A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Characterization and Phylogenetic Analysis of Auxin Transporters
2.3. Gene Structure, Genome Distribution, and Duplication Analysis
2.4. Analysis of Cis-Acting Elements of Auxin Transporters
2.5. Co-Expression Analysis of Auxin Transporters
2.6. Real-Time Quantitative PCR(RT-qPCR) Analysis
3. Results
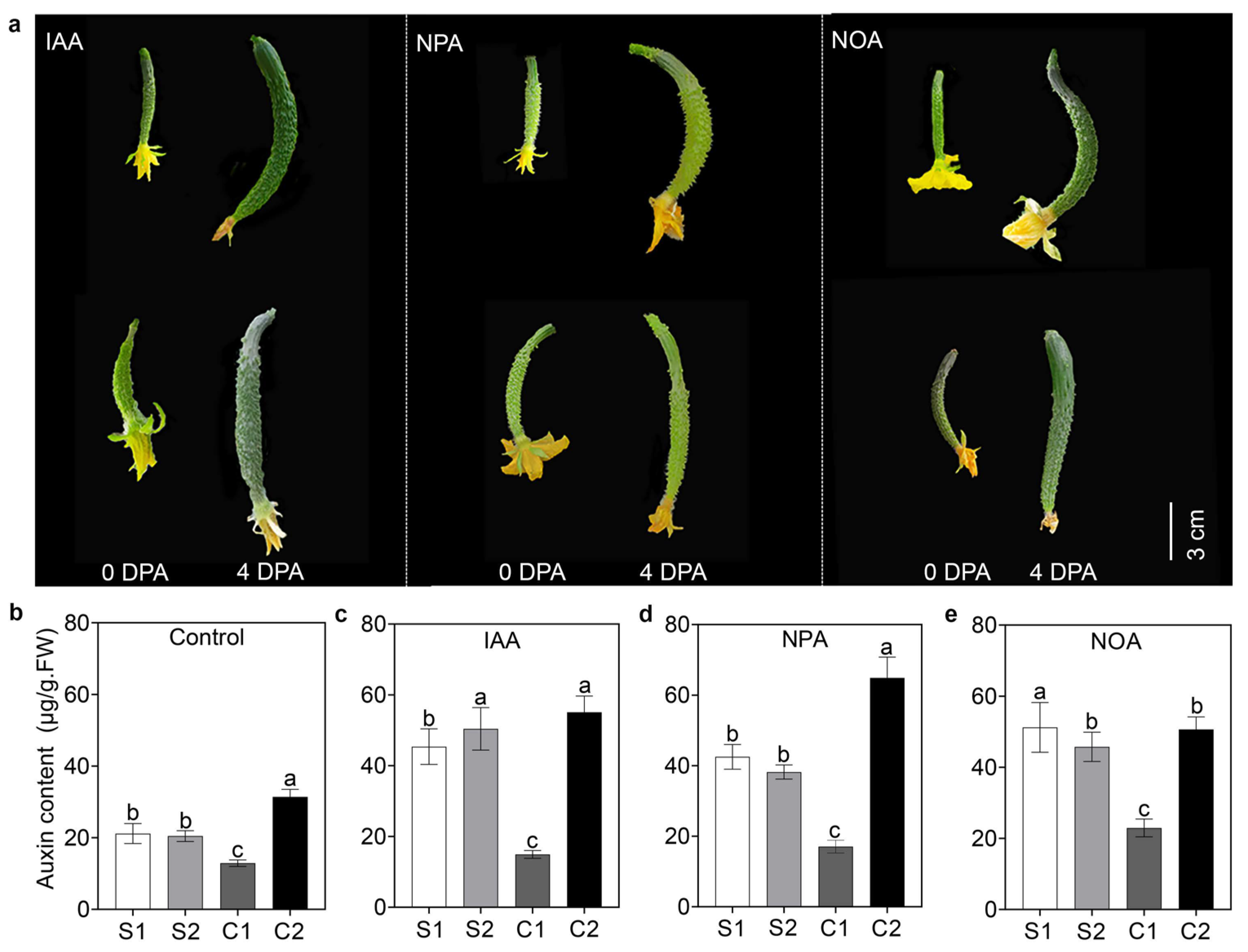
3.1. Auxin Transport Contributed to Asymmetric Auxin Distribution and Resulted in Curved Fruit
3.2. Genome-Wide Identification of Auxin Transporters
3.3. Distribution and Duplication Analysis of Auxin Transporters
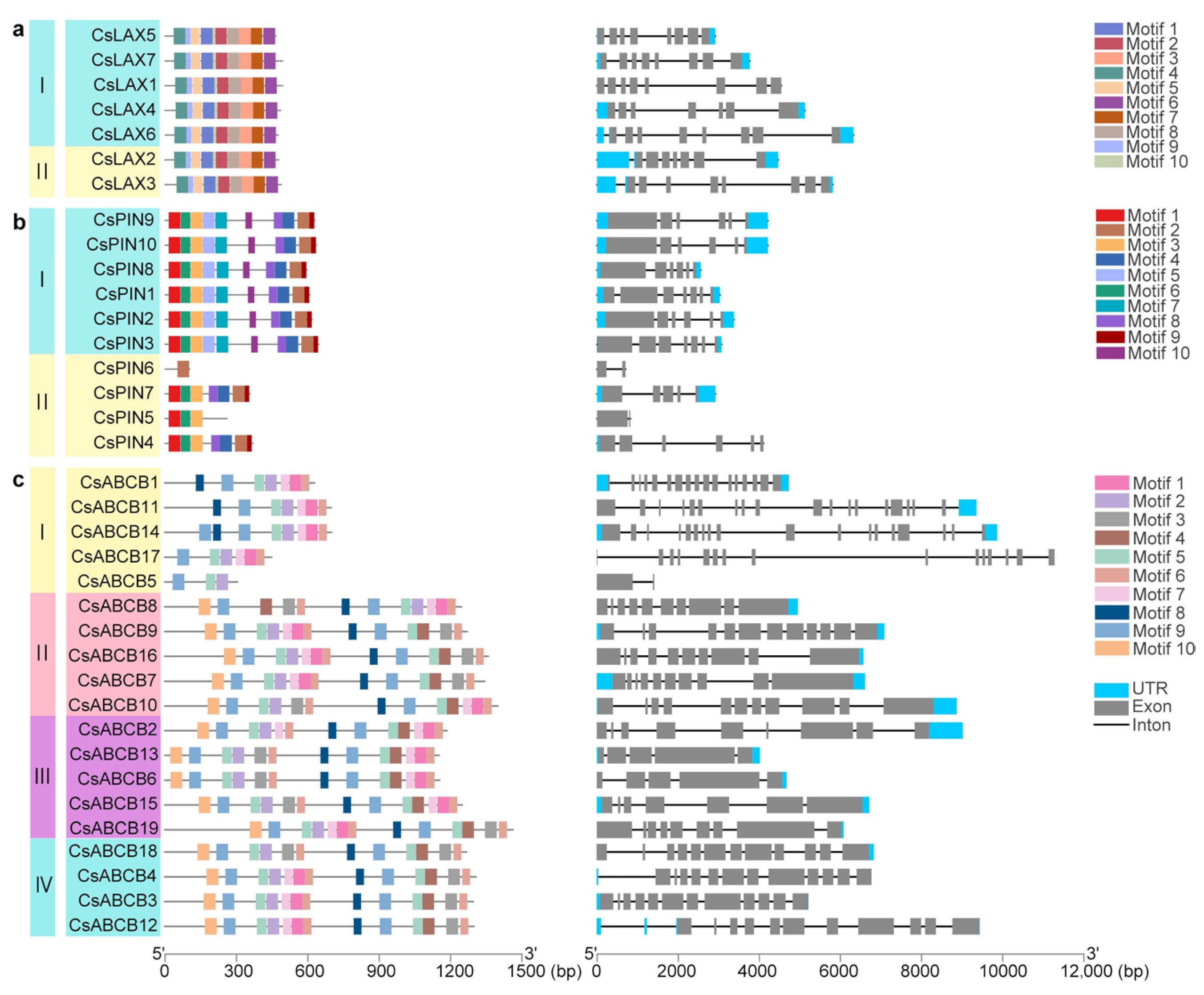
3.4. Analyses of Phylogenetic Relationships, Synteny Relationships, Conservative Motif, and Gene Structure
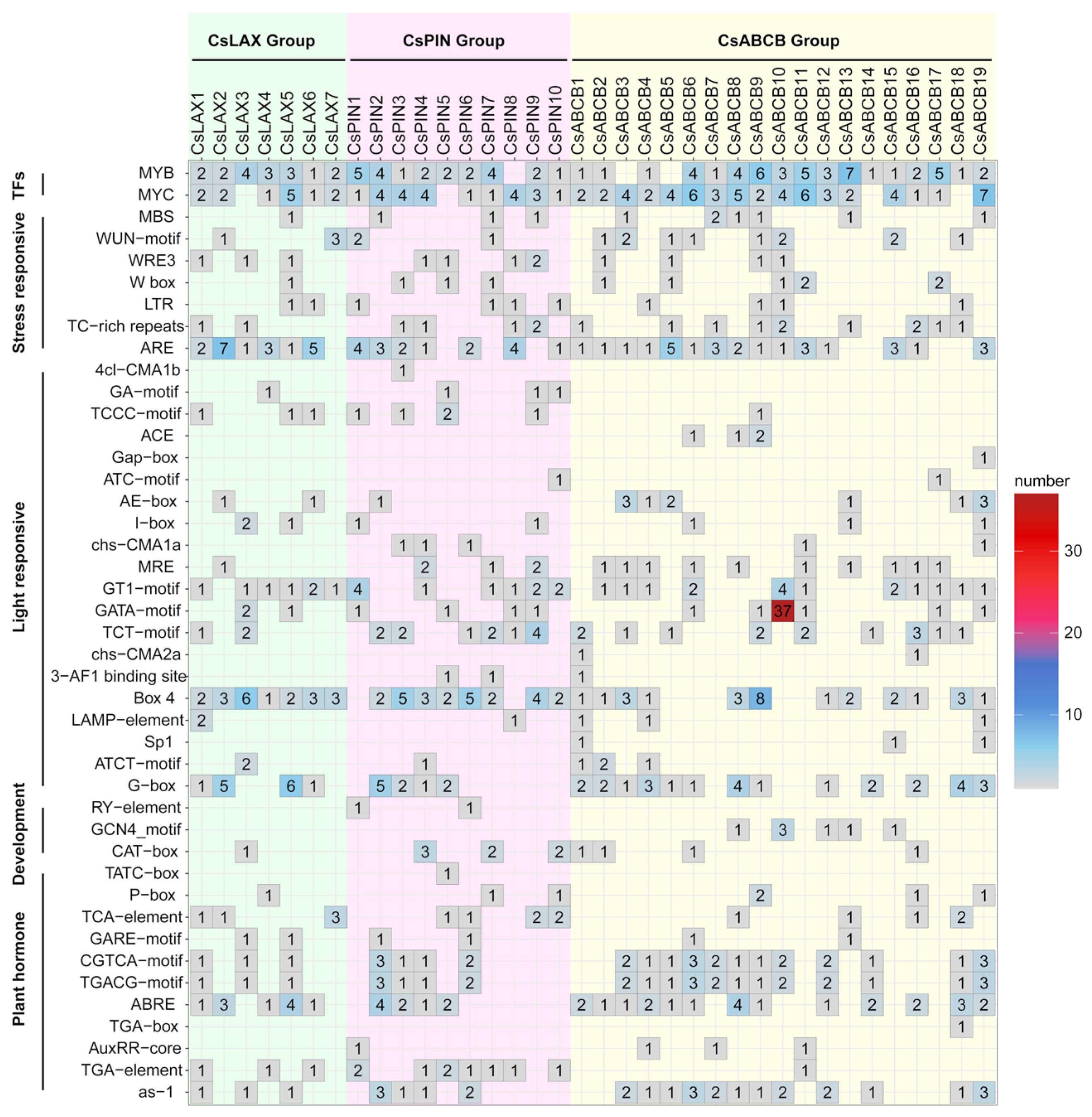
3.5. Analysis of Cis-Regulatory Elements in the Promoter Regions of Auxin Transporters
3.6. Differential Expression of Auxin Transporters Control Fruit Curving
3.7. Potential Roles of Auxin Transporters in Fruit Curving
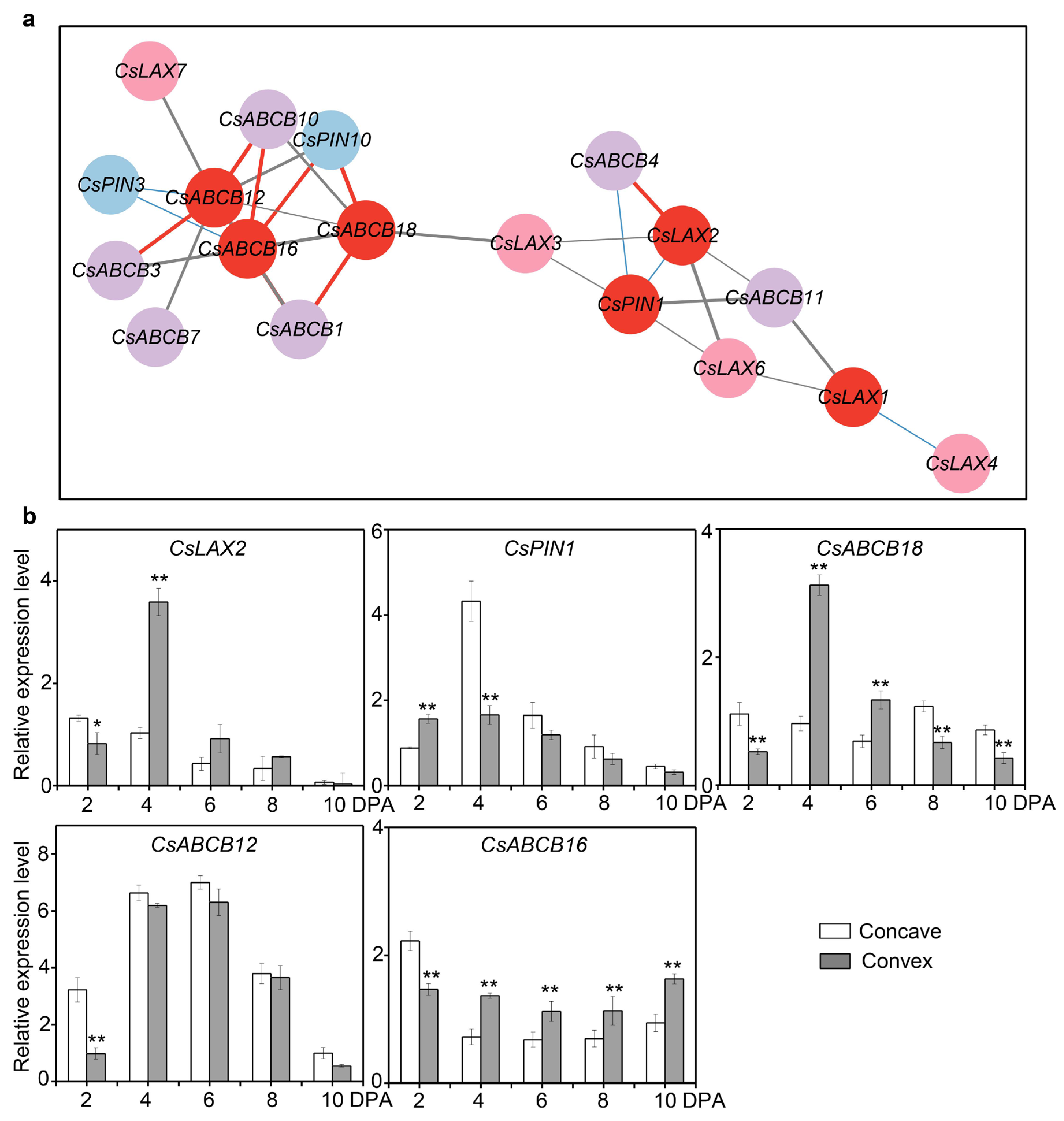
3.8. Co-Expression Analysis of TFs and Transporters
4. Discussion
4.1. Comprehensive Analysis of Auxin Transporters
4.2. Potential Roles of Auxin Transporters
4.3. A Model of Fruit Curving in Cucumber
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hohm, T.; Demarsy, E.; Quan, C.; Allenbach Petrolati, L.; Preuten, T.; Vernoux, T.; Bergmann, S.; Fankhauser, C. Plasma membrane H+-ATP ase regulation is required for auxin gradient formation preceding phototropic growth. Mol. Syst. Biol. 2014, 10, 751. