Design and Characterization of pMyc/pMax Peptide-Coupled Gold Nanosystems for Targeting Myc in Prostate Cancer Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. In Silico Peptide Design and Chemical Synthesis
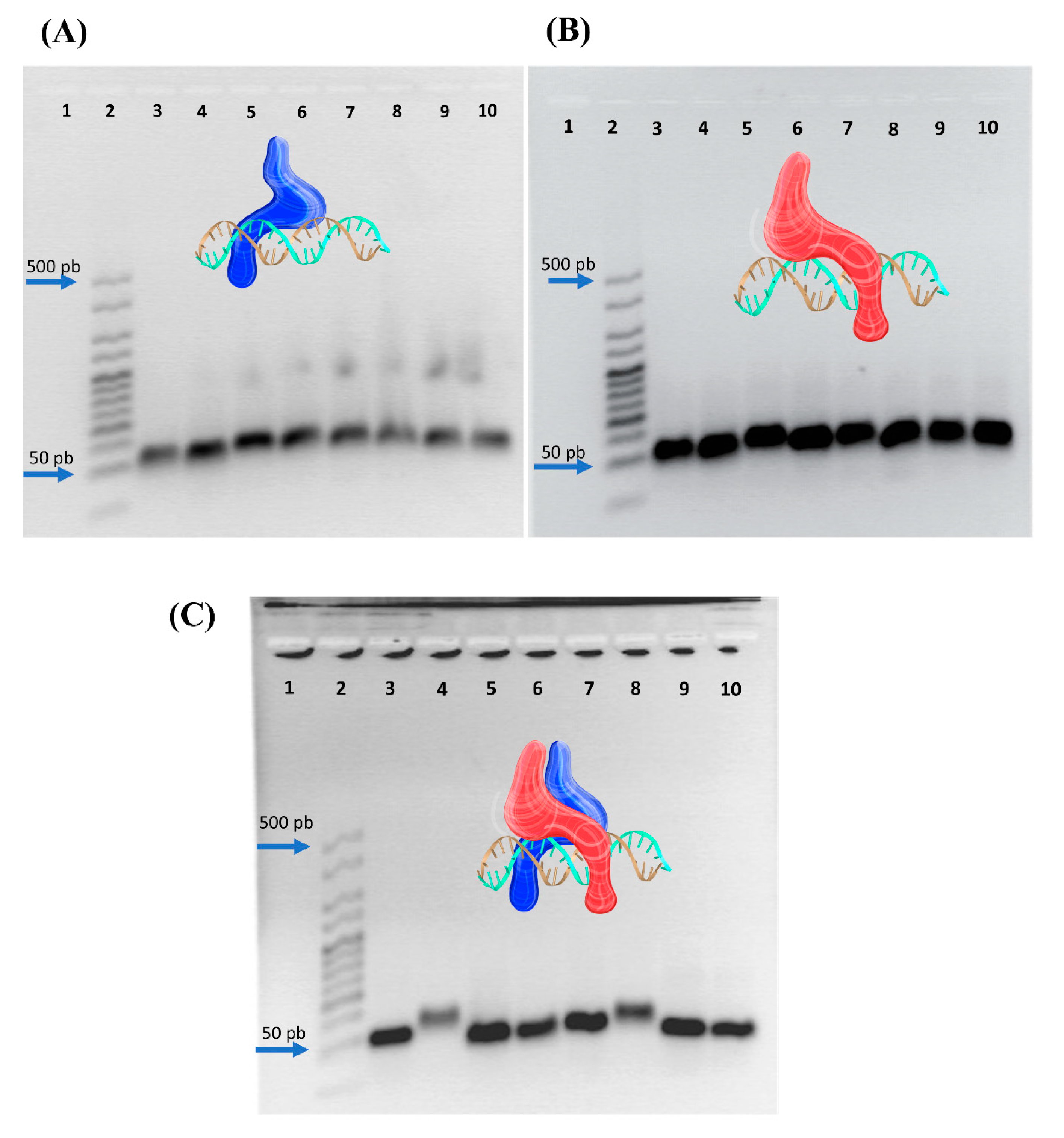
3.2. Peptide–Nucleic Acid Interactions via Electrophoretic Mobility Shift Assay
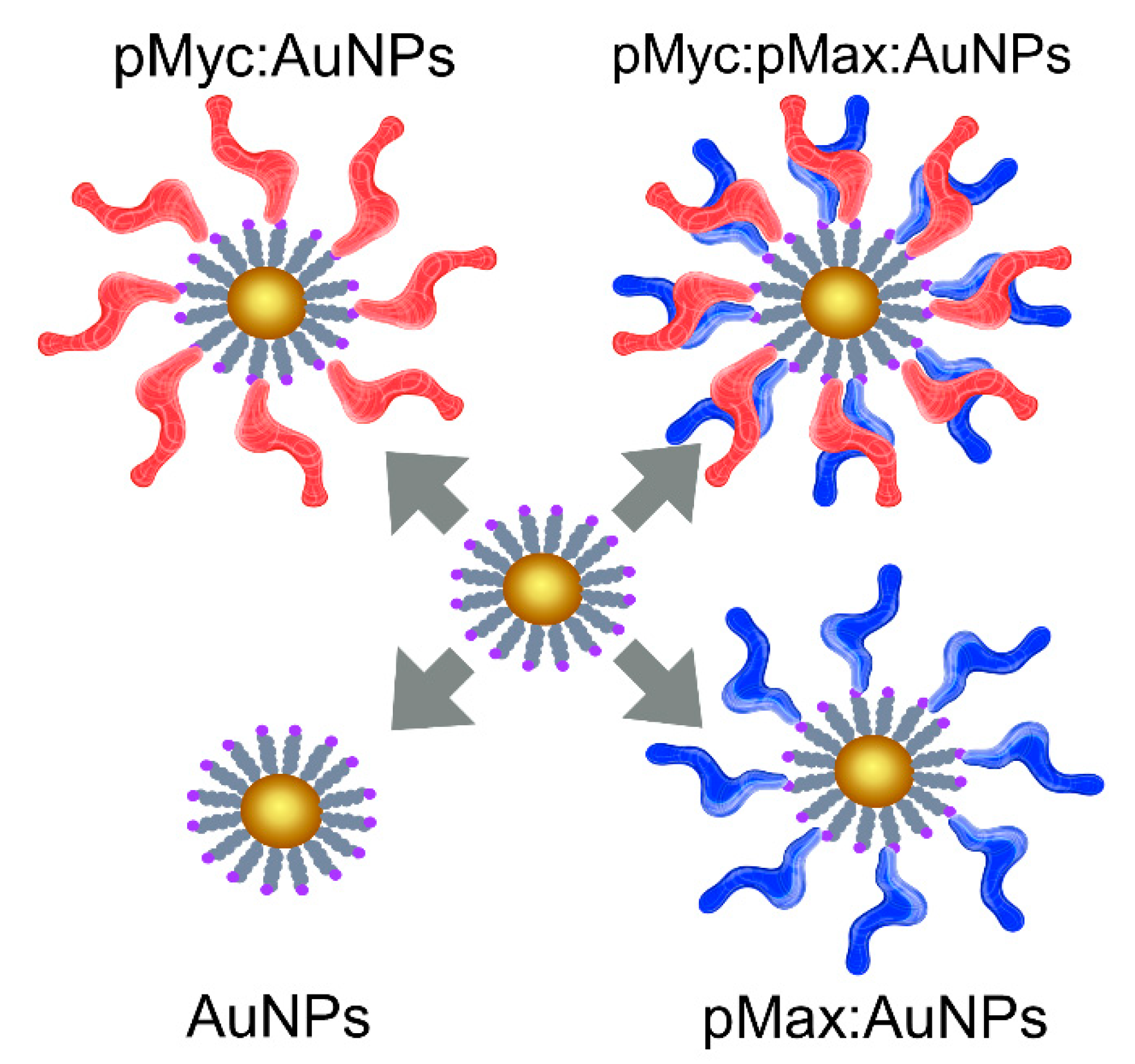
3.3. UV-Vis Analysis of the Nanosystem Assembly
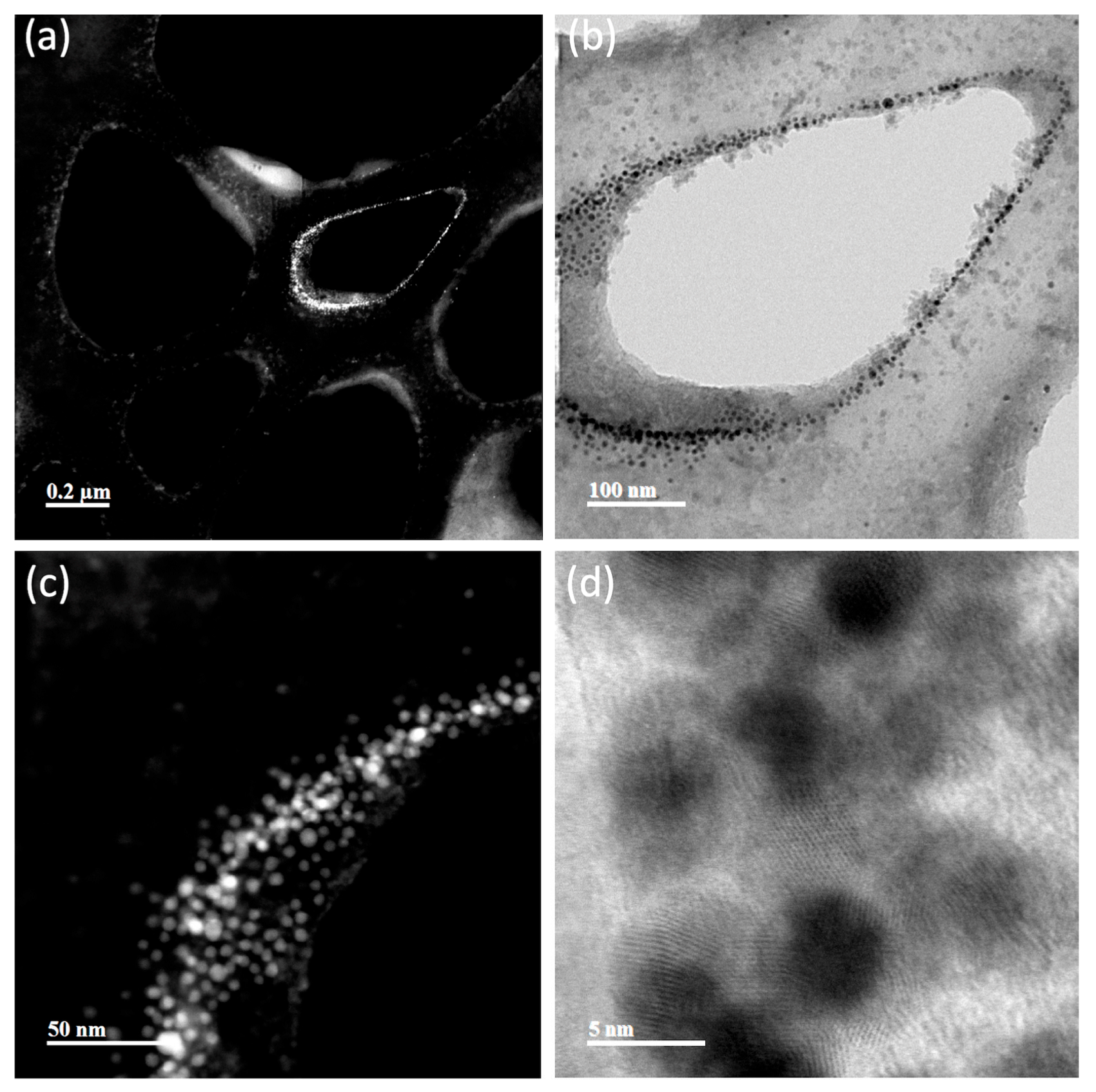
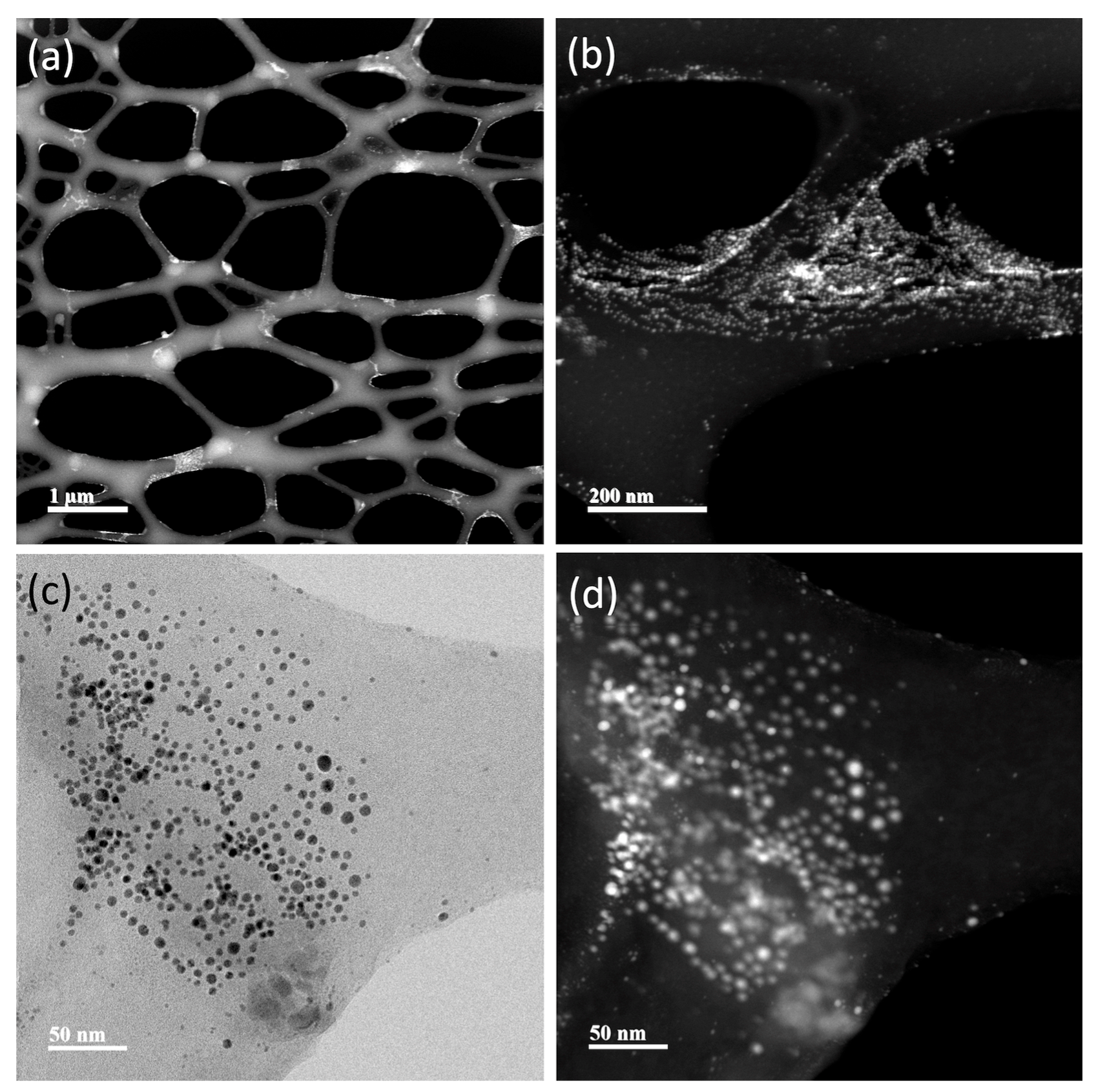
3.4. Nanoparticle Characterization
3.5. Hemolysis Test
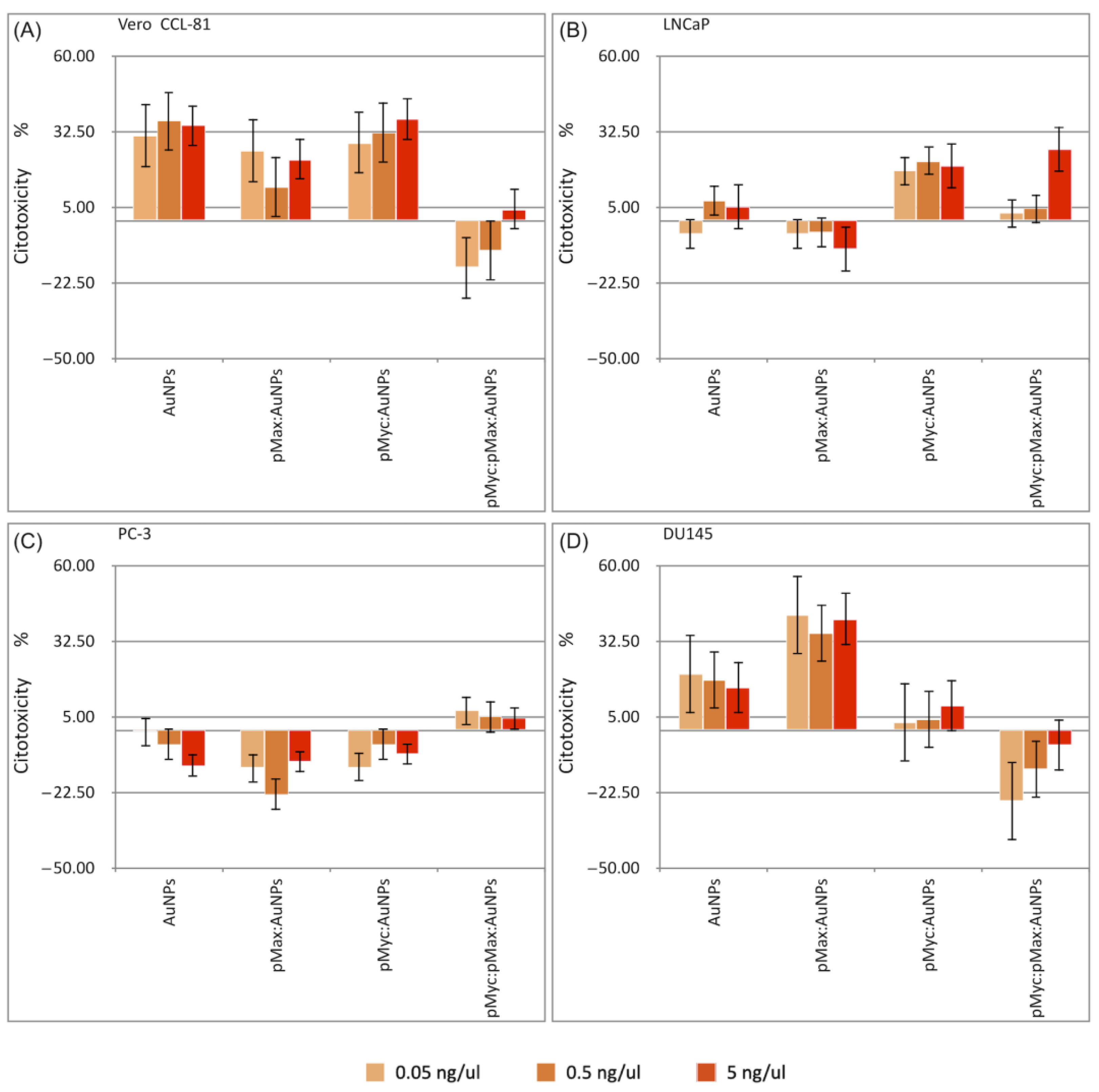
3.6. Cell Viability Assessment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beaulieu, M.E.; Jauset, T.; Massó-Vallés, D.; Martínez-Martín, S.; Rahl, P.; Maltais, L.; Zacarias-Fluck, M.F.; Casacuberta-Serra, S.; Del Pozo, E.S.; Fiore, C.; et al. Intrinsic Cell-Penetrating Activity Propels Omomyc from Proof of Concept to Viable Anti-Myc Therapy. Sci. Transl. Med. 2019, 11, eaar5012. [Google Scholar] [CrossRef]
- Camarda, R.; Zhou, A.Y.; Kohnz, R.A.; Balakrishnan, S.; Mahieu, C.; Anderton, B.; Eyob, H.; Kajimura, S.; Tward, A.; Krings, G.; et al. Inhibition of Fatty Acid Oxidation as a Therapy for MYC-Overexpressing Triple-Negative Breast Cancer. Nat. Med. 2016, 22, 427–432. [Google Scholar] [CrossRef]
- Rebello, R.J.; Pearson, R.B.; Hannan, R.D.; Furic, L. Therapeutic Approaches Targeting MYC-Driven Prostate Cancer. Genes 2017, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Boufaied, N.; Hallal, T.; Feit, A.; de Polo, A.; Luoma, A.M.; Alahmadi, W.; Larocque, J.; Zadra, G.; Xie, Y.; et al. MYC Drives Aggressive Prostate Cancer by Disrupting Transcriptional Pause Release at Androgen Receptor Targets. Nat. Commun. 2022, 13, 2559. [Google Scholar] [CrossRef]
