Graphene Nanoribbons: Prospects of Application in Biomedicine and Toxicity
Abstract
:1. Introduction
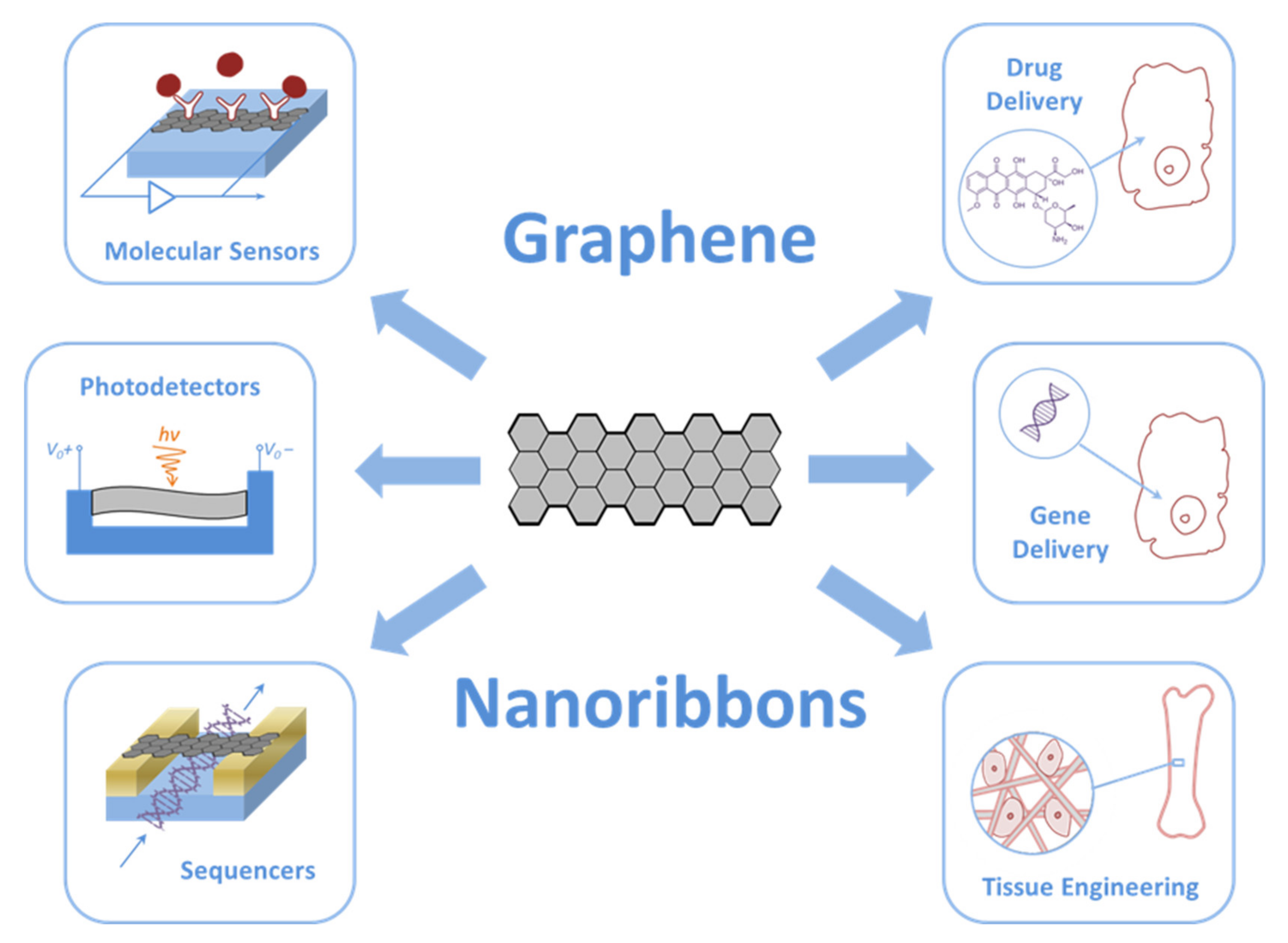
2. GNRs in Biomedicine
2.1. Electronic and Biomedical Devices
2.2. Delivery of Genes and Drugs
3. Biocompatibility and Toxicity
3.1. Biocompatibility
3.2. Toxicity
4. GNRs in the Environment
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hernandez, Y.; Pang, S.; Feng, X.; Müllen, K. 8.16—Graphene and Its Synthesis. In Polymer Science: A Comprehensive Reference; Matyjaszewski, K., Möller, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 415–438. [Google Scholar]
- Nakada, K.; Fujita, M.; Dresselhaus, G.; Dresselhaus, M.S. Edge state in graphene ribbons: Nanometer size effect and edge shape dependence. Phys. Rev. B 1996, 54, 17954–17961. [Google Scholar] [CrossRef] [Green Version]
- Wakabayashi, K.; Fujita, M.; Ajiki, H.; Sigrist, M. Electronic and magnetic properties of nanographite ribbons. Phys. Rev. B 1999, 59, 8271–8282. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Murali, R. Impact of Size Effect on Graphene Nanoribbon Transport. IEEE Electron. Device Lett. 2010, 31, 237–239. [Google Scholar] [CrossRef] [Green Version]
- Kiani, M.J.; Ahmadi, M.T.; Akbari, E.; Karimi, H.; Che Harun, F.K. Graphene Nanoribbon Based Gas Sensor. Key Eng. Mater. 2013, 553, 7–11. [Google Scholar] [CrossRef]
- Berahman, M.; Sheikhi, M.H. Hydrogen sulfide gas sensor based on decorated zigzag graphene nanoribbon with copper. Sens. Actuators B 2015, 219, 338–345. [Google Scholar] [CrossRef]
- Shekhirev, M.; Lipatov, A.; Torres, A.; Vorobeva, N.S.; Harkleroad, A.; Lashkov, A.; Sysoev, V.; Sinitskii, A. Highly Selective Gas Sensors Based on Graphene Nanoribbons Grown by Chemical Vapor Deposition. ACS Appl. Mater. Interfaces 2020, 12, 7392–7402. [Google Scholar] [CrossRef] [PubMed]
- Harada, N.; Sato, S. Electronic properties of NH4-adsorbed graphene nanoribbon as a promising candidate for a gas sensor. AIP Adv. 2016, 6, 055023. [Google Scholar] [CrossRef] [Green Version]
- Han, S.-J.; Jenkins, K.A.; Valdes Garcia, A.; Franklin, A.D.; Bol, A.A.; Haensch, W. High-Frequency Graphene Voltage Amplifier. Nano Lett. 2011, 11, 3690–3693. [Google Scholar] [CrossRef]
- Petrone, N.; Meric, I.; Hone, J.; Shepard, K.L. Graphene Field-Effect Transistors with Gigahertz-Frequency Power Gain on Flexible Substrates. Nano Lett. 2013, 13, 121–125. [Google Scholar] [CrossRef] [Green Version]
- Dimiev, A.M.; Tour, J.M. Graphene Nanoribbons: Production and Applications - Sigma Aldrich. Available online: https://www.sigmaaldrich.com/RU/ru/technical-documents/technical-article/materials-science-and-engineering/microelectronics-and-nanoelectronics/graphene-nanoribbons-production-and-applications (accessed on 4 June 2021).