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Zhang, Y. Adaptive growth: Shaping auxin-mediated root system architecture. Trends Plant Sci. 2020, 25, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, D.; Qiu, Y.; Xiao, Z.; Ji, Y.; Li, W.; Xia, Y.; Wang, Y.; Guo, H. Growth asymmetry precedes differential auxin response during apical hook initiation in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Dindas, J.; Scherzer, S.; Roelfsema, M.R.G.; von Meyer, K.; Müller, H.M.; Al-Rasheid, K.A.S.; Palme, K.; Dietrich, P.; Becker, D.; Bennett, M.J.; et al. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat. Commun. 2018, 9, 1174. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, D.; Zhang, C.; Ye, M.; Kong, N.; Ma, H.; Chen, Q. Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses. Biology 2021, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Maokai, Y.; Cheng, H.; Guo, M.; Liu, Y.; Wang, L.; Chao, S.; Zhang, M.; Lai, L.; Qin, Y. Characterization of auxin transporter AUX, PIN and PILS gene families in pineapple and evaluation of expression profiles during reproductive development and under abiotic stresses. PeerJ 2021, 9, e11410. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Petrášek, J.; Žádníková, P.; Hoyerová, K.; Pešek, B.; Raz, V.; Swarup, R.; Bennett, M.; Zažímalová, E.; Benková, E. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 2010, 137, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Tidy, A.; Abu Bakar, N.; Carrier, D.; Kerr, I.D.; Hodgman, C.; Bennett, M.J.; Swarup, R. Mechanistic insight into the role of AUXIN RESISTANCE4 in trafficking of AUXIN1 and LIKE AUX1-2. Plant Physiol. 2024, 194, 422–433. [Google Scholar] [CrossRef]
- Liu, H.; Luo, Q.; Tan, C.; Song, J.; Zhang, T.; Men, S. Biosynthesis- and transport-mediated dynamic auxin distribution during seed development controls seed size in Arabidopsis. Plant J. 2023, 113, 1259–1277. [Google Scholar] [CrossRef]