- Barsouk, A.; Padala, S.A.; Vakiti, A.; Mohammed, A.; Saginala, K.; Thandra, K.C.; Rawla, P.; Barsouk, A. Epidemiology, Staging and Management of Prostate Cancer. Med. Sci. 2020, 8, 28. [Google Scholar] [CrossRef]
- Gandaglia, G.; Leni, R.; Bray, F.; Fleshner, N.; Freedland, S.J.; Kibel, A.; Stattin, P.; Van Poppel, H.; La Vecchia, C. Epidemiology and Prevention of Prostate Cancer. Eur. Urol. Oncol. 2021, 4, 877–892. [Google Scholar] [CrossRef]
- Iurlaro, R.; León-Annicchiarico, C.L.; Muñoz-Pinedo, C. Regulation of Cancer Metabolism by Oncogenes and Tumor Suppressors. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2014; Volume 542, pp. 59–80. ISBN 9780124166189. [Google Scholar]
- Conacci-Sorrell, M.; McFerrin, L.; Eisenman, R.N. An Overview of MYC and Its Interactome. Cold Spring Harb. Perspect. Med. 2014, 4, a014357. [Google Scholar] [CrossRef] [PubMed]
- Allevato, M.; Bolotin, E.; Grossman, M.; Mane-Padros, D.; Sladek, F.M.; Martinez, E. Sequence-Specific DNA Binding by MYC/MAX to Low-Affinity Non-E-Box Motifs. PLoS ONE 2017, 12, e0180147. [Google Scholar] [CrossRef] [PubMed]
- Carabet, L.A.; Rennie, P.S.; Cherkasov, A. Therapeutic Inhibition of Myc in Cancer. Structural Bases and Computer-Aided Drug Discovery Approaches. Int. J. Mol. Sci. 2019, 20, 120. [Google Scholar] [CrossRef]
- Truong, N.P.; Whittaker, M.R.; Mak, C.W.; Davis, T.P. The Importance of Nanoparticle Shape in Cancer Drug Delivery. Expert Opin. Drug Deliv. 2015, 12, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, B.; Poon, W.; Zhang, Y.N.; Lin, Z.P.; Kingston, B.R.; Tavares, A.J.; Zhang, Y.; Chen, J.; Valic, M.S.; Syed, A.M.; et al. The Dose Threshold for Nanoparticle Tumour Delivery. Nat. Mater. 2020, 19, 1362–1371. [Google Scholar] [CrossRef]
