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The role of ciliate protozoa in the rumen. Front. Microbiol. 2015, 6, 1313. [Google Scholar] [CrossRef] [Green Version]
- Wina, E.; Muetzel, S.; Becker, K. The impact of saponins or saponin-containing plant materials on ruminant production—A review. J. Agric. Food Chem. 2005, 53, 8093–8105. [Google Scholar] [CrossRef]
- Pen, B.; Sar, C.; Mwenya, B.; Kuwaki, K.; Morikawa, R.; Takahashi, J. Effects of Yucca schidigera and Quillaja saponaria extracts on in vitro ruminal fermentation and methane emission. Anim. Feed Sci. Technol. 2006, 129, 175–186. [Google Scholar] [CrossRef]
- Ku-Vera, J.C.; Jiménez-Ocampo, R.; Valencia-Salazar, S.S.; Montoya-Flores, M.D.; Molina-Botero, I.C.; Arango, J.; Gómez-Bravo, C.A.; Aguilar-Pérez, C.F.; Solorio-Sánchez, F.J. Role of secondary plant metabolites on enteric methane mitigation in ruminants. Front. Vet. Sci. 2020, 7, 584. [Google Scholar] [CrossRef] [PubMed]
- Moses, T.; Papadopoulou, K.K.; Osbourn, A. Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 439–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samtiya, M.; Aluko, R.E.; Dhewa, T. Plant food anti-nutritional factors and their reduction strategies: An overview. Food Prod. Process. Nutr. 2020, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Broucek, J. Options to methane production abatement in ruminants: A review. J. Anim. Plant Sci. 2018, 28, 348–364. [Google Scholar]
- Liu, H.; Vaddella, V.; Zhou, D. Effects of chestnut tannins and coconut oil on growth performance, methane emission, ruminal fermentation, and microbial populations in sheep. J. Dairy Sci. 2011, 94, 6069–6077. [Google Scholar] [CrossRef] [Green Version]
- Tavendale, M.H.; Meagher, L.P.; Pacheco, D.; Walker, N.; Attwood, G.T.; Sivakumaran, S. Methane production from in vitro rumen incubations with Lotus pedunculatus and Medicago sativa, and effects of extractable condensed tannin fractions on methanogenesis. Anim. Feed Sci. Technol. 2005, 123–124, 403–419. [Google Scholar] [CrossRef]
- Field, J.A.; Kortekaas, S.; Lettinga, G. The tannin theory of methanogenic toxicity. Biol. Wastes 1989, 29, 241–262. [Google Scholar] [CrossRef]
- McSweeney, C.S.; Palmer, B.; McNeill, D.M.; Krause, D.O. Microbial interactions with tannins: Nutritional consequences for ruminants. Anim. Feed Sci. Technol. 2001, 91, 83–93. [Google Scholar] [CrossRef]
- Tiemann, T.T.; Lascano, C.E.; Wettstein, H.R.; Mayer, A.C.; Kreuzer, M.; Hess, H.D. Effect of the tropical tannin-rich shrub legumes Calliandra calothyrsus and Flemingia macrophylla on methane emission and nitrogen and energy balance in growing lambs. Animal 2008, 2, 790–799. [Google Scholar] [CrossRef] [Green Version]
- Jayanegara, A.; Leiber, F.; Kreuzer, M. Meta-analysis of the relationship between dietary tannin level and methane formation in ruminants from in vivo and in vitro experiments. J. Anim. Physiol. Anim. Nutr. 2012, 96, 365–375. [Google Scholar] [CrossRef]
- Aboagye, I.A.; Beauchemin, K.A. Potential of molecular weight and structure of tannins to reduce methane emissions from ruminants: A review. Animals 2019, 9, 856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasta, V.; Daghio, M.; Cappucci, A.; Buccioni, A.; Serra, A.; Viti, C.; Mele, M. Invited review: Plant polyphenols and rumen microbiota responsible for fatty acid biohydrogenation, fiber digestion, and methane emission: Experimental evidence and methodological approaches. J. Dairy Sci. 2019, 102, 3781–3804. [Google Scholar] [CrossRef] [PubMed]
- McSweeney, C.S.; Palmer, B.; Bunch, R.; Krause, D.O. Effect of the tropical forage calliandra on microbial protein synthesis and ecology in the rumen. J. Appl. Microbiol. 2001, 90, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Obreque-Slier, E.; López-Solís, R.; Peña-Neira, Á.; Zamora-Marín, F. Tannin–protein interaction is more closely associated with astringency than tannin–protein precipitation: Experience with two oenological tannins and a gelatin. Int. J. Food Sci. Technol. 2010, 45, 2629–2636. [Google Scholar] [CrossRef]
- Adamczyk, B.; Simon, J.; Kitunen, V.; Adamczyk, S.; Smolander, A. Tannins and their complex interaction with different organic nitrogen compounds and enzymes: Old paradigms versus recent advances. ChemistryOpen 2017, 6, 610–614. [Google Scholar] [CrossRef]
