The Effect of Chronic Exposure of Graphene Nanoplates on the Viability and Motility of A549 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Nanomaterials
2.2. Cell Culture and Treatment
2.3. Cytotoxicity Assays
2.3.1. WST-1 Assay
2.3.2. alamarBlue™ Assay
2.4. Determination of Number Cell Nuclei by ImageXpress Micro XLS
2.5. Cell Cycle Distribution Analysis
2.6. Annexin V/Propidium Iodide Cell Death Assay
2.7. Chemotactic Migration Using the xCELLigence CIM-Plate-16
2.8. Spontaneous Motility of the A549 Cells
2.9. Analysis of Metabolism
2.10. Analysis of Cell Morphology by Transmission Electron Microscopy (TEM)
2.11. Statistical Analysis
3. Results
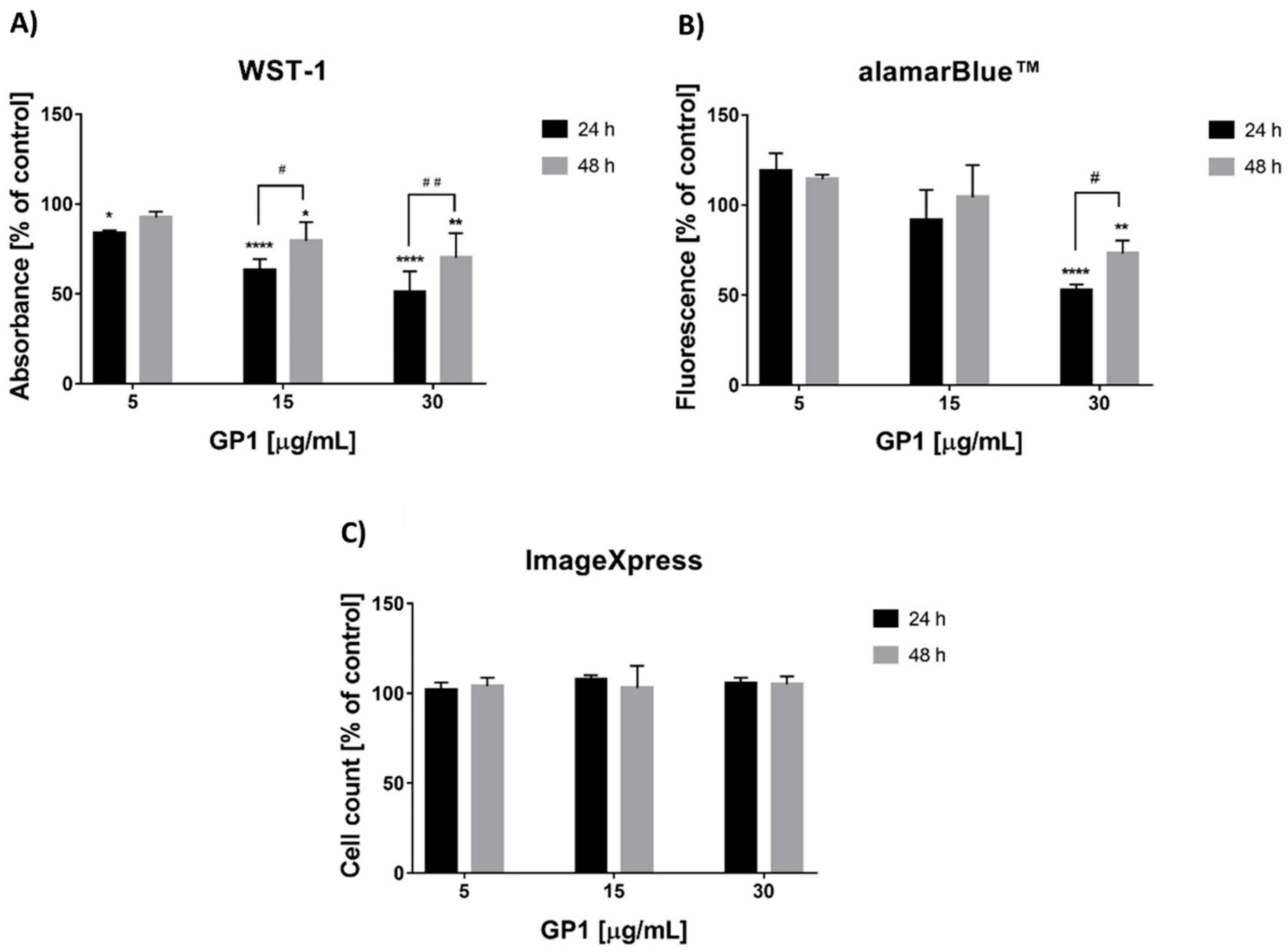
3.1. Short-Term Cytotoxicity of Graphene
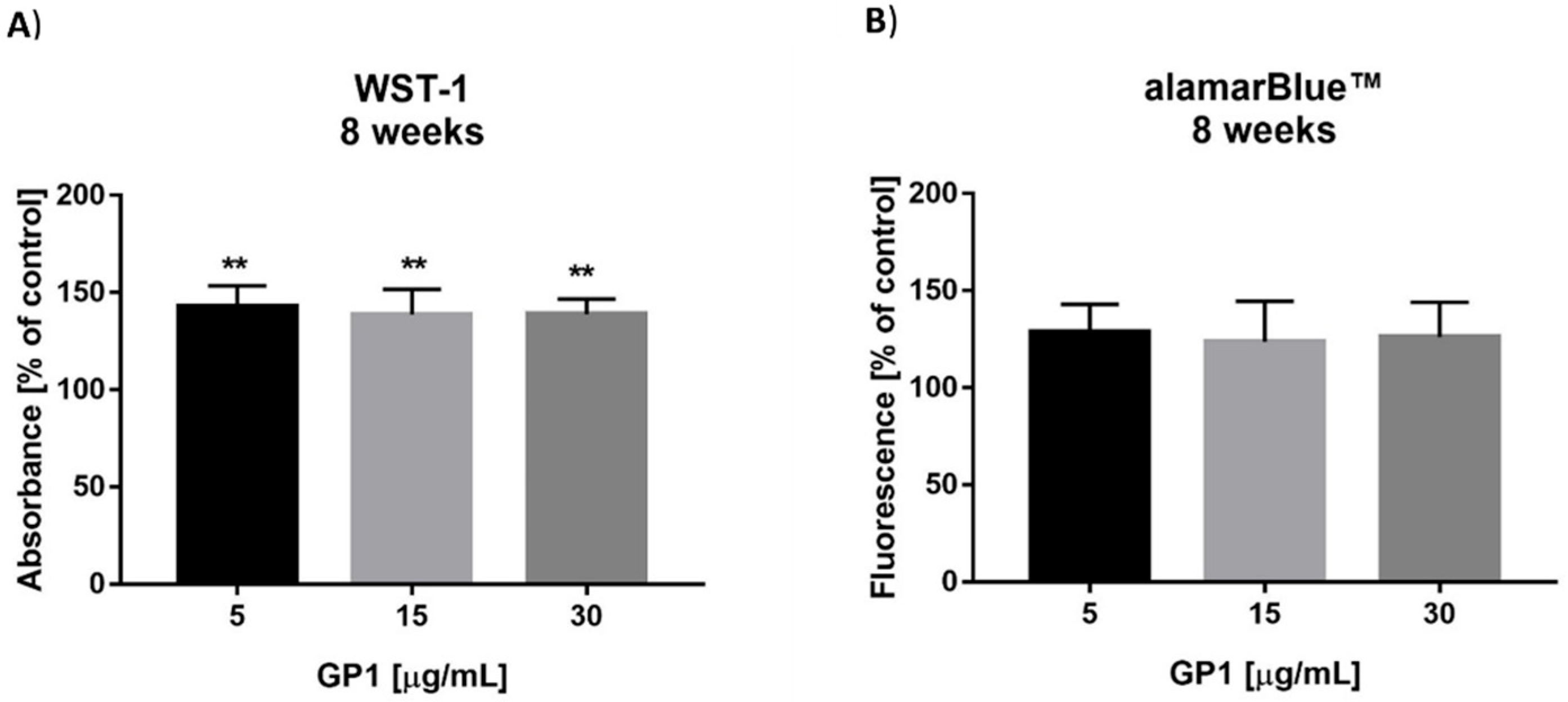
3.2. Long-Term Effects of the GP1 Nanomaterial
3.2.1. In Vitro Cytotoxicity
3.2.2. Analysis of Metabolism
3.2.3. Viability Analysis
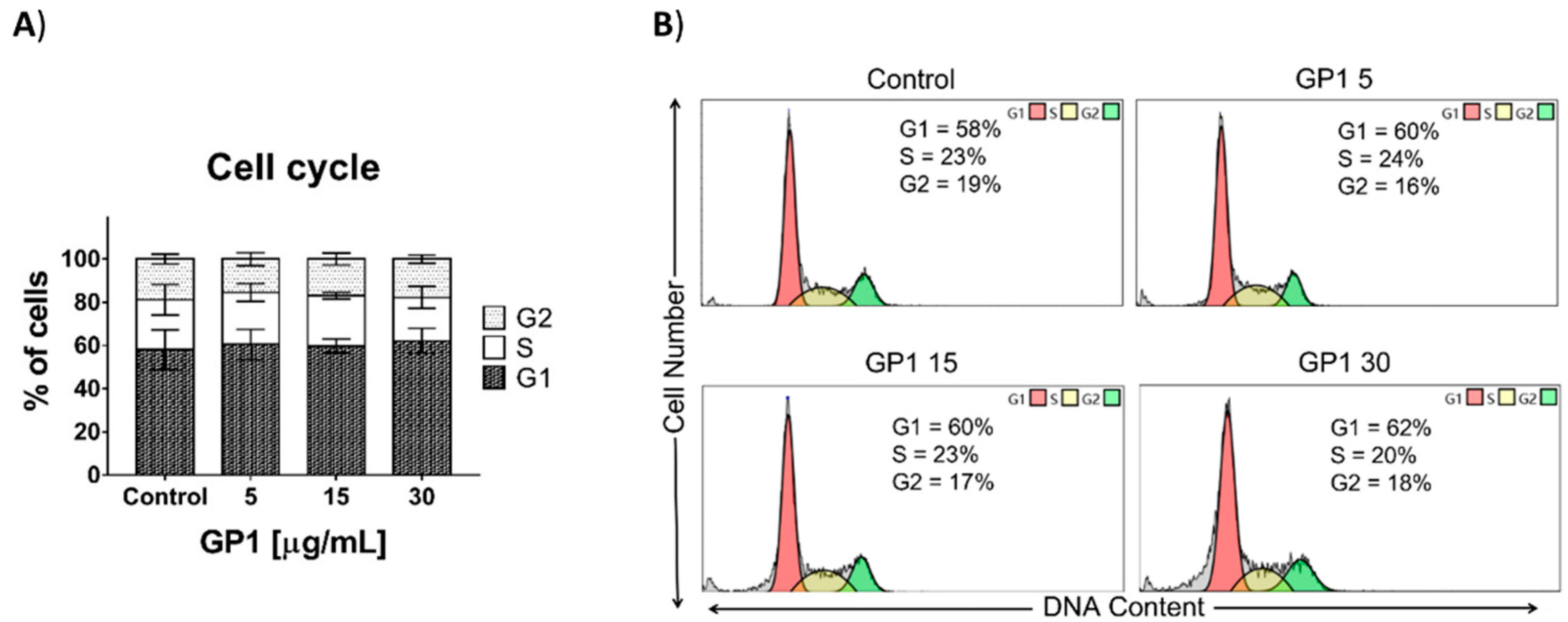
3.2.4. Cell Cycle Distribution Analysis
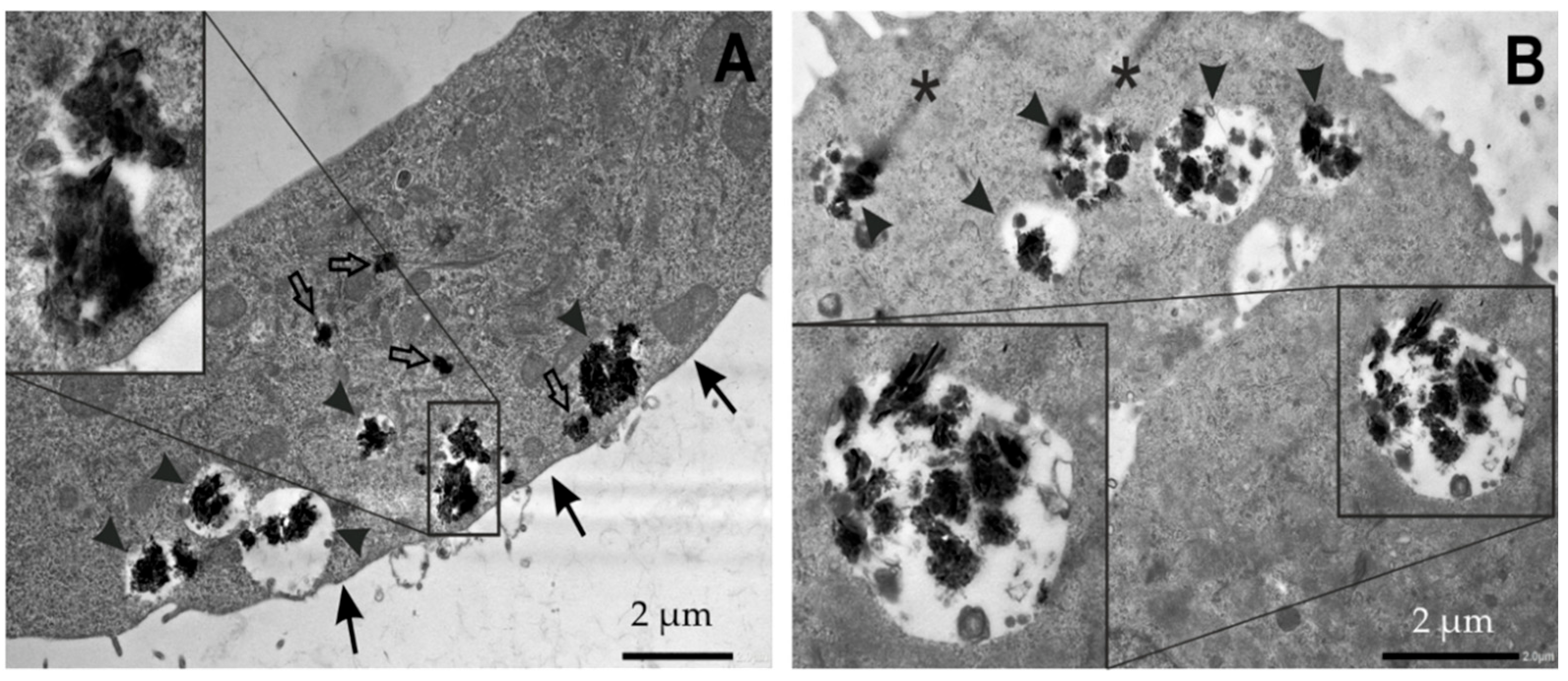
3.2.5. Analysis of Cell Morphology by Transmission Electron Microscopy (TEM)
Acute Graphene Exposure
Long-Term Graphene Exposure
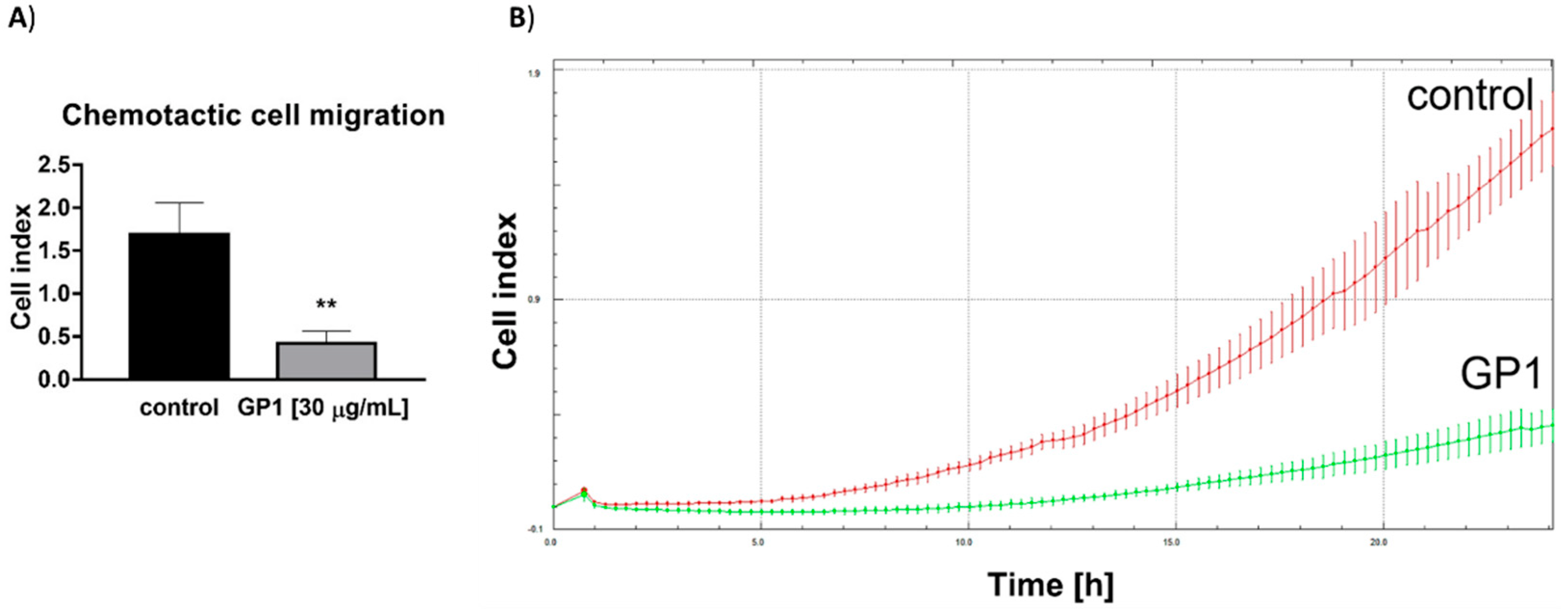
3.2.6. Effect of GP1 on A549 Cell Migration
3.2.7. Effect of GP1 on Spontaneous A549 Cell Motility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sang, M.; Shin, J.; Kim, K.; Yu, K.J. Electronic and Thermal Properties of Graphene and Recent Advances in Graphene Based Electronics Applications. Nanomaterials 2019, 9, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmoudi, T.; Wang, Y.; Hahn, Y.B. Graphene and its derivatives for solar cells application. Nano Energy 2018, 47, 51–65. [Google Scholar] [CrossRef]
- Fahimi, A.; Jurewicz, I.; Smith, R.J.; Sharrock, C.S.; Bradley, D.A.; Henley, S.J.; Coleman, J.N.; Dalton, A.B. Density controlled conductivity of pristine graphene films. Carbon 2013, 64, 435–443. [Google Scholar] [CrossRef]