- Zhang, Y.; Berman, A.; Shani, E. Plant Hormone Transport and Localization: Signaling Molecules on the Move. Annu. Rev. Plant Biol. 2023, 74, 453–479. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; Brockington, S.F.; Rothfels, C.; Graham, S.W.; Stevenson, D.; Kutchan, T.; Rolf, M.; Thomas, P.; Wong, G.K.-S.; Leyser, O.; et al. Paralogous Radiations of PIN Proteins with Multiple Origins of Noncanonical PIN Structure. Mol. Biol. Evol. 2014, 31, 2042–2060. [Google Scholar] [CrossRef] [PubMed]
- Hammes, U.Z.; Murphy, A.S.; Schwechheimer, C. Auxin transporters—A biochemical view. Cold Spring Harbor Perspect. Biol. 2022, 14, a039875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rodriguez, L.; Li, L.; Zhang, X.; Friml, J. Functional innovations of PIN auxin transporters mark crucial evolutionary transitions during rise of flowering plants. Sci. Adv. 2020, 6, eabc8895. [Google Scholar] [CrossRef] [PubMed]
- Žádníková, P.; Petrášek, J.; Marhavý, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejátko, J. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef]
- Wang, H.; Ouyang, Q.; Yang, C.; Zhang, Z.; Hou, D.; Liu, H.; Xu, H. Mutation of OsPIN1b by CRISPR/Cas9 Reveals a Role for Auxin Transport in Modulating Rice Architecture and Root Gravitropism. Int. J. Mol. Sci. 2022, 23, 8965. [Google Scholar] [CrossRef] [PubMed]
- Ung, K.L.; Winkler, M.; Schulz, L.; Kolb, M.; Janacek, D.P.; Dedic, E.; Stokes, D.L.; Hammes, U.Z.; Pedersen, B.P. Structures and mechanism of the plant PIN-FORMED auxin transporter. Nature 2022, 609, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xia, J.; Hong, J.; Zhang, C.; Wei, H.; Ying, W.; Sun, C.; Sun, L.; Mao, Y.; Gao, Y.; et al. Structural insights into auxin recognition and efflux by Arabidopsis PIN1. Nature 2022, 609, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Aryal, B.; Di Donato, M.; Hao, P. A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 2017, 58, 1601–1614. [Google Scholar] [CrossRef]