- Malik, P.K.; Kolte, A.P.; Baruah, L.; Saravanan, M.; Bakshi, B.; Bhatta, R. Enteric methane mitigation in sheep through leaves of selected tanniniferous tropical tree species. Livest. Sci. 2017, 200, 29–34. [Google Scholar] [CrossRef]
- Salami, S.A.; Valenti, B.; Bella, M.; O’Grady, M.N.; Luciano, G.; Kerry, J.P.; Jones, E.; Priolo, A.; Newbold, C.J. Characterisation of the ruminal fermentation and microbiome in lambs supplemented with hydrolysable and condensed tannins. FEMS Microbiol. Ecol. 2018, 94, fiy061. [Google Scholar] [CrossRef]
- Cardoso-Gutierrez, E.; Aranda-Aguirre, E.; Robles-Jimenez, L.E.; Castelán-Ortega, O.A.; Chay-Canul, A.J.; Foggi, G.; Angeles-Hernandez, J.C.; Vargas-Bello-Pérez, E.; González-Ronquillo, M. Effect of tannins from tropical plants on methane production from ruminants: A systematic review. Vet. Anim. Sci. 2021, 14, 100214. [Google Scholar] [CrossRef]
- Animut, G.; Puchala, R.; Goetsch, A.L.; Patra, A.K.; Sahlu, T.; Varel, V.H.; Wells, J. Methane emission by goats consuming diets with different levels of condensed tannins from lespedeza. Anim. Feed Sci. Technol. 2008, 144, 212–227. [Google Scholar] [CrossRef]
- Waghorn, G. Beneficial and detrimental effects of dietary condensed tannins for sustainable sheep and goat production—Progress and challenges. Anim. Feed Sci. Technol. 2008, 147, 116–139. [Google Scholar] [CrossRef]
- Smith, A.H.; Zoetendal, E.; Mackie, R.I. Bacterial mechanisms to overcome inhibitory effects of dietary tannins. Microb. Ecol. 2005, 50, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Oskoueian, E.; Abdullah, N.; Oskoueian, A. Effects of flavonoids on rumen fermentation activity, methane production, and microbial population. BioMed Res. Int. 2013, 2013, 349129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formato, M.; Cimmino, G.; Brahmi-Chendouh, N.; Piccolella, S.; Pacifico, S. Polyphenols for livestock feed: Sustainable perspectives for animal husbandry? Molecules 2022, 27, 7752. [Google Scholar] [CrossRef] [PubMed]
- Olagaray, K.E.; Bradford, B.J. Plant flavonoids to improve productivity of ruminants—A review. Anim. Feed Sci. Technol. 2019, 251, 21–36. [Google Scholar] [CrossRef]
- Seradj, A.R.; Abecia, L.; Crespo, J.; Villalba, D.; Fondevila, M.; Balcells, J. The effect of Bioflavex® and its pure flavonoid components on in vitro fermentation parameters and methane production in rumen fluid from steers given high concentrate diets. Anim. Feed Sci. Technol. 2014, 197, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Shen, Y.; Wang, C.; Ding, L.; Zhao, F.; Wang, M.; Fu, J.; Wang, H. Megasphaera elsdenii lactate degradation pattern shifts in rumen acidosis models. Front. Microbiol. 2019, 10, 162. [Google Scholar] [CrossRef] [Green Version]
- Sinz, S.; Kunz, C.; Liesegang, A.; Braun, U.; Marquardt, S.; Soliva, R.C.; Kreuzer, M. In vitro bioactivity of various pure flavonoids in ruminal fermentation, with special reference to methane formation. Czech J. Anim. Sci. 2018, 63, 293–304. [Google Scholar] [CrossRef] [Green Version]
- María Carpena, R.; Cristina, C.; Bernabe, N.-E.; Eliana, P.; Maria, F.-C.; Filipa, S.R.; Jesus, S.-G.; Isabel, C.F.R.F.; Miguel, A.P.; Lillian, B. Flavonoids: A group of potential food additives with beneficial health effects. In Natural Food Additives; Miguel, A.P., Paz, O., Eds.; IntechOpen: Rijeka, Croatia, 2021; p. 4. [Google Scholar]
- Machmüller, A.; Soliva, C.R.; Kreuzer, M. Methane-suppressing effect of myristic acid in sheep as affected by dietary calcium and forage proportion. Br. J. Nutr. 2003, 90, 529–540. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Zeitz, J.O.; Meile, L.; Kreuzer, M.; Schwarm, A. Influence of pH and the degree of protonation on the inhibitory effect of fatty acids in the ruminal methanogen Methanobrevibacter ruminantium strain M1. J. Appl. Microbiol. 2015, 119, 1482–1493. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Meile, L.; Kreuzer, M.; Zeitz, J.O. The effect of saturated fatty acids on methanogenesis and cell viability of Methanobrevibacter ruminantium. Archaea 2013, 2013, 106916. [Google Scholar] [CrossRef] [Green Version]