- Ou, L.L.; Song, B.; Liang, H.M.; Liu, J.; Feng, X.L.; Deng, B.; Sun, T.; Shao, L.Q. Toxicity of graphene-family nanoparticles: A general review of the origins and mechanisms. Part. Fibre Toxicol. 2016, 13, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Perez, L.; Herranz, M.A.; Martin, N. The chemistry of pristine graphene. Chem. Commun. 2013, 49, 3721–3735. [Google Scholar] [CrossRef]
- Yan, M.D. Pristine graphene: Functionalization, fabrication, and nanocomposite materials. Xvi Meet. Phys. 2018, 1143. [Google Scholar] [CrossRef]
- Magne, T.M.; de Oliveira Vieira, T.; Alencar, L.M.R.; Junior, F.F.M.; Gemini-Piperni, S.; Carneiro, S.V.; Fechine, L.; Freire, R.M.; Golokhvast, K.; Metrangolo, P.; et al. Graphene and its derivatives: Understanding the main chemical and medicinal chemistry roles for biomedical applications. J. Nanostructure Chem. 2021, 9, 1–35. [Google Scholar] [CrossRef]
- Woltornist, S.J.; Oyer, A.J.; Carrillo, J.M.Y.; Dobrynin, A.V.; Adamson, D.H. Conductive Thin Films of Pristine Graphene by Solvent Interface Trapping. Acs Nano. 2013, 7, 7062–7066. [Google Scholar] [CrossRef]
- Yang, K.; Feng, L.Z.; Shi, X.Z.; Liu, Z. Nano-graphene in biomedicine: Theranostic applications. Chem. Soc. Rev. 2013, 42, 530–547. [Google Scholar] [CrossRef]
- Yang, K.; Zhang, S.; Zhang, G.; Sun, X.; Lee, S.T.; Liu, Z. Graphene in mice: Ultrahigh in vivo tumor uptake and efficient photothermal therapy. Nano Lett. 2010, 10, 3318–3323. [Google Scholar] [CrossRef]
- Guo, X.; Mei, N. Assessment of the toxic potential of graphene family nanomaterials. J. Food Drug Anal. 2014, 22, 105–115. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Li, Y.; Tjong, S.C. Graphene Nanomaterials: Synthesis, Biocompatibility, and Cytotoxicity. Int. J. Mol. Sci. 2018, 19, 3564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Sung, J.H.; Song, K.S.; Kim, J.K.; Choi, B.S.; Yu, I.J.; Park, J.D. Derivation of occupational exposure limits for multi-walled carbon nanotubes and graphene using subchronic inhalation toxicity data and a multi-path particle dosimetry model. Toxicol. Res. 2019, 8, 580–586. [Google Scholar] [CrossRef]
- Pelin, M.; Sosa, S.; Prato, M.; Tubaro, A. Occupational exposure to graphene based nanomaterials: Risk assessment. Nanoscale 2018, 10, 15894–15903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spinazze, A.; Cattaneo, A.; Borghi, F.; Del Buono, L.; Campagnolo, D.; Rovelli, S.; Cavallo, D.M. Probabilistic approach for the risk assessment of nanomaterials: A case study for graphene nanoplatelets. Int. J. Hyg. Environ. Health 2019, 222, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Ursini, C.L.; Fresegna, A.M.; Ciervo, A.; Maiello, R.; Del Frate, V.; Folesani, G.; Galetti, M.; Poli, D.; Buresti, G.; Di Cristo, L.; et al. Occupational exposure to graphene and silica nanoparticles. Part II: Pilot study to identify a panel of sensitive biomarkers of genotoxic, oxidative and inflammatory effects on suitable biological matrices. Nanotoxicology 2021, 15, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Falagan-Lotsch, P.; Grzincic, E.M.; Murphy, C.J. One low-dose exposure of gold nanoparticles induces long-term changes in human cells. Proc. Natl. Acad. Sci. USA 2016, 113, 13318–13323. [Google Scholar] [CrossRef] [Green Version]
- Mohammadinejad, R.; Moosavi, M.A.; Tavakol, S.; Vardar, D.O.; Hosseini, A.; Rahmati, M.; Dini, L.; Hussain, S.; Mandegary, A.; Klionsky, D.J. Necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy 2019, 15, 4–33. [Google Scholar] [CrossRef] [Green Version]
- Mrakovcic, M.; Absenger, M.; Riedl, R.; Smole, C.; Roblegg, E.; Frohlich, L.F.; Frohlich, E. Assessment of long-term effects of nanoparticles in a microcarrier cell culture system. PLoS ONE 2013, 8, e56791. [Google Scholar] [CrossRef] [Green Version]