- Que, F.; Zhu, Y.; Liu, Q.; Wei, Q.; Ramakrishnan, M. Genome-Wide Identification, Expansion, Evolution, and Expression Analysis Reveals ABCB Genes Important for Secondary Cell Wall Development in Moso Bamboo (Phyllostachys edulis). Agronomy 2023, 13, 1828. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Q.; Wang, X.; Mei, J.; Sharma, A.; Tripathi, D.K.; Yuan, H.; Zheng, B. Genome-wide identification and expression profiles of ABCB gene family in Chinese hickory (Carya cathayensis Sarg.) during grafting. Plant Physiol. Biochem. 2021, 168, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Kubeš, M.; Yang, H.; Richter, G.L.; Cheng, Y.; Młodzińska, E.; Wang, X.; Blakeslee, J.J.; Carraro, N.; Petrášek, J.; Zažímalová, E. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 2012, 69, 640–654. [Google Scholar] [CrossRef]
- Cho, M.; Lee, S.H.; Cho, H.-T. P-Glycoprotein4 displays auxin efflux transporter–like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 2007, 19, 3930–3943. [Google Scholar] [CrossRef]
- Wu, G.; Cameron, J.N.; Ljung, K.; Spalding, E.P. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. Cell Mol. Biol. 2010, 62, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Mazzella, M.A.; Casal, J.J.; Muschietti, J.P.; Fox, A.R. Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 2014, 5, 52. [Google Scholar] [CrossRef]
- Jenness, M.K.; Tayengwa, R.; Bate, G.A.; Tapken, W.; Zhang, Y.; Pang, C.; Murphy, A.S. Loss of multiple ABCB auxin transporters recapitulates the major twisted dwarf 1 phenotypes in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 840260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nasser, V.; Pisanty, O.; Omary, M.; Wulff, N.; Di Donato, M.; Tal, I.; Hauser, F.; Hao, P.; Roth, O. A transportome-scale amiRNA-based screen identifies redundant roles of Arabidopsis ABCB6 and ABCB20 in auxin transport. Nat. Commun. 2018, 9, 4204. [Google Scholar] [CrossRef]