- Han, M.Y.; Özyilmaz, B.; Zhang, Y.; Kim, P. Energy Band-Gap Engineering of Graphene Nanoribbons. Phys. Rev. Lett. 2007, 98, 206805. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Dai, H. Etching and narrowing of graphene from the edges. Nat. Chem. 2010, 2, 661–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abramova, V.; Slesarev, A.S.; Tour, J.M. Meniscus-Mask Lithography for Narrow Graphene Nanoribbons. ACS Nano 2013, 7, 6894–6898. [Google Scholar] [CrossRef]
- Chen, L.; Hernandez, Y.; Feng, X.; Müllen, K. From Nanographene and Graphene Nanoribbons to Graphene Sheets: Chemical Synthesis. Angew. Chem. Int. Ed. 2012, 51, 7640–7654. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Ruffieux, P.; Jaafar, R.; Bieri, M.; Braun, T.; Blankenburg, S.; Muoth, M.; Seitsonen, A.P.; Saleh, M.; Feng, X.; et al. Atomically precise bottom-up fabrication of graphene nanoribbons. Nature 2010, 466, 470–473. [Google Scholar] [CrossRef]
- Narita, A.; Feng, X.; Hernandez, Y.; Jensen, S.A.; Bonn, M.; Yang, H.; Verzhbitskiy, I.A.; Casiraghi, C.; Hansen, M.R.; Koch, A.H.R.; et al. Synthesis of structurally well-defined and liquid-phase-processable graphene nanoribbons. Nat. Chem. 2014, 6, 126–132. [Google Scholar] [CrossRef] [Green Version]
- Vo, T.H.; Shekhirev, M.; Kunkel, D.A.; Morton, M.D.; Berglund, E.; Kong, L.; Wilson, P.M.; Dowben, P.A.; Enders, A.; Sinitskii, A. Large-scale solution synthesis of narrow graphene nanoribbons. Nat. Commun. 2014, 5, 3189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosynkin, D.V.; Lu, W.; Sinitskii, A.; Pera, G.; Sun, Z.; Tour, J.M. Highly Conductive Graphene Nanoribbons by Longitudinal Splitting of Carbon Nanotubes Using Potassium Vapor. ACS Nano 2011, 5, 968–974. [Google Scholar] [CrossRef]
- Dimiev, A.; Lu, W.; Zeller, K.; Crowgey, B.; Kempel, L.C.; Tour, J.M. Low-Loss, High-Permittivity Composites Made from Graphene Nanoribbons. ACS Appl. Mater. Interfaces 2011, 3, 4657–4661. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Zhang, L.; Wang, X.; Diankov, G.; Dai, H. Narrow graphene nanoribbons from carbon nanotubes. Nature 2009, 458, 877–880. [Google Scholar] [CrossRef]
- Celis, A.; Nair, M.N.; Taleb-Ibrahimi, A.; Conrad, E.H.; Berger, C.; de Heer, W.A.; Tejeda, A. Graphene nanoribbons: Fabrication, properties and devices. J. Phys. D: Appl. Phys. 2016, 49, 143001. [Google Scholar] [CrossRef]
- Guo, X.; Baumgarten, M.; Müllen, K. Designing π-conjugated polymers for organic electronics. Prog. Polym. Sci. 2013, 38, 1832–1908. [Google Scholar] [CrossRef]
- Talirz, L.; Ruffieux, P.; Fasel, R. On-Surface Synthesis of Atomically Precise Graphene Nanoribbons. Adv. Mater. 2016, 28, 6222–6231. [Google Scholar] [CrossRef]
- Perkins, W.S. Synthesis and Applications of Graphene Nanoribbons and Heterostructures from Molecular Precursors. Ph.D. Thesis, University of California, Berkeley, CA, USA, 2017. [Google Scholar]
- Narita, A.; Wang, X.-Y.; Feng, X.; Müllen, K. New advances in nanographene chemistry. Chem. Soc. Rev. 2015, 44, 6616–6643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magda, G.Z.; Jin, X.; Hagymási, I.; Vancsó, P.; Osváth, Z.; Nemes-Incze, P.; Hwang, C.; Biró, L.P.; Tapasztó, L. Room-temperature magnetic order on zigzag edges of narrow graphene nanoribbons. Nature 2014, 514, 608–611. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-X.; Zhou, M.; Li, X.; Li, S.-Y.; Wu, X.; Duan, W.; He, L. Energy gaps of atomically precise armchair graphene sidewall nanoribbons. Phys. Rev. B 2016, 93, 241403. [Google Scholar] [CrossRef] [Green Version]
- Merino-Díez, N.; Garcia-Lekue, A.; Carbonell-Sanromà, E.; Li, J.; Corso, M.; Colazzo, L.; Sedona, F.; Sánchez-Portal, D.; Pascual, J.I.; de Oteyza, D.G. Width-Dependent Band Gap in Armchair Graphene Nanoribbons Reveals Fermi Level Pinning on Au(111). ACS Nano 2017, 11, 11661–11668. [Google Scholar] [CrossRef]
- Narita, A.; Chen, Z.; Chen, Q.; Müllen, K. Solution and on-surface synthesis of structurally defined graphene nanoribbons as a new family of semiconductors. Chem. Sci. 2019, 10, 964–975. [Google Scholar] [CrossRef] [Green Version]
- de Sousa Araújo Cassiano, T.; Monteiro, F.F.; Evaristo de Sousa, L.; Magela e Silva, G.; de Oliveira Neto, P.H. Smooth gap tuning strategy for cove-type graphene nanoribbons. RSC Adv. 2020, 10, 26937–26943. [Google Scholar] [CrossRef]
- Johnson, A.P.; Sabu, C.; Swamy, N.K.; Anto, A.; Gangadharappa, H.V.; Pramod, K. Graphene nanoribbon: An emerging and efficient flat molecular platform for advanced biosensing. Biosens. Bioelectron. 2021, 184, 113245. [Google Scholar] [CrossRef]