- Longoria-García, S.; Sánchez-Domínguez, C.N.; Gallardo-Blanco, H.L. Recent Applications of Cell-Penetrating Peptide Guidance of Nanosystems in Breast and Prostate Cancer (Review). Oncol. Lett. 2022, 23, 103. [Google Scholar] [CrossRef] [PubMed]
- Mout, R.; Ray, M.; Tay, T.; Sasaki, K.; Yesilbag Tonga, G.; Rotello, V.M. General Strategy for Direct Cytosolic Protein Delivery via Protein–Nanoparticle Co-Engineering. ACS Nano 2017, 11, 6416–6421. [Google Scholar] [CrossRef]
- Ma, N.; Ma, C.; Li, C.; Wang, T.; Tang, Y.; Wang, H.; Mou, X.; Chen, Z.; He, N. Influence of Nanoparticle Shape, Size, and Surface Functionalization on Cellular Uptake. J. Nanosci. Nanotechnol. 2013, 13, 6485–6498. [Google Scholar] [CrossRef] [PubMed]
- Longoria-García, S.; Sánchez-Domínguez, C.N.; Sánchez-Domínguez, M.; Delgado-Balderas, J.R.; Gallardo-Blanco, H.L. PMyc and PMax Peptides Nanosystems and the Potential Treatment of Prostate Cancer, In Vitro Assays. Mater. Proc. 2023, 14, 46. [Google Scholar] [CrossRef]
- Elahi, N.; Kamali, M.; Baghersad, M.H. Recent Biomedical Applications of Gold Nanoparticles: A Review. Talanta 2018, 184, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Salesa, B.; Ferrús-Manzano, P.; Tuñón-Molina, A.; Cano-Vicent, A.; Assis, M.; Andrés, J.; Serrano-Aroca, Á. Study of Biological Properties of Gold Nanoparticles: Low Toxicity, No Proliferative Activity, No Ability to Induce Cell Gene Expression and No Antiviral Activity. Chem. Biol. Interact. 2023, 382, 110646. [Google Scholar] [CrossRef]