- Chen, J.; Hu, Y.; Hao, P.; Tsering, T.; Xia, J.; Zhang, Y.; Roth, O.; Njo, M.F.; Sterck, L.; Hu, Y. ABCB-mediated shootward auxin transport feeds into the root clock. EMBO Rep. 2023, 24, e56271. [Google Scholar] [CrossRef]
- Li, S.; Wang, C.; Zhou, X.; Liu, D.; Liu, C.; Luan, J.; Qin, Z.; Xin, M. The curvature of cucumber fruits is associated with spatial variation in auxin accumulation and expression of a YUCCA biosynthesis gene. Hortic. Res. 2020, 7, 35. [Google Scholar] [CrossRef]
- Jahn, L.; Hofmann, U.; Ludwig-Müller, J. Indole-3-Acetic Acid Is Synthesized by the Endophyte Cyanodermella asteris via a Tryptophan-Dependent and -Independent Way and Mediates the Interaction with a Non-Host Plant. Int. J. Mol. Sci. 2021, 22, 2651. [Google Scholar] [CrossRef]
- Weiler, E.; Jourdan, P.; Conrad, W. Levels of indole-3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 1981, 153, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv 2022. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xin, M.; Zhou, X.; Liu, C.; Li, S.; Liu, D.; Xu, Y.; Qin, Z. The novel ethylene-responsive factor CsERF025 affects the development of fruit bending in cucumber. Plant Mol. Biol. 2017, 95, 519–531. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Li, Y.; Yang, S.; Shi, M.; Zhang, S.; Wu, S.; Chen, Y.; Li, W.; Tian, W.-M. HbARF2 and HbARF16. 3 function as negative regulators for the radial trunk growth of rubber tree. Ind. Crops Prod. 2020, 158, 112978. [Google Scholar] [CrossRef]
- Gou, H.; Nai, G.; Lu, S.; Ma, W.; Chen, B.; Mao, J. Genome-wide identification and expression analysis of PIN gene family under phytohormone and abiotic stresses in Vitis vinifera L. Physiol. Mol. Biol. Plants 2022, 28, 1905–1919. [Google Scholar] [CrossRef]