- Klevenhusen, F.; Zeitz, J.O.; Duval, S.; Kreuzer, M.; Soliva, C.R. Garlic oil and its principal component diallyl disulfide fail to mitigate methane, but improve digestibility in sheep. Anim. Feed Sci. Technol. 2011, 166–167, 356–363. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; McGinn, S.M.; Petit, H.V. Methane abatement strategies for cattle: Lipid supplementation of diets. Can. J. Anim. Sci. 2007, 87, 431–440. [Google Scholar] [CrossRef]
- McGinn, S.M.; Beauchemin, K.A.; Coates, T.; Colombatto, D. Methane emissions from beef cattle: Effects of monensin, sunflower oil, enzymes, yeast, and fumaric acid. J. Anim. Sci. 2004, 82, 3346–3356. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K. The effect of dietary fats on methane emissions, and its other effects on digestibility, rumen fermentation and lactation performance in cattle: A meta-analysis. Livest. Sci. 2013, 155, 244–254. [Google Scholar] [CrossRef]
- Jordan, E.; Lovett, D.K.; Monahan, F.J.; Callan, J.; Flynn, B.; O’Mara, F.P. Effect of refined coconut oil or copra meal on methane output and on intake and performance of beef heifers. J. Anim. Sci. 2006, 84, 162–170. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Ungerfeld, E.M.; Abdalla, A.L.; Alvarez, C.; Arndt, C.; Becquet, P.; Benchaar, C.; Berndt, A.; Mauricio, R.M.; McAllister, T.A.; et al. Invited review: Current enteric methane mitigation options. J. Dairy Sci. 2022, 105, 9297–9326. [Google Scholar] [CrossRef]
- Hassanat, F.; Benchaar, C. Methane emissions of manure from dairy cows fed red clover- or corn silage-based diets supplemented with linseed oil. J. Dairy Sci. 2019, 102, 11766–11776. [Google Scholar] [CrossRef]
- Grainger, C.; Beauchemin, K.A. Can enteric methane emissions from ruminants be lowered without lowering their production? Anim. Feed Sci. Technol. 2011, 166–167, 308–320. [Google Scholar] [CrossRef]
- Arndt, C.; Hristov, A.N.; Price, W.J.; McClelland, S.C.; Pelaez, A.M.; Cueva, S.F.; Oh, J.; Dijkstra, J.; Bannink, A.; Bayat, A.R.; et al. Full adoption of the most effective strategies to mitigate methane emissions by ruminants can help meet the 1.5 °C target by 2030 but not 2050. Proc. Natl. Acad. Sci. USA 2022, 119, e2111294119. [Google Scholar] [CrossRef]
- Palmquist, D.L.; Jenkins, T.C. A 100-year review: Fat feeding of dairy cows. J. Dairy Sci. 2017, 100, 10061–10077. [Google Scholar] [CrossRef] [Green Version]
- Falleh, H.; Ben Jemaa, M.; Saada, M.; Ksouri, R. Essential oils: A promising eco-friendly food preservative. Food Chem. 2020, 330, 127268. [Google Scholar] [CrossRef] [PubMed]
- Kalogianni, A.I.; Lazou, T.; Bossis, I.; Gelasakis, A.I. Natural phenolic compounds for the control of oxidation, bacterial spoilage, and foodborne pathogens in meat. Foods 2020, 9, 794. [Google Scholar] [CrossRef] [PubMed]
- Busquet, M.; Calsamiglia, S.; Ferret, A.; Carro, M.D.; Kamel, C. Effect of garlic oil and four of its compounds on rumen microbial fermentation. J. Dairy Sci. 2005, 88, 4393–4404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soliva, C.R.; Amelchanka, S.L.; Duval, S.M.; Kreuzer, M. Ruminal methane inhibition potential of various pure compounds in comparison with garlic oil as determined with a rumen simulation technique (Rusitec). Br. J. Nutr. 2011, 106, 114–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belanche, A.; Newbold, C.J.; Morgavi, D.P.; Bach, A.; Zweifel, B.; Yáñez-Ruiz, D.R. A Meta-analysis describing the effects of the essential oils blend agolin ruminant on performance, rumen fermentation and methane emissions in dairy Cows. Animals 2020, 10, 620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benchaar, C.; Calsamiglia, S.; Chaves, A.V.; Fraser, G.R.; Colombatto, D.; McAllister, T.A.; Beauchemin, K.A. A review of plant-derived essential oils in ruminant nutrition and production. Anim. Feed Sci. Technol. 2008, 145, 209–228. [Google Scholar] [CrossRef]
- Saro, C.; Hohenester, U.M.; Bernard, M.; Lagrée, M.; Martin, C.; Doreau, M.; Boudra, H.; Popova, M.; Morgavi, D.P. Effectiveness of interventions to modulate the rumen microbiota composition and function in pre-ruminant and ruminant lambs. Front. Microbiol. 2018, 9, 1273. [Google Scholar] [CrossRef]
- Mucha, W.; Witkowska, D. The applicability of essential oils in different stages of production of animal-based foods. Molecules 2021, 26, 3798. [Google Scholar] [CrossRef]