- Amendola, V.; Pilot, R.; Frasconi, M.; Maragò, O.M.; Iatì, M.A. Surface Plasmon Resonance in Gold Nanoparticles: A Review. J. Phys. Condens. Matter 2017, 29, 203002. [Google Scholar] [CrossRef]
- Mangadlao, J.D.; Wang, X.; McCleese, C.; Escamilla, M.; Ramamurthy, G.; Wang, Z.; Govande, M.; Basilion, J.P.; Burda, C. Prostate-Specific Membrane Antigen Targeted Gold Nanoparticles for Theranostics of Prostate Cancer. ACS Nano 2018, 12, 3714–3725. [Google Scholar] [CrossRef]
- Hori, K.; Higashida, S.; Osaki, T.; Kawano, T.; Inaba, H.; Matsuura, K.; Iwasaki, T. Intracellular Delivery and Photothermal Therapeutic Effects of Polyhistidine Peptide-Modified Gold Nanoparticles. J. Biotechnol. 2022, 354, 34–44. [Google Scholar] [CrossRef]
- Jeong, W.-J.; Bu, J.; Kubiatowicz, L.J.; Chen, S.S.; Kim, Y.S.; Hong, S. Peptide–Nanoparticle Conjugates: A next Generation of Diagnostic and Therapeutic Platforms? Nano Converg. 2018, 5, 38. [Google Scholar] [CrossRef]
- Wang, X.; Qian, X.; Beitler, J.J.; Chen, Z.G.; Khuri, F.R.; Lewis, M.M.; Shin, H.J.C.; Nie, S.; Shin, D.M. Detection of Circulating Tumor Cells in Human Peripheral Blood Using Surface-Enhanced Raman Scattering Nanoparticles. Cancer Res. 2011, 71, 1526–1532. [Google Scholar] [CrossRef]
- Guerrero, S.; Herance, J.R.; Rojas, S.; Mena, J.F.; Gispert, J.D.; Acosta, G.A.; Albericio, F.; Kogan, M.J. Synthesis and In Vivo Evaluation of the Biodistribution of a 18F-Labeled Conjugate Gold-Nanoparticle-Peptide with Potential Biomedical Application. Bioconj. Chem. 2012, 23, 399–408. [Google Scholar] [CrossRef]