- Shende, P.; Pathan, N. Graphene nanoribbons: A state-of-the-art in health care. Int. J. Pharm. 2021, 595, 120269. [Google Scholar] [CrossRef]
- Johnson, A.P.; Gangadharappa, H.V.; Pramod, K. Graphene nanoribbons: A promising nanomaterial for biomedical applications. J. Controlled Release 2020, 325, 141–162. [Google Scholar] [CrossRef]
- Shende, P.; Augustine, S.; Prabhakar, B. A review on graphene nanoribbons for advanced biomedical applications. Carbon Lett. 2020, 30, 465–475. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Soroshnia, S.; Hashemi, S.A.; Babapoor, A.; Younes, G.; Amani, A. Graphene Nano-ribbon Based high potential and Efficiency for DNA, Cancer therapy and drug delivery applications. Drug Metab. Rev. 2019, 51, 1–35. [Google Scholar] [CrossRef]
- Cai, J.; Pignedoli, C.A.; Talirz, L.; Ruffieux, P.; Söde, H.; Liang, L.; Meunier, V.; Berger, R.; Li, R.; Feng, X.; et al. Graphene nanoribbon heterojunctions. Nat. Nanotech. 2014, 9, 896–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, P.B.; Pedramrazi, Z.; Madani, A.; Chen, Y.-C.; Oteyza, D.G.d.; Chen, C.; Fischer, F.R.; Crommie, M.F.; Bokor, J. Bottom-up graphene nanoribbon field-effect transistors. Appl. Phys. Lett. 2013, 103, 253114. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Xie, L.; Lee, K.K.; Hu, Z.; Tan, S.; Chen, W.; Sow, C.H.; Chen, K.; Liu, Y.; Wee, A.T.S. Controllable unzipping for intramolecular junctions of graphene nanoribbons and single-walled carbon nanotubes. Nat. Commun. 2013, 4, 1374. [Google Scholar] [CrossRef] [Green Version]
- Freitag, M.; Low, T.; Zhu, W.; Yan, H.; Xia, F.; Avouris, P. Photocurrent in graphene harnessed by tunable intrinsic plasmons. Nat. Commun. 2013, 4, 1951. [Google Scholar] [CrossRef] [Green Version]
- Ryzhii, V.; Ryabova, N.; Ryzhii, M.; Baryshnikov, N.V.; Karasik, V.E.; Mitin, V.; Otsuji, T. Terahertz and infrared photodetectors based on multiple graphene layer and nanoribbon structures. Opto-Electron. Rev. 2012, 20, 15–25. [Google Scholar] [CrossRef]
- Johnson, J.L.; Behnam, A.; Pearton, S.J.; Ural, A. Hydrogen Sensing Using Pd-Functionalized Multi-Layer Graphene Nanoribbon Networks. Adv. Mater. 2010, 22, 4877–4880. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.; Sharifi, M.J. Graphene nanoribbon photodetectors based on an asymmetric potential barrier: A new concept and a new structure. J. Comput. Electron. 2018, 17, 531–539. [Google Scholar] [CrossRef]
- Yu, X.; Dong, Z.; Liu, Y.; Liu, T.; Tao, J.; Zeng, Y.; Yang, J.K.W.; Wang, Q.J. A high performance, visible to mid-infrared photodetector based on graphene nanoribbons passivated with HfO2. Nanoscale 2016, 8, 327–332. [Google Scholar] [CrossRef]
- Isaeva, O.G.; Katkov, V.L.; Osipov, V.A. DNA sequencing through graphene nanogap: A model of sequential electron transport. Eur. Phys. J. B 2014, 87, 272. [Google Scholar] [CrossRef]
- Chen, W.; Liu, G.-C.; Ouyang, J.; Gao, M.-J.; Liu, B.; Zhao, Y.-D. Graphene nanopores toward DNA sequencing: A review of experimental aspects. Sci. China Chem. 2017, 60, 721–729. [Google Scholar] [CrossRef]
- Wu, X.; Mu, F.; Wang, Y.; Zhao, H. Graphene and Graphene-Based Nanomaterials for DNA Detection: A Review. Molecules 2018, 23, 2050. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Fengwen, M.; Zhao, H. Synthesis and potential applications of nanoporous graphene: A review. Proc. Nat. Res. Soc. 2018, 2, 02003. [Google Scholar] [CrossRef]
- Arjmandi-Tash, H.; Belyaeva, L.A.; Schneider, G.F. Single molecule detection with graphene and other two-dimensional materials: Nanopores and beyond. Chem. Soc. Rev. 2016, 45, 476–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, T.; Zhang, B.; Prezhdo, O.V. Detection of Nucleic Acids with Graphene Nanopores: Ab Initio Characterization of a Novel Sequencing Device. Nano Lett. 2010, 10, 3237–3242. [Google Scholar] [CrossRef] [PubMed]
- Traversi, F.; Raillon, C.; Benameur, S.M.; Liu, K.; Khlybov, S.; Tosun, M.; Krasnozhon, D.; Kis, A.; Radenovic, A. Detecting the translocation of DNA through a nanopore using graphene nanoribbons. Nat. Nanotech. 2013, 8, 939–945. [Google Scholar] [CrossRef]
- Min, S.K.; Kim, W.Y.; Cho, Y.; Kim, K.S. Fast DNA sequencing with a graphene-based nanochannel device. Nat. Nanotech. 2011, 6, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Agah, S.; Zheng, M.; Pasquali, M.; Kolomeisky, A.B. DNA sequencing by nanopores: Advances and challenges. J. Phys. D: Appl. Phys. 2016, 49, 413001. [Google Scholar] [CrossRef] [Green Version]