- Benchaar, C.; Greathead, H. Essential oils and opportunities to mitigate enteric methane emissions from ruminants. Anim. Feed Sci. Technol. 2011, 166–167, 338–355. [Google Scholar] [CrossRef]
- Wallace, R.J. Antimicrobial properties of plant secondary metabolites. Proc. Nutr. Soc. 2004, 63, 621–629. [Google Scholar] [CrossRef]
- Amin, N.; Tagliapietra, F.; Arango, S.; Guzzo, N.; Bailoni, L. Free and microencapsulated essential oils incubated in vitro: Ruminal stability and fermentation parameters. Animals 2021, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Machado, L.; Magnusson, M.; Paul, N.A.; Kinley, R.; de Nys, R.; Tomkins, N. Identification of bioactives from the red seaweed Asparagopsis taxiformis that promote antimethanogenic activity in vitro. J. Appl. Phycol. 2016, 28, 3117–3126. [Google Scholar] [CrossRef]
- Li, X.; Norman, H.C.; Kinley, R.D.; Laurence, M.; Wilmot, M.; Bender, H.; de Nys, R.; Tomkins, N. Asparagopsis taxiformis decreases enteric methane production from sheep. Anim. Prod. Sci. 2018, 58, 681–688. [Google Scholar] [CrossRef]
- Roque, B.M.; Salwen, J.K.; Kinley, R.; Kebreab, E. Inclusion of asparagopsis armata in lactating dairy cows’ diet reduces enteric methane emission by over 50 percent. J. Clean. Prod. 2019, 234, 132–138. [Google Scholar] [CrossRef]
- Roque, B.M.; Venegas, M.; Kinley, R.D.; de Nys, R.; Duarte, T.L.; Yang, X.; Kebreab, E. Red seaweed (Asparagopsis taxiformis) supplementation reduces enteric methane by over 80 percent in beef steers. PLoS ONE 2021, 16, e0247820. [Google Scholar] [CrossRef] [PubMed]
- Stefenoni, H.A.; Räisänen, S.E.; Cueva, S.F.; Wasson, D.E.; Lage, C.F.A.; Melgar, A.; Fetter, M.E.; Smith, P.; Hennessy, M.; Vecchiarelli, B.; et al. Effects of the macroalga Asparagopsis taxiformis and oregano leaves on methane emission, rumen fermentation, and lactational performance of dairy cows. J. Dairy Sci. 2021, 104, 4157–4173. [Google Scholar] [CrossRef]
- Kinley, R.D.; Martinez-Fernandez, G.; Matthews, M.K.; de Nys, R.; Magnusson, M.; Tomkins, N.W. Mitigating the carbon footprint and improving productivity of ruminant livestock agriculture using a red seaweed. J. Clean. Prod. 2020, 259, 120836. [Google Scholar] [CrossRef]
- Lean, I.J.; Golder, H.M.; Grant, T.M.D.; Moate, P.J. A meta-analysis of effects of dietary seaweed on beef and dairy cattle performance and methane yield. PLoS ONE 2021, 16, e0249053. [Google Scholar] [CrossRef]
- Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological profile for Bromoform and Chlorodibromomethane; U.S. Department of Health and Human Services, Public Health Service: Atlanta, GA, USA, 2005.
- Nilsson, J.; Martin, M. Exploratory environmental assessment of large-scale cultivation of seaweed used to reduce enteric methane emissions. Sustain. Prod. Consum. 2022, 30, 413–423. [Google Scholar] [CrossRef]
- Yáñez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 2015, 6, 1133. [Google Scholar] [CrossRef] [Green Version]
- Chaucheyras, F.; Fonty, G.; Bertin, G.; Gouet, P. In vitro H2 utilization by a ruminal acetogenic bacterium cultivated alone or in association with an archaea methanogen is stimulated by a probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1995, 61, 3466–3467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alazzeh, A.Y.; Sultana, H.; Beauchemin, K.A.; Wang, Y.; Holo, H.; Harstad, O.M.; McAllister, T.A. Using strains of Propionibacteria to mitigate methane emissions in vitro. Acta Agric. Scand. Sect. A Anim. Sci. 2012, 62, 263–272. [Google Scholar] [CrossRef]
- Vyas, D.; Alazzeh, A.; McGinn, S.M.; McAllister, T.A.; Harstad, O.M.; Holo, H.; Beauchemin, K.A. Enteric methane emissions in response to ruminal inoculation of Propionibacterium strains in beef cattle fed a mixed diet. Anim. Prod. Sci. 2016, 56, 1035–1040. [Google Scholar] [CrossRef] [Green Version]
- Vyas, D.; McGeough, E.J.; Mohammed, R.; McGinn, S.M.; McAllister, T.A.; Beauchemin, K.A. Effects of Propionibacterium strains on ruminal fermentation, nutrient digestibility and methane emissions in beef cattle fed a corn grain finishing diet. Animal 2014, 8, 1807–1815. [Google Scholar] [CrossRef]
- Henderson, G.; Naylor, G.E.; Leahy, S.C.; Janssen, P.H. Presence of novel, potentially homoacetogenic bacteria in the rumen as determined by analysis of formyltetrahydrofolate synthetase sequences from ruminants. Appl. Environ. Microbiol. 2010, 76, 2058–2066. [Google Scholar] [CrossRef] [Green Version]