- Li, N.; Yang, H.; Yu, Z.; Li, Y.; Pan, W.; Wang, H.; Tang, B. Nuclear-Targeted SiRNA Delivery for Long-Term Gene Silencing. Chem. Sci. 2017, 8, 2816–2822. [Google Scholar] [CrossRef]
- Niu, J.; Chu, Y.; Huang, Y.-F.; Chong, Y.-S.; Jiang, Z.-H.; Mao, Z.-W.; Peng, L.-H.; Gao, J.-Q. Transdermal Gene Delivery by Functional Peptide-Conjugated Cationic Gold Nanoparticle Reverses the Progression and Metastasis of Cutaneous Melanoma. ACS Appl. Mater. Interfaces 2017, 9, 9388–9401. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Choi, S.-R.; Heo, J.H. Simultaneous Stabilization and Functionalization of Gold Nanoparticles via Biomolecule Conjugation: Progress and Perspectives. ACS Appl. Mater. Interfaces 2021, 13, 42311–42328. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Geer, L.Y.; Chappey, C.; Kans, J.A.; Bryant, S.H. Cn3D: Sequence and Structure Views for Entrez. Trends Biochem. Sci. 2000, 25, 300–302. [Google Scholar] [CrossRef]
- Pucci, F.; Kwasigroch, J.M.; Rooman, M. SCooP: An Accurate and Fast Predictor of Protein Stability Curves as a Function of Temperature. Bioinformatics 2017, 33, 3415–3422. [Google Scholar] [CrossRef]
- Ream, J.A.; Lewis, L.K.; Lewis, K.A. Rapid Agarose Gel Electrophoretic Mobility Shift Assay for Quantitating Protein: RNA Interactions. Anal. Biochem. 2016, 511, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R.; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e16. [Google Scholar] [CrossRef]
- Nair, S.K.; Burley, S.K. X-Ray Structures of Myc-Max and Mad-Max Recognizing DNA. Cell 2003, 112, 193–205. [Google Scholar] [CrossRef]