- Heerema, S.J.; Vicarelli, L.; Pud, S.; Schouten, R.N.; Zandbergen, H.W.; Dekker, C. Probing DNA Translocations with Inplane Current Signals in a Graphene Nanoribbon with a Nanopore. ACS Nano 2018, 12, 2623–2633. [Google Scholar] [CrossRef] [Green Version]
- Puster, M.; Rodríguez-Manzo, J.A.; Balan, A.; Drndić, M. Toward Sensitive Graphene Nanoribbon–Nanopore Devices by Preventing Electron Beam-Induced Damage. ACS Nano 2013, 7, 11283–11289. [Google Scholar] [CrossRef]
- Puster, M.; Balan, A.; Rodríguez-Manzo, J.A.; Danda, G.; Ahn, J.-H.; Parkin, W.; Drndić, M. Cross-Talk Between Ionic and Nanoribbon Current Signals in Graphene Nanoribbon-Nanopore Sensors for Single-Molecule Detection. Small 2015, 11, 6309–6316. [Google Scholar] [CrossRef] [Green Version]
- Saha, K.K.; Drndić, M.; Nikolić, B.K. DNA Base-Specific Modulation of Microampere Transverse Edge Currents through a Metallic Graphene Nanoribbon with a Nanopore. Nano Lett. 2012, 12, 50–55. [Google Scholar] [CrossRef] [Green Version]
- Paulechka, E.; Wassenaar, T.A.; Kroenlein, K.; Kazakov, A.; Smolyanitsky, A. Nucleobase-functionalized graphene nanoribbons for accurate high-speed DNA sequencing. Nanoscale 2016, 8, 1861–1867. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, B.A.G.; Pérez-Caro, M.; Alencar, R.S.; Filho, A.G.S.; Aguiar, J.A. Graphene nanoribbons and iron oxide nanoparticles composite as a potential candidate in DNA sensing applications. J. Appl. Phys. 2020, 127, 044901. [Google Scholar] [CrossRef]
- Feng, Q.; Zhao, X.; Guo, Y.; Liu, M.; Wang, P. Stochastic DNA walker for electrochemical biosensing sensitized with gold nanocages@graphene nanoribbons. Biosens. Bioelectron. 2018, 108, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Mehmeti, E.; Stanković, D.M.; Chaiyo, S.; Zavasnik, J.; Žagar, K.; Kalcher, K. Wiring of glucose oxidase with graphene nanoribbons: An electrochemical third generation glucose biosensor. Microchim. Acta 2017, 184, 1127–1134. [Google Scholar] [CrossRef]
- Rostami, S.; Niroumand, R.; Jabbari, A. Enhanced LSPR performance of graphene nanoribbons-silver nanoparticles hybrid as a colorimetric sensor for sequential detection of dopamine and glutathione. Anal. Chim. Acta 2020, 1120, 11–23. [Google Scholar] [CrossRef]
- Sainz, R.; del Pozo, M.; Vilas-Varela, M.; Castro-Esteban, J.; Pérez Corral, M.; Vázquez, L.; Blanco, E.; Peña, D.; Martín-Gago, J.A.; Ellis, G.J.; et al. Chemically synthesized chevron-like graphene nanoribbons for electrochemical sensors development: Determination of epinephrine. Sci. Rep. 2020, 10, 14614. [Google Scholar] [CrossRef] [PubMed]
- Lalwani, G.; Cai, X.; Nie, L.; Wang, L.; Sitharaman, B. Graphene-based contrast agents for photoacoustic and thermoacoustic tomography. Photoacoustics 2013, 1, 62–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurunathan, S.; Kim, J.-H. Synthesis, toxicity, biocompatibility, and biomedical applications of graphene and graphene-related materials. Int. J. Nanomed. 2016, 11, 1927–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, F.-P.; Peng, S.-L.; Zhang, H.; Weng, L.-B.; Xu, H. A biosensor based on graphene nanoribbon with nanopores: A first-principles devices-design. Chin. Phys. B 2011, 20, 058504. [Google Scholar] [CrossRef]
- Avdoshenko, S.M.; Nozaki, D.; Gomes da Rocha, C.; González, J.W.; Lee, M.H.; Gutierrez, R.; Cuniberti, G. Dynamic and Electronic Transport Properties of DNA Translocation through Graphene Nanopores. Nano Lett. 2013, 13, 1969–1976. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, H.; Algaragholy, L.; Pope, T.; Bailey, S.; Visontai, D.; Manrique, D.; Ferrer, J.; Garcia-Suarez, V.; Sangtarash, S.; Lambert, C.J. Graphene Sculpturene Nanopores for DNA Nucleobase Sensing. J. Phys. Chem. B 2014, 118, 6908–6914. [Google Scholar] [CrossRef]
- Jampilek, J.; Kralova, K. Advances in Drug Delivery Nanosystems Using Graphene-Based Materials and Carbon Nanotubes. Materials 2021, 14, 1059. [Google Scholar] [CrossRef] [PubMed]
- Janani, K.; Thiruvadigal, D.J. Density functional study on covalent functionalization of zigzag graphene nanoribbon through l-Phenylalanine and boron doping: Effective nanocarriers in drug delivery applications. Appl. Surf. Sci. 2018, 449, 815–822. [Google Scholar] [CrossRef]
- Mullick Chowdhury, S.; Zafar, S.; Tellez, V.; Sitharaman, B. Graphene Nanoribbon-Based Platform for Highly Efficacious Nuclear Gene Delivery. ACS Biomater. Sci. Eng. 2016, 2, 798–808. [Google Scholar] [CrossRef]
- Dong, H.; Ding, L.; Yan, F.; Ji, H.; Ju, H. The use of polyethylenimine-grafted graphene nanoribbon for cellular delivery of locked nucleic acid modified molecular beacon for recognition of microRNA. Biomaterials 2011, 32, 3875–3882. [Google Scholar] [CrossRef]