- Sazinsky, M.H.; Lippard, S.J. Methane monooxygenase: Functionalizing methane at iron and copper. Met. Ions Life Sci. 2015, 15, 205–256. [Google Scholar] [CrossRef]
- De Raphélis-Soissan, V.; Li, L.; Godwin, I.R.; Barnett, M.C.; Perdok, H.B.; Hegarty, R.S. Use of nitrate and Propionibacterium acidipropionici to reduce methane emissions and increase wool growth of Merino sheep. Anim. Prod. Sci. 2014, 54, 1860–1866. [Google Scholar] [CrossRef]
- Jeyanathan, J.; Martin, C.; Morgavi, D.P. The use of direct-fed microbials for mitigation of ruminant methane emissions: A review. Animal 2014, 8, 250–261. [Google Scholar] [CrossRef] [Green Version]
- Kittelmann, S.; Pinares-Patiño, C.S.; Seedorf, H.; Kirk, M.R.; McEwan, J.C.; Janssen, P.H. Natural variation in methane emission of sheep fed on a lucerne pellet diet is unrelated to rumen ciliate community type. Microbiology 2016, 162, 459–465. [Google Scholar] [CrossRef]
- Eadie, J.M. Inter-relationships between certain rumen ciliate protozoa. Microbiology 1962, 29, 579–588. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Kalscheur, K.F.; Huhtanen, P.; Faciola, A.P. Effects of ruminal protozoa on methane emissions in ruminants—A meta-analysis. J. Dairy Sci. 2022, 105, 7482–7491. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Faciola, A.P. Evaluating strategies to reduce ruminal protozoa and their impacts on nutrient utilization and animal performance in ruminants—A meta-analysis. Front. Microbiol. 2019, 10, 2648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guyader, J.; Eugène, M.; Nozière, P.; Morgavi, D.P.; Doreau, M.; Martin, C. Influence of rumen protozoa on methane emission in ruminants: A meta-analysis approach. Animal 2014, 8, 1816–1825. [Google Scholar] [CrossRef] [PubMed]
- Eugène, M.; Archimède, H.; Sauvant, D. Quantitative meta-analysis on the effects of defaunation of the rumen on growth, intake and digestion in ruminants. Livest. Prod. Sci. 2004, 85, 81–97. [Google Scholar] [CrossRef]
- Nguyen, S.H.; Nguyen, H.D.T.; Hegarty, R.S. Defaunation and its impacts on ruminal fermentation, enteric methane production and animal productivity. Livest. Res. Rural Dev. 2020, 32, 60. [Google Scholar]
- Hristov, A.N.; Oh, J.; Firkins, J.L.; Dijkstra, J.; Kebreab, E.; Waghorn, G.; Makkar, H.P.; Adesogan, A.T.; Yang, W.; Lee, C.; et al. Special topics—Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J. Anim. Sci. 2013, 91, 5045–5069. [Google Scholar] [CrossRef] [Green Version]
- Tong, F.; Wang, T.; Gao, N.L.; Liu, Z.; Cui, K.; Duan, Y.; Wu, S.; Luo, Y.; Li, Z.; Yang, C.; et al. The microbiome of the buffalo digestive tract. Nat. Commun. 2022, 13, 823. [Google Scholar] [CrossRef]
- Morvan, B.; Bonnemoy, F.; Fonty, G.; Gouet, P. Quantitative determination of H2-utilizing acetogenic and sulfate-reducing bacteria and methanogenic archaea from digestive tract of different mammals. Curr. Microbiol. 1996, 32, 129–133. [Google Scholar] [CrossRef]
- Ungerfeld, E.M. Corrigendum: Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: A meta-analysis. Front. Microbiol. 2015, 6, 538. [Google Scholar] [CrossRef] [Green Version]
- Ungerfeld, E.M.; Aedo, M.F.; Martínez, E.D.; Saldivia, M. Inhibiting methanogenesis in rumen batch cultures did not increase the recovery of metabolic hydrogen in microbial amino acids. Microorganisms 2019, 7, 115. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Marin, S.B.; Betancur-Murillo, C.L.; Isaza, G.A.; Mesa, H.; Jovel, J. Lower methane emissions were associated with higher abundance of ruminal Prevotella in a cohort of Colombian buffalos. BMC Microbiology 2020, 20, 364. [Google Scholar] [CrossRef] [PubMed]
- Bandarupalli, V.V.K.; St-Pierre, B. Identification of a candidate starch utilizing strain of prevotella albensis from bovine rumen. Microorganisms 2020, 8, 2005. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, X.; Yang, Y.; Zou, C.; Yang, Y.; Lin, B. A comparative study on rumen ecology of water buffalo and cattle calves under similar feeding regime. Vet. Med. Sci. 2020, 6, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Subharat, S.; Shu, D.; Zheng, T.; Buddle, B.M.; Kaneko, K.; Hook, S.; Janssen, P.H.; Wedlock, D.N. Vaccination of sheep with a methanogen protein provides insight into levels of antibody in saliva needed to target ruminal methanogens. PLoS ONE 2016, 11, e0159861. [Google Scholar] [CrossRef]