- Svadlakova, T.; Hubatka, F.; Turanek Knotigova, P.; Kulich, P.; Masek, J.; Kotoucek, J.; Macak, J.; Motola, M.; Kalbac, M.; Kolackova, M.; et al. Proinflammatory Effect of Carbon-Based Nanomaterials: In Vitro Study on Stimulation of Inflammasome NLRP3 via Destabilisation of Lysosomes. Nanomaterials 2020, 10, 418. [Google Scholar] [CrossRef] [Green Version]
- Svadlakova, T.; Kolackova, M.; Vankova, R.; Karakale, R.; Malkova, A.; Kulich, P.; Hubatka, F.; Turanek-Knotigova, P.; Kratochvilova, I.; Raska, M.; et al. Carbon-Based Nanomaterials Increase Reactivity of Primary Monocytes towards Various Bacteria and Modulate Their Differentiation into Macrophages. Nanomaterials 2021, 11, 2510. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cechakova, L.; Ondrej, M.; Pavlik, V.; Jost, P.; Cizkova, D.; Bezrouk, A.; Pejchal, J.; Amaravadi, R.K.; Winkler, J.D.; Tichy, A. A Potent Autophagy Inhibitor (Lys05) Enhances the Impact of Ionizing Radiation on Human Lung Cancer Cells H1299. Int. J. Mol. Sci. 2019, 20, 5881. [Google Scholar] [CrossRef] [Green Version]
- Barosova, H.; Meldrum, K.; Karakocak, B.B.; Balog, S.; Doak, S.H.; Petri-Fink, A.; Clift, M.J.D.; Rothen-Rutishauser, B. Inter-laboratory variability of A549 epithelial cells grown under submerged and air-liquid interface conditions. Toxicol. In Vitro 2021, 75, 105178. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.A.; Oster, C.G.; Mayer, M.M.; Avery, M.L.; Audus, K.L. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell. Res. 1998, 243, 359–366. [Google Scholar] [CrossRef]
- Halim, N.H.A.; Zakaria, N.; Satar, N.A.; Yahaya, B.H. Isolation and Characterization of Cancer Stem Cells of the Non-Small-Cell Lung Cancer (A549) Cell Line. Methods Mol. Biol. 2016, 1516, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Yang, S.T.; Liu, J.H.; Dong, E.; Wang, Y.; Cao, A.; Liu, Y.; Wang, H. In vitro toxicity evaluation of graphene oxide on A549 cells. Toxicol. Lett. 2011, 200, 201–210. [Google Scholar] [CrossRef]