- Chowdhury, S.M.; Surhland, C.; Sanchez, Z.; Chaudhary, P.; Suresh Kumar, M.A.; Lee, S.; Peña, L.A.; Waring, M.; Sitharaman, B.; Naidu, M. Graphene nanoribbons as a drug delivery agent for lucanthone mediated therapy of glioblastoma multiforme. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Dasari Shareena, T.P.; McShan, D.; Dasmahapatra, A.K.; Tchounwou, P.B. A Review on Graphene-Based Nanomaterials in Biomedical Applications and Risks in Environment and Health. Nano-Micro Lett. 2018, 10, 53. [Google Scholar] [CrossRef]
- Mullick Chowdhury, S.; Manepalli, P.; Sitharaman, B. Graphene nanoribbons elicit cell specific uptake and delivery via activation of epidermal growth factor receptor enhanced by human papillomavirus E5 protein. Acta Biomater. 2014, 10, 4494–4504. [Google Scholar] [CrossRef] [Green Version]
- Wieduwilt, M.J.; Moasser, M.M. The epidermal growth factor receptor family: Biology driving targeted therapeutics. Cell. Mol. Life Sci. 2008, 65, 1566–1584. [Google Scholar] [CrossRef] [Green Version]
- Engelking, L.R. Chapter 59: Sphingolipids. In Textbook of Veterinary Physiological Chemistry (Third Edition); Academic Press: San Diego, CA, USA, 2014; pp. 378–383. [Google Scholar]
- Goñi, F.M.; Alonso, A. Biophysics of sphingolipids I. Membrane properties of sphingosine, ceramides and other simple sphingolipids. Biochim. Biophys. Acta 2006, 1758, 1902–1921. [Google Scholar] [CrossRef] [Green Version]
- Padrón, J.M. Sphingolipids in anticancer therapy. Curr. Med. Chem. 2006, 13, 755–770. [Google Scholar] [CrossRef]
- Saddoughi, S.A.; Song, P.; Ogretmen, B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell. Biochem. 2008, 49, 413–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ségui, B.; Andrieu-Abadie, N.; Jaffrézou, J.P.; Benoist, H.; Levade, T. Sphingolipids as modulators of cancer cell death: Potential therapeutic targets. Biochim. Biophys. Acta 2006, 1758, 2104–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suhrland, C.; Truman, J.P.; Obeid, L.M.; Sitharaman, B. Oxidized graphene nanoparticles as a delivery system for the pro-apoptotic sphingolipid C(6) ceramide. J. Biomed. Mater. Res. Part. A 2019, 107, 25–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suhrland, C.; Truman, J.P.; Obeid, L.M.; Sitharaman, B. Delivery of long chain C(16) and C(24) ceramide in HeLa cells using oxidized graphene nanoribbons. J. Biomed. Mater. Res. Part. B, Appl. Biomater. 2020, 108, 1141–1156. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Wan, W.; Li, L.; Dong, Y.; Zhao, Z.; Qiu, J. Multifunctional nitrogen-doped graphene nanoribbon aerogels for superior lithium storage and cell culture. Nanoscale 2016, 8, 2159–2167. [Google Scholar] [CrossRef]
- Chowdhury, S.M.; Fang, J.; Sitharaman, B. Interaction of graphene nanoribbons with components of the blood vascular system. Future Sci. OA 2015, 1, 3. [Google Scholar] [CrossRef] [Green Version]
- Silva, M.; Caridade, S.G.; Vale, A.C.; Cunha, E.; Sousa, M.P.; Mano, J.F.; Paiva, M.C.; Alves, N.M. Biomedical films of graphene nanoribbons and nanoflakes with natural polymers. RSC Adv. 2017, 7, 27578–27594. [Google Scholar] [CrossRef] [Green Version]
- Foreman, H.-C.C.; Lalwani, G.; Kalra, J.; Krug, L.T.; Sitharaman, B. Gene delivery to mammalian cells using a graphene nanoribbon platform. J. Mater. Chem. B 2017, 5, 2347–2354. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-J.; Lin, C.-W.; Yang, H.-W.; Lin, K.-J.; Wey, S.-P.; Sun, C.-L.; Wei, K.-C.; Yen, T.-C.; Lin, C.-I.; Ma, C.-C.M.; et al. Biodistribution of PEGylated graphene oxide nanoribbons and their application in cancer chemo-photothermal therapy. Carbon 2014, 74, 83–95. [Google Scholar] [CrossRef]
- Mbeh, D.A.; Akhavan, O.; Javanbakht, T.; Mahmoudi, M.; Yahia, L.H. Cytotoxicity of protein corona-graphene oxide nanoribbons on human epithelial cells. Appl. Surf. Sci. 2014, 320, 596–601. [Google Scholar] [CrossRef]
- Mullick Chowdhury, S.; Lalwani, G.; Zhang, K.; Yang, J.Y.; Neville, K.; Sitharaman, B. Cell specific cytotoxicity and uptake of graphene nanoribbons. Biomaterials 2013, 34, 283–293. [Google Scholar] [CrossRef] [Green Version]
- Mari, E.; Mardente, S.; Morgante, E.; Tafani, M.; Lococo, E.; Fico, F.; Valentini, F.; Zicari, A. Graphene Oxide Nanoribbons Induce Autophagic Vacuoles in Neuroblastoma Cell Lines. Int. J. Mol. Sci. 2016, 17, 1995. [Google Scholar] [CrossRef] [Green Version]