- Cook, S.R.; Maiti, P.K.; Chaves, A.V.; Benchaar, C.; Beauchemin, K.A.; McAllister, T.A. Avian (IgY) anti-methanogen antibodies for reducing ruminal methane production: In vitro assessment of their effects. Aust. J. Exp. Agric. 2008, 48, 260–264. [Google Scholar] [CrossRef]
- Baca-González, V.; Asensio-Calavia, P.; González-Acosta, S.; Pérez de la Lastra, J.M.; Morales de la Nuez, A. Are vaccines the solution for methane emissions from ruminants? A systematic review. Vaccines 2020, 8, 460. [Google Scholar] [CrossRef]
- Baker, S.K.; Perth, W. Method for Improving Utilization of Nutrients by Ruminant or Ruminant-like Animals. US6036950A, 14 March 2000. [Google Scholar]
- Clark, H.; Wright, A.-D.; Joblin, K.; Molano, G.; Cavanagh, A.; Peters, J. Field testing an Australian developed anti-methanogen vaccine in growing ewe lambs. In Proceedings of the Workshop on the Science of Atmospheric Trace Gases, Wellington, New Zealand, 18–19 March 2004; Clarkson, T.S., Ed.; pp. 107–108. [Google Scholar]
- Wedlock, D.N.; Janssen, P.H.; Leahy, S.C.; Shu, D.; Buddle, B.M. Progress in the development of vaccines against rumen methanogens. Animal 2013, 7 (Suppl. 2), 244–252. [Google Scholar] [CrossRef] [Green Version]
- Williams, Y.J.; Rea, S.M.; Popovski, S.; Pimm, C.L.; Williams, A.J.; Toovey, A.F.; Skillman, L.C.; Wright, A.D. Reponses of sheep to a vaccination of entodinial or mixed rumen protozoal antigens to reduce rumen protozoal numbers. Br. J. Nutr. 2008, 99, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Wright, A.D.; Kennedy, P.; O’Neill, C.J.; Toovey, A.F.; Popovski, S.; Rea, S.M.; Pimm, C.L.; Klein, L. Reducing methane emissions in sheep by immunization against rumen methanogens. Vaccine 2004, 22, 3976–3985. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, X.; Xue, B.; Peng, Q.; Wang, Z.; Yan, T.; Wang, L. Immunization against rumen methanogenesis by vaccination with a new recombinant protein. PLoS ONE 2015, 10, e0140086. [Google Scholar] [CrossRef] [Green Version]
- Williams, Y.J.; Popovski, S.; Rea, S.M.; Skillman, L.C.; Toovey, A.F.; Northwood, K.S.; Wright, A.D. A vaccine against rumen methanogens can alter the composition of archaeal populations. Appl. Environ. Microbiol. 2009, 75, 1860–1866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khampa, S.; Wanapat, M. Manipulation of rumen fermentation with organic acids supplementation in ruminants raised in the tropics. Pak. J. Nutr. 2007, 6, 20–27. [Google Scholar]
- Alvarez-Hess, P.S.; Little, S.M.; Moate, P.J.; Jacobs, J.L.; Beauchemin, K.A.; Eckard, R.J. A partial life cycle assessment of the greenhouse gas mitigation potential of feeding 3-nitrooxypropanol and nitrate to cattle. Agric. Syst. 2019, 169, 14–23. [Google Scholar] [CrossRef]
- Duin, E.C.; Wagner, T.; Shima, S.; Prakash, D.; Cronin, B.; Yáñez-Ruiz, D.R.; Duval, S.; Rümbeli, R.; Stemmler, R.T.; Thauer, R.K.; et al. Mode of action uncovered for the specific reduction of methane emissions from ruminants by the small molecule 3-nitrooxypropanol. Proc. Natl. Acad. Sci. USA 2016, 113, 6172–6177. [Google Scholar] [CrossRef] [Green Version]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Bampidis, V.; Azimonti, G.; Bastos, M.D.L.; Christensen, H.; Dusemund, B.; Fašmon Durjava, M.; Kouba, M.; López-Alonso, M. Safety and efficacy of a feed additive consisting of 3-nitrooxypropanol (Bovaer® 10) for ruminants for milk production and reproduction (DSM Nutritional Products Ltd.). EFSA J. 2021, 19, e06905. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Beauchemin, K.A.; Dong, R. A review of 3-nitrooxypropanol for enteric methane mitigation from ruminant livestock. Animals 2021, 11, 3540. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, K.; Nan, X.; Cai, M.; Yang, L.; Xiong, B.; Zhao, Y. Synergistic effects of 3-nitrooxypropanol with fumarate in the regulation of propionate formation and methanogenesis in dairy cows in vitro. Appl. Environ. Microbiol. 2022, 88, e0190821. [Google Scholar] [CrossRef]
- Dijkstra, J.; Bannink, A.; France, J.; Kebreab, E.; van Gastelen, S. Short communication: Antimethanogenic effects of 3-nitrooxypropanol depend on supplementation dose, dietary fiber content, and cattle type. J. Dairy Sci. 2018, 101, 9041–9047. [Google Scholar] [CrossRef] [Green Version]