- Jin, C.; Wang, F.; Tang, Y.; Zhang, X.; Wang, J.; Yang, Y. Distribution of graphene oxide and TiO2-graphene oxide composite in A549 cells. Biol. Trace. Elem. Res. 2014, 159, 393–398. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, W.; Huang, X.; Sun, Y.; Tian, S.; Cai, P. Reduced graphene oxide triggered epithelial-mesenchymal transition in A549 cells. Sci. Rep. 2018, 8, 15188. [Google Scholar] [CrossRef]
- Wu, B.; Wu, J.L.; Liu, S.; Shen, Z.Y.; Chen, L.; Zhang, X.X.; Ren, H.Q. Combined effects of graphene oxide and zinc oxide nanoparticle on human A549 cells: Bioavailability, toxicity and mechanisms. Environ. Sci. Nano 2019, 6, 635–645. [Google Scholar] [CrossRef]
- Carreño, E.A.; Alberto, A.V.P.; de Souza, C.A.M.; de Mello, H.L.; Henriques-Pons, A.; Anastacio Alves, L. Considerations and Technical Pitfalls in the Employment of the MTT Assay to Evaluate Photosensitizers for Photodynamic Therapy. Appl. Sci. 2021, 11, 2603. [Google Scholar] [CrossRef]
- Jiao, G.Z.; He, X.; Li, X.; Qiu, J.Q.; Xu, H.Y.; Zhang, N.; Liu, S.M. Limitations of MTT and CCK-8 assay for evaluation of graphene cytotoxicity. Rsc Adv. 2015, 5, 53240–53244. [Google Scholar] [CrossRef]
- Malkova, A.; Svadlakova, T.; Singh, A.; Kolackova, M.; Vankova, R.; Borsky, P.; Holmannova, D.; Karas, A.; Borska, L.; Fiala, Z. In Vitro Assessment of the Genotoxic Potential of Pristine Graphene Platelets. Nanomaterials 2021, 11, 2210. [Google Scholar] [CrossRef] [PubMed]