- Khim Chng, E.L.; Chua, C.K.; Pumera, M. Graphene oxide nanoribbons exhibit significantly greater toxicity than graphene oxide nanoplatelets. Nanoscale 2014, 6, 10792–10797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullick Chowdhury, S.; Dasgupta, S.; McElroy, A.E.; Sitharaman, B. Structural disruption increases toxicity of graphene nanoribbons. J. Appl. Toxicol. 2014, 34, 1235–1246. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E.; Emamy, H.; Akhavan, F. Genotoxicity of graphene nanoribbons in human mesenchymal stem cells. Carbon 2013, 54, 419–431. [Google Scholar] [CrossRef]
- Ou, L.; Song, B.; Liang, H.; Liu, J.; Feng, X.; Deng, B.; Sun, T.; Shao, L. Toxicity of graphene-family nanoparticles: A general review of the origins and mechanisms. Part. Fibre Toxicol. 2016, 13, 57. [Google Scholar] [CrossRef] [Green Version]
- Talukdar, Y.; Rashkow, J.; Lalwani, G.; Kanakia, S.; Sitharaman, B. The effects of graphene nanostructures on mesenchymal stem cells. Biomaterials 2014, 35, 4863–4877. [Google Scholar] [CrossRef] [Green Version]
- Akhavan, O.; Ghaderi, E.; Emamy, H. Nontoxic concentrations of PEGylated graphene nanoribbons for selective cancer cell imaging and photothermal therapy. J. Mater. Chem. 2012, 22, 20626–20633. [Google Scholar] [CrossRef]
- Gurcan, C.; Taheri, H.; Bianco, A.; Delogu, L.G.; Yilmazer, A. A closer look at the genotoxicity of graphene based materials. J. Phys. Mater. 2019, 3, 014007. [Google Scholar] [CrossRef]
- Madannejad, R.; Shoaie, N.; Jahanpeyma, F.; Darvishi, M.H.; Azimzadeh, M.; Javadi, H. Toxicity of carbon-based nanomaterials: Reviewing recent reports in medical and biological systems. Chem. Biol. Interact. 2019, 307, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Ema, M.; Gamo, M.; Honda, K. A review of toxicity studies of single-walled carbon nanotubes in laboratory animals. Regul. Toxicol. Pharm. 2016, 74, 42–63. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, Y.; Horie, M.; Kobayashi, N.; Shinohara, N.; Shimada, M. Inhalation Toxicity Assessment of Carbon-Based Nanoparticles. Acc. Chem. Res. 2013, 46, 770–781. [Google Scholar] [CrossRef] [PubMed]
- De Maio, F.; Palmieri, V.; De Spirito, M.; Delogu, G.; Papi, M. Carbon nanomaterials: A new way against tuberculosis. Expert Rev. Med. Devices 2019, 16, 863–875. [Google Scholar] [CrossRef]
- Raja, I.S.; Song, S.J.; Kang, M.S.; Lee, Y.B.; Kim, B. Toxicity of Zero- and One-Dimensional Carbon. Nanomaterials 2019, 9, 1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalwani, G.; Xing, W.; Sitharaman, B. Enzymatic degradation of oxidized and reduced graphene nanoribbons by lignin peroxidase. J. Mater. Chem. B 2014, 2, 6354–6362. [Google Scholar] [CrossRef] [Green Version]
- Lalwani, G.; D’Agati, M.; Khan, A.M.; Sitharaman, B. Toxicology of graphene-based nanomaterials. Adv. Drug Delivery Rev. 2016, 105, 109–144. [Google Scholar] [CrossRef] [Green Version]
- Al-Jumaili, A.; Alancherry, S.; Bazaka, K.; Jacob, M.V. Review on the Antimicrobial Properties of Carbon Nanostructures. Materials 2017, 10, 1066. [Google Scholar] [CrossRef] [PubMed]
- Maas, M. Carbon Nanomaterials as Antibacterial Colloids. Materials 2016, 9, 617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maleki Dizaj, S.; Mennati, A.; Jafari, S.; Khezri, K.; Adibkia, K. Antimicrobial activity of carbon-based nanoparticles. Adv. Pharm Bull. 2015, 5, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, X.; Chen, J.; Han, H.; Yuan, Z. Evaluation and mechanism of antifungal effects of carbon nanomaterials in controlling plant fungal pathogen. Carbon 2014, 68, 798–806. [Google Scholar] [CrossRef]
- Hao, Y.; Cao, X.; Ma, C.; Zhang, Z.; Zhao, N.; Ali, A.; Hou, T.; Xiang, Z.; Zhuang, J.; Wu, S.; et al. Potential Applications and Antifungal Activities of Engineered Nanomaterials against Gray Mold Disease Agent Botrytis cinerea on Rose Petals. Front. Plant Sci. 2017, 8, 1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghafari, P.; St-Denis, C.H.; Power, M.E.; Jin, X.; Tsou, V.; Mandal, H.S.; Bols, N.C.; Tang, X.S. Impact of carbon nanotubes on the ingestion and digestion of bacteria by ciliated protozoa. Nat. Nanotech. 2008, 3, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Basiuk, E.V.; Ochoa-Olmos, O.E.; De la Mora-Estrada, L.F. Ecotoxicological effects of carbon nanomaterials on algae, fungi and plants. J. Nanosci. Nanotech. 2011, 11, 3016–3038. [Google Scholar] [CrossRef]
- Freixa, A.; Acuña, V.; Sanchís, J.; Farré, M.; Barceló, D.; Sabater, S. Ecotoxicological effects of carbon based nanomaterials in aquatic organisms. Sci. Total Environ. 2018, 619–620, 328–337. [Google Scholar] [CrossRef]