- Soucek, L.; Helmer-Citterich, M.; Sacco, A.; Jucker, R.; Cesareni, G.; Nasi, S. Design and Properties of a Myc Derivative That Efficiently Homodimerizes. Oncogene 1998, 17, 2463–2472. [Google Scholar] [CrossRef]
- Chen, H.; Liu, H.; Qing, G. Targeting Oncogenic Myc as a Strategy for Cancer Treatment. Signal Transduct. Target. Ther. 2018, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Calo-Lapido, R.; Penas, C.; Jiménez-Balsa, A.; Vázquez, M.E.; Mascareñas, J.L. A Chemical Approach for the Synthesis of the DNA-Binding Domain of the Oncoprotein MYC. Org. Biomol. Chem. 2019, 17, 6748–6752. [Google Scholar] [CrossRef]
- Chen, B.; Hsu, R.; Li, Z.; Kogut, P.C.; Du, Q.; Rouser, K.; Camoretti-Mercado, B.; Solway, J. Upstream Stimulatory Factor 1 Activates GATA5 Expression through an E-Box Motif. Biochem. J. 2012, 446, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Reth, M. Matching Cellular Dimensions with Molecular Sizes. Nat. Immunol. 2013, 14, 765–767. [Google Scholar] [CrossRef]
- Cirri, A.; Silakov, A.; Lear, B.J. Ligand Control over the Electronic Properties within the Metallic Core of Gold Nanoparticles. Angew. Chem. Int. Ed. 2015, 54, 11750–11753. [Google Scholar] [CrossRef]
- Gamrad, L.; Rehbock, C.; Westendorf, A.M.; Buer, J.; Barcikowski, S.; Hansen, W. Efficient Nucleic Acid Delivery to Murine Regulatory T Cells by Gold Nanoparticle Conjugates. Sci. Rep. 2016, 6, 28709. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ma, H.; Zhang, X.; Huang, K.; Jin, S.; Liu, J.; Wei, T.; Cao, W.; Zou, G.; Liang, X.-J. Gold Nanoparticles Functionalized with Therapeutic and Targeted Peptides for Cancer Treatment. Biomaterials 2012, 33, 1180–1189. [Google Scholar] [CrossRef]