- Villaécija-Aguilar, J.A.; Körösy, C.; Maisch, L.; Hamon-Josse, M.; Petrich, A.; Magosch, S.; Chapman, P.; Bennett, T.; Gutjahr, C. KAI2 promotes Arabidopsis root hair elongation at low external phosphate by controlling local accumulation of AUX1 and PIN2. Curr. Biol. 2022, 32, 228–236. e223. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Giehl, R.F.H.; Hartmann, A.; Estevez, J.M.; Bennett, M.J.; von Wirén, N. A spatially concerted epidermal auxin signaling framework steers the root hair foraging response under low nitrogen. Curr. Biol. 2023, 33, 3926–3941.e3925. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jiang, L.; Che, G.; Pan, Y.; Li, Y.; Hou, Y.; Zhao, W.; Zhong, Y.; Ding, L.; Yan, S. A functional allele of CsFUL1 regulates fruit length through repressing CsSUP and inhibiting auxin transport in cucumber. Plant Cell 2019, 31, 1289–1307. [Google Scholar] [CrossRef]
- Dong, X.; Ma, C.; Xu, T.; Reid, M.S.; Jiang, C.-Z.; Li, T. Auxin response and transport during induction of pedicel abscission in tomato. Hortic. Res. 2021, 8, 192. [Google Scholar] [CrossRef]
- Teale, W.D.; Pasternak, T.; Dal Bosco, C.; Dovzhenko, A.; Kratzat, K.; Bildl, W.; Schwörer, M.; Falk, T.; Ruperti, B.; V Schaefer, J.; et al. Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J. 2021, 40, e104416. [Google Scholar] [CrossRef]
- Mellor, N.L.; Voß, U.; Ware, A.; Janes, G.; Barrack, D.; Bishopp, A.; Bennett, M.J.; Geisler, M.; Wells, D.M.; Band, L.R. Systems approaches reveal that ABCB and PIN proteins mediate co-dependent auxin efflux. Plant Cell 2022, 34, 2309–2327. [Google Scholar] [CrossRef]
- Hajný, J.; Tan, S.; Friml, J. Auxin canalization: From speculative models toward molecular players. Curr. Opin. Plant Biol. 2022, 65, 102174. [Google Scholar] [CrossRef]