- Saxena, P.; Sangela, V.; Ranjan, S.; Dutta, V.; Dasgupta, N.; Phulwaria, M.; Rathore, D.S.; Harish. Aquatic nanotoxicology: Impact of carbon nanomaterials on algal flora. Energy Ecol. Environ. 2020, 5, 240–252. [Google Scholar] [CrossRef]
- Zaytseva, O.; Neumann, G. Carbon nanomaterials: Production, impact on plant development, agricultural and environmental applications. Chem. Biol. Technol. Agric. 2016, 3, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Zhang, R.; Fang, X.; Song, T.; Cai, X.; Liu, H.; Du, S. Toxic effects of graphene on the growth and nutritional levels of wheat (Triticum aestivum L.): Short- and long-term exposure studies. J. Hazard. Mater. 2016, 317, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jin, L.; Wang, Y.; Kong, Y.; Wang, D. Prolonged exposure to multi-walled carbon nanotubes dysregulates intestinal mir-35 and its direct target MAB-3 in nematode Caenorhabditis elegans. Sci. Rep. 2019, 9, 12144. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, J.; Wang, D. A MicroRNA-Mediated Insulin Signaling Pathway Regulates the Toxicity of Multi-Walled Carbon Nanotubes in Nematode Caenorhabditis elegans. Sci. Rep. 2016, 6, 23234. [Google Scholar] [CrossRef] [Green Version]
- Walczynska, M.; Jakubowski, W.; Wasiak, T.; Kadziola, K.; Bartoszek, N.; Kotarba, S.; Siatkowska, M.; Komorowski, P.; Walkowiak, B. Toxicity of silver nanoparticles, multiwalled carbon nanotubes, and dendrimers assessed with multicellular organism Caenorhabditis elegans. Toxicol. Mech. Meth. 2018, 28, 432–439. [Google Scholar] [CrossRef]
- Bai, X.; Zhao, S.; Duo, L. Impacts of carbon nanomaterials on the diversity of microarthropods in turfgrass soil. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Hassan, A. Toxic and benefit effects on human and animal health Book 2. In Carbon Nanomaterials; Elsevier: Amsterdam, The Netherlands, 2019; p. 25. [Google Scholar]
- Liang, G.; Yin, L.; Zhang, J.; Liu, R.; Zhang, T.; Ye, B.; Pu, Y. Effects of subchronic exposure to multi-walled carbon nanotubes on mice. J. Toxicol. Environ. Health. Part A 2010, 73, 463–470. [Google Scholar] [CrossRef]
- Mohammadi, E.; Zeinali, M.; Mohammadi-Sardoo, M.; Iranpour, M.; Behnam, B.; Mandegary, A. The effects of functionalization of carbon nanotubes on toxicological parameters in mice. Hum. Exp. Toxicol. 2020, 39, 1147–1167. [Google Scholar] [CrossRef]
- Porter, D.W.; Hubbs, A.F.; Mercer, R.R.; Wu, N.; Wolfarth, M.G.; Sriram, K.; Leonard, S.; Battelli, L.; Schwegler-Berry, D.; Friend, S.; et al. Mouse pulmonary dose- and time course-responses induced by exposure to multi-walled carbon nanotubes. Toxicology 2010, 269, 136–147. [Google Scholar] [CrossRef]
- Jackson, P.; Jacobsen, N.R.; Baun, A.; Birkedal, R.; Kühnel, D.; Jensen, K.A.; Vogel, U.; Wallin, H. Bioaccumulation and ecotoxicity of carbon nanotubes. Chem. Cent. J. 2013, 7, 154. [Google Scholar] [CrossRef] [Green Version]
- Xie, P.; Yang, S.-T.; He, T.; Yang, S.; Tang, X.-H. Bioaccumulation and Toxicity of Carbon Nanoparticles Suspension Injection in Intravenously Exposed Mice. Int. J. Mol. Sci. 2017, 18, 2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarma, S.; Bhattacharya, I.; Brar, S.; Tyagi, R.; Surampalli, R. Carbon Nanotube-Bioaccumulation and Recent Advances in Environmental Monitoring. Crit. Rev. Environ. Sci.Technol. 2015, 45, 905–938. [Google Scholar] [CrossRef]
- Smirnova, E.; Gusev, A.; Zaytseva, O.; Sheina, O.; Tkachev, A.; Kuznetsova, E.; Lazareva, E.; Onishchenko, G.; Feofanov, A.; Kirpichnikov, M. Uptake and accumulation of multiwalled carbon nanotubes change the morphometric and biochemical characteristics of Onobrychis arenaria seedlings. Front. Chem. Sci. Eng. 2012, 6, 132–138. [Google Scholar] [CrossRef]
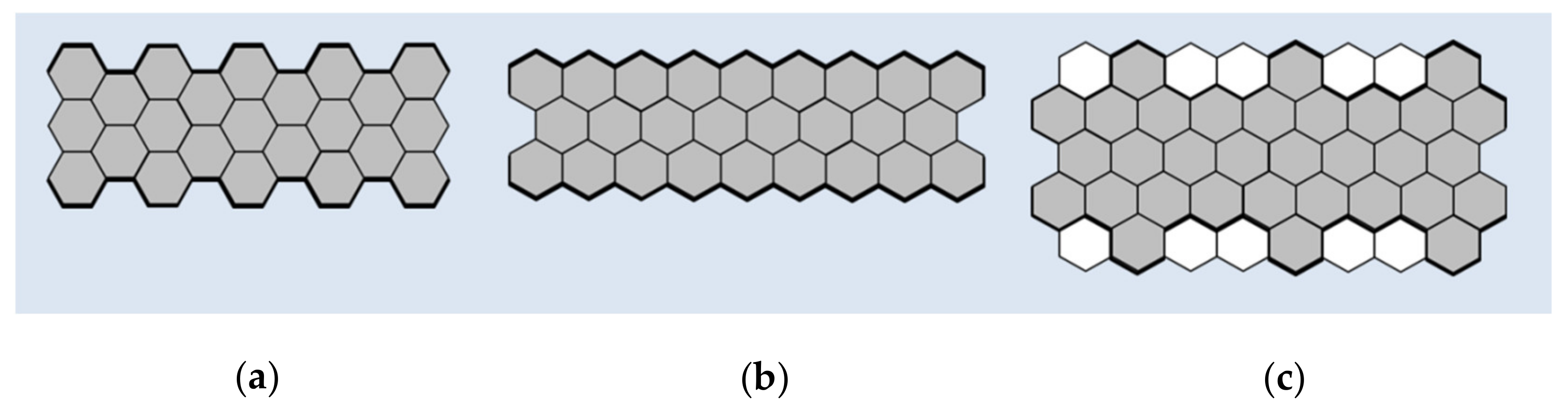


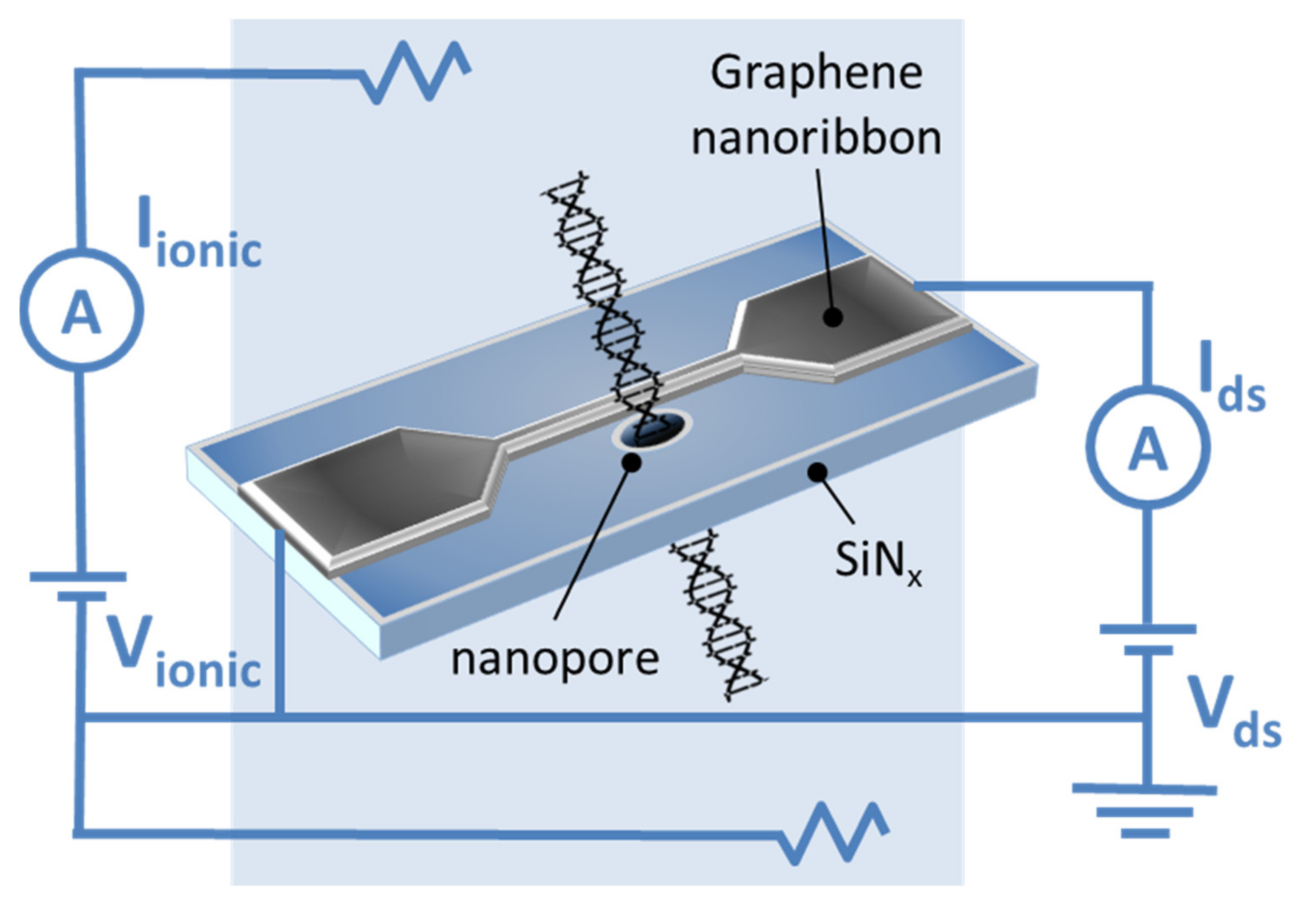
| Functionalized Material | Devices | Detection Method | Detection Limits | References |
|---|---|---|---|---|
| GNRs with nanopores | DNA sequenser | Electrochemical | - | [51] |
| GNRs with nanopores | DNA sequenser | Electrochemical | - | [54] |
| GNRs with nanopores | DNA sequenser | Electrochemical | - | [55,56] |
| Metallic GNRs with zigzag edges (ZGNRs) with a nanopore in its inner part | DNA sequenser | Electrochemical | - | [57] |
| Cytosine-functionalized GNRs | DNA sequenser | Electrochemical | - | [58] |
| GNRs decorated with iron oxide (Fe3O4) nanoparticles | DNA sensor | Electrochemical | - | [59] |
| GNRs with gold nanocages (AuNCs@GNRs) | DNA sensor | Electrochemical | 1 fM–100 pM | [60] |
| Template enzyme glucose oxidase with GNRs | Electrochemical biosensor of glucose | Electrochemical | 20 mg/L | [61] |