- Hernández, J.; Benedito, J.L.; Abuelo, A.; Castillo, C. Ruminal acidosis in feedlot: From aetiology to prevention. Sci. World J. 2014, 2014, 702572. [Google Scholar] [CrossRef] [Green Version]
- Foley, P.A.; Kenny, D.A.; Callan, J.J.; Boland, T.M.; O’Mara, F.P. Effect of DL-malic acid supplementation on feed intake, methane emission, and rumen fermentation in beef cattle. J. Anim. Sci. 2009, 87, 1048–1057. [Google Scholar] [CrossRef]
- Mohammed, N.; Lila, Z.A.; Ajisaka, N.; Hara, K.; Mikuni, K.; Hara, K.; Kanda, S.; Itabashi, H. Inhibition of ruminal microbial methane production by beta-cyclodextrin iodopropane, malate and their combination in vitro. J. Anim. Physiol. Anim. Nutr. 2004, 88, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimia, S.H.; Mohini, M.; Singhal, K.K.; Miri, V.H.; Tyagi, A.K. Evaluation of complementary effects of 9,10-anthraquinone and fumaric acid on methanogenesis and ruminal fermentation in vitro. Arch. Anim. Nutr. 2011, 65, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Van Zijderveld, S.M.; Gerrits, W.J.; Dijkstra, J.; Newbold, J.R.; Hulshof, R.B.; Perdok, H.B. Persistency of methane mitigation by dietary nitrate supplementation in dairy cows. J. Dairy Sci. 2011, 94, 4028–4038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fewtrell, L. Drinking-water nitrate, methemoglobinemia, and global burden of disease: A discussion. Environ. Health Perspect. 2004, 112, 1371–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hook, S.E.; Steele, M.A.; Northwood, K.S.; Wright, A.D.; McBride, B.W. Impact of high-concentrate feeding and low ruminal pH on methanogens and protozoa in the rumen of dairy cows. Microb. Ecol. 2011, 62, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Schären, M.; Drong, C.; Kiri, K.; Riede, S.; Gardener, M.; Meyer, U.; Hummel, J.; Urich, T.; Breves, G.; Dänicke, S. Differential effects of monensin and a blend of essential oils on rumen microbiota composition of transition dairy cows. J. Dairy Sci. 2017, 100, 2765–2783. [Google Scholar] [CrossRef] [Green Version]
- National Research Council. Nutrient Requirements of Beef Cattle, 7th ed.; National Academies Press: Washington, DC, USA, 2000.
- EFSA Panel on Additives and Products or Substances used in Animal Feed. Opinion of the scientific panel on additives and products or substances used in animal feed on the safety and efficacy of the coccidiostat Elancoban® (monensin sodium) as a feed additive for calves for rearing and cattle for fattening in accordance with Regulation (EC) No 1831/2003. EFSA J. 2006, 4, 387. [Google Scholar]
- Ranga Niroshan Appuhamy, J.A.D.; Strathe, A.B.; Jayasundara, S.; Wagner-Riddle, C.; Dijkstra, J.; France, J.; Kebreab, E. Anti-methanogenic effects of monensin in dairy and beef cattle: A meta-analysis. J. Dairy Sci. 2013, 96, 5161–5173. [Google Scholar] [CrossRef] [Green Version]
- Guan, H.; Wittenberg, K.M.; Ominski, K.H.; Krause, D.O. Efficacy of ionophores in cattle diets for mitigation of enteric methane1. J. Anim. Sci. 2006, 84, 1896–1906. [Google Scholar] [CrossRef] [Green Version]
- Hook, S.E.; Northwood, K.S.; Wright, A.D.; McBride, B.W. Long-term monensin supplementation does not significantly affect the quantity or diversity of methanogens in the rumen of the lactating dairy cow. Appl. Environ. Microbiol. 2009, 75, 374–380. [Google Scholar] [CrossRef] [Green Version]
- Hook, S.E.; Wright, A.-D.G.; McBride, B.W. Methanogens: Methane producers of the rumen and mitigation strategies. Archaea 2010, 2010, 945785. [Google Scholar] [CrossRef] [Green Version]
- Odongo, N.E.; Bagg, R.; Vessie, G.; Dick, P.; Or-Rashid, M.M.; Hook, S.E.; Gray, J.T.; Kebreab, E.; France, J.; McBride, B.W. Long-term effects of feeding monensin on methane production in lactating dairy cows. J. Dairy Sci. 2007, 90, 1781–1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Committee for Medicinal Products for Veterinary Use. Monensin (Cattle, Including Dairy Cows). European Medicines Agency Veterinary Medicines and Inspections, 2007. Available online: https://www.ema.europa.eu/en/medicines/ema_group_types/ema_document-maximum_residue_limits_report?page=4 (accessed on 10 February 2023).