- Matea, C.T.; Mocan, T.; Tabaran, F.; Iancu, C.; Mocan, L.C. Rational Design of Gold Nanocarrier for the Delivery of JAG-1 Peptide. J. Nanobiotechnol. 2015, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Satriano, C.; Munzone, A.; Cucci, L.M.; Giacomelli, C.; Trincavelli, M.L.; Martini, C.; Rizzarelli, E.; La Mendola, D. Angiogenin-Mimetic Peptide Functionalised Gold Nanoparticles for Cancer Therapy Applications. Microchem. J. 2018, 136, 157–163. [Google Scholar] [CrossRef]
- Taha, A.A.; AL-Jawad, S.M.H.; AL-Barram, L.F.A. Improvement of Cancer Therapy by TAT Peptide Conjugated Gold Nanoparticles. J. Clust. Sci. 2019, 30, 403–414. [Google Scholar] [CrossRef]
- Chang, T.; Wang, L.; Zhao, K.; Ge, Y.; He, M.; Li, G. Duplex Identification of Staphylococcus Aureus by Aptamer and Gold Nanoparticles. J. Nanosci. Nanotechnol. 2016, 16, 5513–5519. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, Y.; Yang, Y.; Wu, J.; Cao, M.; Sun, L. One-Pot Synthesis of Functional Peptide-Modified Gold Nanoparticles for Gene Delivery. Colloids Surf. A Physicochem. Eng. Asp. 2022, 640, 128491. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, W.; Zhou, H.; Zhang, Q.; Zhang, S. ATP-Triggered Intracellular In Situ Aggregation of a Gold-Nanoparticle-Equipped Triple-Helix Molecular Switch for Fluorescence Imaging and Photothermal Tumor Therapy. Langmuir 2022, 38, 3755–3764. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Yin, Y. Colloidal Nanoparticle Clusters: Functional Materials by Design. Chem. Soc. Rev. 2012, 41, 6874. [Google Scholar] [CrossRef]
- ASTM-F756-17; Standard Practice for Assessment of Hemolytic Properties of Materials. ASTM: West Conshohocken, PA, USA, 2017.