- Hu, J.; Su, H.; Cao, H.; Wei, H.; Fu, X.; Jiang, X.; Song, Q.; He, X.; Xu, C.; Luo, K. AUXIN RESPONSE FACTOR7 integrates gibberellin and auxin signaling via interactions between DELLA and AUX/IAA proteins to regulate cambial activity in poplar. Plant Cell 2022, 34, 2688–2707. [Google Scholar] [CrossRef]
- Wang, H.-Z.; Yang, K.-Z.; Zou, J.-J.; Zhu, L.-L.; Xie, Z.D.; Morita, M.T.; Tasaka, M.; Friml, J.; Grotewold, E.; Beeckman, T. Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat. Commun. 2015, 6, 8822. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, Y.; Sun, Y.; Xie, Z.; Luo, Y.; Long, Q.; Feng, J.; Liu, X.; Wang, B.; He, D. Natural variations of OsAUX5, a target gene of OsWRKY78, control the neutral essential amino acid content in rice grains. Mol. Plant 2023, 16, 322–336. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Hou, Y.; Sun, Y.; Chen, X.; Wang, G.; Wang, H.; Zhu, B.; Du, X. The maize WRKY transcription factor ZmWRKY64 confers cadmium tolerance in Arabidopsis and maize (Zea mays L.). Plant Cell Rep. 2024, 43, 44. [Google Scholar] [CrossRef]
- Béziat, C.; Barbez, E.; Feraru, M.I.; Lucyshyn, D.; Kleine-Vehn, J. Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat. Plants 2017, 3, 17105. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, K.; Zhang, L.; Fan, L.; Zhou, X.; Li, S. A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving. Agriculture 2024, 14, 657. https://doi.org/10.3390/agriculture14050657
Lu K, Zhang L, Fan L, Zhou X, Li S. A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving. Agriculture. 2024; 14(5):657. https://doi.org/10.3390/agriculture14050657
Chicago/Turabian StyleLu, Ke, La Zhang, Lianxue Fan, Xiuyan Zhou, and Shengnan Li. 2024. "A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving" Agriculture 14, no. 5: 657. https://doi.org/10.3390/agriculture14050657