- Pickering, N.K.; Dodds, K.G.; Blair, H.T.; Hickson, R.E.; Johnson, P.L.; McEwan, J.C. Genetic parameters for production traits in New Zealand dual-purpose sheep, with an emphasis on dagginess1. J. Anim. Sci. 2012, 90, 1411–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pszczola, M.; Rzewuska, K.; Mucha, S.; Strabel, T. Heritability of methane emissions from dairy cows over a lactation measured on commercial farms. J. Anim. Sci. 2017, 95, 4813–4819. [Google Scholar] [CrossRef] [PubMed]
- Rowe, S.; Hickey, S.; Jonker, A.; Hess, M.; Janssen, P.; Johnson, T.; Bryson, B.; Knowler, K.; Pinares-Patino, C.; Bain, W.; et al. Selection for divergent methane yield in New Zealand sheep—A ten year perspective. In Proceedings of the 23rd Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG), Armidale, NSW, Australia, 27 October–1 November 2019; pp. 306–309. [Google Scholar]
- Pinares-Patiño, C.S.; Hickey, S.M.; Young, E.A.; Dodds, K.G.; MacLean, S.; Molano, G.; Sandoval, E.; Kjestrup, H.; Harland, R.; Hunt, C.; et al. Heritability estimates of methane emissions from sheep. Animal 2013, 7 (Suppl. 2), 316–321. [Google Scholar] [CrossRef] [Green Version]
- Sypniewski, M.; Strabel, T.; Pszczola, M. Genetic variability of methane production and concentration measured in the breath of polish holstein-friesian cattle. Animals 2021, 11, 3175. [Google Scholar] [CrossRef]
- Manzanilla-Pech, C.I.V.; L⊘vendahl, P.; Mansan Gordo, D.; Difford, G.F.; Pryce, J.E.; Schenkel, F.; Wegmann, S.; Miglior, F.; Chud, T.C.; Moate, P.J.; et al. Breeding for reduced methane emission and feed-efficient Holstein cows: An international response. J. Dairy Sci. 2021, 104, 8983–9001. [Google Scholar] [CrossRef]
- Rowe, J.B.; Gill, S.; Banks, R.G.; van der Werf, J.H.J. Genomics for the Australian sheep industry: From design to delivery. Proc. Assoc. Adv. Anim. Breed. Genet. 2013, 20, 14–17. [Google Scholar]
- Bain, W.; Bezuidenhout, L.; Jopson, N.; Pinares-Patino, C.; McEwan, J. Rumen differences between sheep identified as being low or high methane emitters. In Proceedings of the 10th World Congress on Genetics Applied to Livestock, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Hickey, S.M.; Bain, W.E.; Bilton, T.P.; Greer, G.J.; Elmes, S.; Bryson, B.; Pinares-Patiño, C.S.; Wing, J.; Jonker, A.; Young, E.A.; et al. Impact of breeding for reduced methane emissions in New Zealand sheep on maternal and health traits. Front. Genet. 2022, 13, 2165. [Google Scholar] [CrossRef]
- De Haas, Y.; Veerkamp, R.F.; de Jong, G.; Aldridge, M.N. Selective breeding as a mitigation tool for methane emissions from dairy cattle. Animal 2021, 15, 100294. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency (USEPA). Inventory of US Greenhouse Gas Emissions and Sinks 1990–2019; United States Environmental Protection Agency (USEPA): Washington, DC, USA, 2021.
- FAO. World Food and Agriculture—Statistical Yearbook; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- FAO. FAOSTAT Statistical Database; FAO: Rome, Italy, 2022. [Google Scholar]
- USDA’s Foreign Agricultural Service (FAS). 2022. Available online: https://gain.fas.usda.gov/#/search (accessed on 5 December 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Króliczewska, B.; Pecka-Kiełb, E.; Bujok, J. Strategies Used to Reduce Methane Emissions from Ruminants: Controversies and Issues. Agriculture 2023, 13, 602. https://doi.org/10.3390/agriculture13030602
Króliczewska B, Pecka-Kiełb E, Bujok J. Strategies Used to Reduce Methane Emissions from Ruminants: Controversies and Issues. Agriculture. 2023; 13(3):602. https://doi.org/10.3390/agriculture13030602
Chicago/Turabian StyleKróliczewska, Bożena, Ewa Pecka-Kiełb, and Jolanta Bujok. 2023. "Strategies Used to Reduce Methane Emissions from Ruminants: Controversies and Issues" Agriculture 13, no. 3: 602. https://doi.org/10.3390/agriculture13030602