- Rampersad, S.N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Bachmann, K.A.; Bailer, A.J.; Bolger, P.M.; Borak, J.; Cai, L.; Cedergreen, N.; Cherian, M.G.; Chiueh, C.C.; Clarkson, T.W.; et al. Biological stress response terminology: Integrating the concepts of adaptive response and preconditioning stress within a hormetic dose-response framework. Toxicol. Appl. Pharmacol. 2007, 222, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, V.C.; Jachak, A.; Hurt, R.H.; Kane, A.B. Biological interactions of graphene-family nanomaterials: An interdisciplinary review. Chem. Res. Toxicol. 2012, 25, 15–34. [Google Scholar] [CrossRef] [Green Version]
- Kucki, M.; Diener, L.; Bohmer, N.; Hirsch, C.; Krug, H.F.; Palermo, V.; Wick, P. Uptake of label-free graphene oxide by Caco-2 cells is dependent on the cell differentiation status. J. Nanobiotechnol. 2017, 15, 46. [Google Scholar] [CrossRef]
- Li, Y.F.; Yuan, H.Y.; von dem Bussche, A.; Creighton, M.; Hurt, R.H.; Kane, A.B.; Gao, H.J. Graphene microsheets enter cells through spontaneous membrane penetration at edge asperities and corner sites. Proc. Natl. Acad. Sci. USA 2013, 110, 12295–12300. [Google Scholar] [CrossRef] [Green Version]
- Jaworski, S.; Sawosz, E.; Grodzik, M.; Winnicka, A.; Prasek, M.; Wierzbicki, M.; Chwalibog, A. In vitro evaluation of the effects of graphene platelets on glioblastoma multiforme cells. Int. J. Nanomed. 2013, 8, 413–420. [Google Scholar] [CrossRef] [Green Version]
- Wan, B.; Wang, Z.X.; Lv, Q.Y.; Dong, P.X.; Zhao, L.X.; Yang, Y.; Guo, L.H. Single-walled carbon nanotubes and graphene oxides induce autophagosome accumulation and lysosome impairment in primarily cultured murine peritoneal macrophages. Toxicol. Lett. 2013, 221, 118–127. [Google Scholar] [CrossRef]
- Chen, G.Y.; Chen, C.L.; Tuan, H.Y.; Yuan, P.X.; Li, K.C.; Yang, H.J.; Hu, Y.C. Graphene oxide triggers toll-like receptors/autophagy responses in vitro and inhibits tumor growth in vivo. Adv. Healthc. Mater. 2014, 3, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Y.; Yang, H.J.; Lu, C.H.; Chao, Y.C.; Hwang, S.M.; Chen, C.L.; Lo, K.W.; Sung, L.Y.; Luo, W.Y.; Tuan, H.Y.; et al. Simultaneous induction of autophagy and toll-like receptor signaling pathways by graphene oxide. Biomaterials 2012, 33, 6559–6569. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Wang, J.; Nie, Y.; Du, H.; Dai, H.; Wang, J.; Wang, M.; Chen, S.; Hei, T.K.; et al. Graphene Oxide Attenuates the Cytotoxicity and Mutagenicity of PCB 52 via Activation of Genuine Autophagy. Environ. Sci. Technol. 2016, 50, 3154–3164. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Lee, G.H.; Han, B.S.; Lee, B.S.; Lee, S.; Cho, M.H.; Kim, J.H.; Kim, D.W. Toxic response of graphene nanoplatelets in vivo and in vitro. Arch. Toxicol. 2015, 89, 1557–1568. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Song, W.C.; Kim, B.; Oh, J.W.; Park, G. Nano-Graphene Oxide-Promoted Epithelial-Mesenchymal Transition of Human Retinal Pigment Epithelial Cells through Regulation of Phospholipase D Signaling. Nanomaterials 2021, 11, 2546. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kravchuk, P.; Krishnaprasad, A.; Roy, T.; Kang, E.H. Graphene Enhances Actin Filament Assembly Kinetics and Modulates NIH-3T3 Fibroblast Cell Spreading. Int. J. Mol. Sci. 2022, 23, 509. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.; Basu, S.; Singha, S.; Patra, H.K. Inner-View of Nanomaterial Incited Protein Conformational Changes: Insights into Designable Interaction. Research 2018, 2018, 9712832. [Google Scholar] [CrossRef] [Green Version]
- Sasidharan, A.; Panchakarla, L.S.; Chandran, P.; Menon, D.; Nair, S.; Rao, C.N.; Koyakutty, M. Differential nano-bio interactions and toxicity effects of pristine versus functionalized graphene. Nanoscale 2011, 3, 2461–2464. [Google Scholar] [CrossRef]
- Zhou, H.J.; Zhao, K.; Li, W.; Yang, N.; Liu, Y.; Chen, C.Y.; Wei, T.T. The interactions between pristine graphene and macrophages and the production of cytokines/chemokines via TLR- and NF-kappa B-related signaling pathways. Biomaterials 2012, 33, 6933–6942. [Google Scholar] [CrossRef]