| Hybrid GNR/Ag NPs | Plasmonic sensing platform for the sequential colorimetric detection of dopamine and glutathione | Colorimetric | 0.46 μM for dopamine and 1.2 μM for glutathione | [62] |
| Chevron-like GNRs | Electrochemical sensor of epinephrine | Differential pulse voltammetry | 2.1 × 10−6 M | [63] |
| Functionalized Material | Component for Delivery | Cells | Effect | References |
|---|---|---|---|---|
| O-GNRs (20–60 μg/mL) | Enhanced green fluorescence protein plasmid or siRNA against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | HeLa and HUVEC | Concentration- and time-dependent increase in gene delivery and gene transfection efficiencies up to 96–98% | [71] |
| GNR-based nanocarrier grafted by polyethyleneimine | Locked nucleic acid modified by molecular beacon (LNA-m-MB) | HeLa | The efficient transfer of LNA-m-MB into cells for the recognition of the target miRNA has been demonstrated. | [72] |
| O-GNRs coated by PEG-DSPE | Antitumor drug Lucanthone | U251 | Uptake by U251 cells exceeding 67% and 60% in APE-1-overexpressing U251 post 24 h | [73] |
| O-GNRs coated by PEG-DSPE | Doxorubicin- | HeLa | Epidermal growth factor receptors (EGFRs) are activated and are taken up in significant amounts in cells with high EGFR expression. | [75] |
| O-GNRs (100 µg/mL) | C6 ceramide | HeLa | Decrease in cell viability by 93%. O-GNRs without C6 ceramide did not significantly reduce cell viability. | [82] |
| O-GNRs coated by PEG-DSPE (5–40 μg/mL) | C16 and C24 ceramides | HeLa | Significant biological effects in cells in conjunction with C6 ceramide and UV irradiation treatment. O-GNRs themselves have a number of significant biological effects that interfere with the ability of long-chain ceramides to sensitize or protect cells from pro-apoptotic stressors. | [83] |
| Material | Physical-Chemical Properties and Functionalization | Object | Dose and Exposure Time | Effect | References |
|---|---|---|---|---|---|
| GNRs | Nitrogen-doped GNR aerogels | Human medulloblastoma (DAOY) | - | Biocompatible sample | [84] |
| GNRs | Multilayer films consisting of chitosan, alginate and 2.5 wt % GNRs | Mouse fibroblasts (L929) | 1, 3, and 7 days | Cytocompatible sample | [86] |
| O-GNRs | - | Adhesive epithelial cells (HEK293T) and non-adherent cells (A20) | Up to 100 mg/mL; 24 and 48 h | No effect | [87] |
| O-GNRs | Functionalized with albumin | Human epithelial cells | 100 μg/mL | High cytotoxicity. Inhibition of proliferation and induction of apoptosis | [89] |
| O-GNRs | Functionalized with PEG-1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino (polyethylene glycol)] (DSPE) | Cervical cancer cells (HeLa), mouse fibroblasts (NIH-3T3), and breast cancer cells (MCF7) | 10–400 μg/mL; 12–48 h | Dose-, time-, and cell-dependent effects. MCF7 or SKBR3 were 100% viable up to 48 h at 10 μg/mL and reduced viability to 78% at 400 μg/mL. For HeLa cells, a 5–25% decrease in viability was observed even at a low concentration of 10 μg/mL. | [90] |
| O-GNRs | Functionalized with PEG-DSPE | Erythrocytes, endothelial cells | 20, 80, and 160 μg/mL | The concentration-dependent deformation of erythrocytes did not lead to hemolysis. The uptake of nanomaterials by endothelial cells and a concentration-dependent decrease in their viability | [85] |
| O-GNRs | - | Human neuroblastoma SK-N-BE (2) and SH-SY5Y | Increased reactive oxidative stress (ROS) production and the induction of autophagy within hours of exposure | [91] | |
| rO-GNRs | - | Human mesenchymal stem cells (hMSCs) | 1 and 10 μg/mL; 1 h | Significant cytotoxic effects. rO-GNRs can enter cells and cause DNA fragmentation and chromosomal aberrations even at low concentrations | [94,95] |
| rO-GNRs | Functionalized with polyethylene glycol (r O-GNR–PEG) | Human glioblastoma (U87MG) | 100 μg/mL; 24 h | More than 72% of cell death and more than 29% of DNA fragmentation | [97,98] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakharova, O.V.; Mastalygina, E.E.; Golokhvast, K.S.; Gusev, A.A. Graphene Nanoribbons: Prospects of Application in Biomedicine and Toxicity. Nanomaterials 2021, 11, 2425. https://doi.org/10.3390/nano11092425
Zakharova OV, Mastalygina EE, Golokhvast KS, Gusev AA. Graphene Nanoribbons: Prospects of Application in Biomedicine and Toxicity. Nanomaterials. 2021; 11(9):2425. https://doi.org/10.3390/nano11092425
Chicago/Turabian StyleZakharova, Olga V., Elena E. Mastalygina, Kirill S. Golokhvast, and Alexander A. Gusev. 2021. "Graphene Nanoribbons: Prospects of Application in Biomedicine and Toxicity" Nanomaterials 11, no. 9: 2425. https://doi.org/10.3390/nano11092425