- Li, T.; Oloyede, A.; Gu, Y.T. Adhesive characteristics of low dimensional carbon nanomaterial on actin. Appl. Phys. Lett. 2014, 104, 023702. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Zhang, B.; Zheng, J.; Yu, M.; Zhou, T.; Zhao, K.; Jia, Y.; Gao, X.; Chen, C.; Wei, T. The inhibition of migration and invasion of cancer cells by graphene via the impairment of mitochondrial respiration. Biomaterials 2014, 35, 1597–1607. [Google Scholar] [CrossRef] [PubMed]
- Sonnemann, K.J.; Bement, W.M. Wound repair: Toward understanding and integration of single-cell and multicellular wound responses. Annu. Rev. Cell. Dev. Biol. 2011, 27, 237–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Suspension Diluted in | Particle Size (nm) (Z-Average) | PdI | Average ζ-Potential (mV) |
|---|---|---|---|
| Sterile water | 179 ± 103 | 0.188 | −42.5 ± 6.9 |
| Full medium | 208 ± 103 | 0.203 | −8.52 ± 15.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šestáková, B.; Schröterová, L.; Bezrouk, A.; Čížková, D.; Elkalaf, M.; Havelek, R.; Rudolf, E.; Králová, V. The Effect of Chronic Exposure of Graphene Nanoplates on the Viability and Motility of A549 Cells. Nanomaterials 2022, 12, 2074. https://doi.org/10.3390/nano12122074
Šestáková B, Schröterová L, Bezrouk A, Čížková D, Elkalaf M, Havelek R, Rudolf E, Králová V. The Effect of Chronic Exposure of Graphene Nanoplates on the Viability and Motility of A549 Cells. Nanomaterials. 2022; 12(12):2074. https://doi.org/10.3390/nano12122074
Chicago/Turabian StyleŠestáková, Blanka, Ladislava Schröterová, Aleš Bezrouk, Dana Čížková, Moustafa Elkalaf, Radim Havelek, Emil Rudolf, and Věra Králová. 2022. "The Effect of Chronic Exposure of Graphene Nanoplates on the Viability and Motility of A549 Cells" Nanomaterials 12, no. 12: 2074. https://doi.org/10.3390/nano12122074