- Jabir, M.S.; Taha, A.A.; Sahib, U.I.; Taqi, Z.J.; Al-Shammari, A.M.; Salman, A.S. Novel of Nano Delivery System for Linalool Loaded on Gold Nanoparticles Conjugated with CALNN Peptide for Application in Drug Uptake and Induction of Cell Death on Breast Cancer Cell Line. Mater. Sci. Eng. C 2019, 94, 949–964. [Google Scholar] [CrossRef]
- Verimli, N.; Demiral, A.; Yılmaz, H.; Çulha, M.; Erdem, S.S. Design of Dense Brush Conformation Bearing Gold Nanoparticles as Theranostic Agent for Cancer. Appl. Biochem. Biotechnol. 2019, 189, 709–728. [Google Scholar] [CrossRef]
- Akrami, M.; Balalaie, S.; Hosseinkhani, S.; Alipour, M.; Salehi, F.; Bahador, A.; Haririan, I. Tuning the Anticancer Activity of a Novel Pro-Apoptotic Peptide Using Gold Nanoparticle Platforms. Sci. Rep. 2016, 6, 31030. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, A.; Qayoom, I.; Teotia, A.K.; Gupta, S.; Padmanabhan, P.; Dev, A.; Kumar, A. Biofabrication of Gold Nanoparticles with Bone Remodeling Potential: An in Vitro and in Vivo Assessment. J. Nanopart. Res. 2020, 22, 152. [Google Scholar] [CrossRef]
- Sani, A.; Cao, C.; Cui, D. Toxicity of Gold Nanoparticles (AuNPs): A Review. Biochem. Biophys. Rep. 2021, 26, 100991. [Google Scholar] [CrossRef]
- Chuang, S.-M.; Lee, Y.-H.; Liang, R.-Y.; Roam, G.-D.; Zeng, Z.-M.; Tu, H.-F.; Wang, S.-K.; Chueh, P.J. Extensive Evaluations of the Cytotoxic Effects of Gold Nanoparticles. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2013, 1830, 4960–4973. [Google Scholar] [CrossRef]
- Chueh, P.J.; Liang, R.-Y.; Lee, Y.-H.; Zeng, Z.-M.; Chuang, S.-M. Differential Cytotoxic Effects of Gold Nanoparticles in Different Mammalian Cell Lines. J. Hazard. Mater. 2014, 264, 303–312. [Google Scholar] [CrossRef]
- Venkatpurwar, V.; Mali, V.; Bodhankar, S.; Pokharkar, V. In Vitro Cytotoxicity and In Vivo Sub-Acute Oral Toxicity Assessment of Porphyran Reduced Gold Nanoparticles. Toxicol. Environ. Chem. 2012, 94, 1357–1367. [Google Scholar] [CrossRef]
- Cho, T.J.; MacCuspie, R.I.; Gigault, J.; Gorham, J.M.; Elliott, J.T.; Hackley, V.A. Highly Stable Positively Charged Dendron-Encapsulated Gold Nanoparticles. Langmuir 2014, 30, 3883–3893. [Google Scholar] [CrossRef] [PubMed]
- Carabet, L.A.; Lallous, N.; Leblanc, E.; Ban, F.; Morin, H.; Lawn, S.; Ghaidi, F.; Lee, J.; Mills, I.G.; Gleave, M.E.; et al. Computer-Aided Drug Discovery of Myc-Max Inhibitors as Potential Therapeutics for Prostate Cancer. Eur. J. Med. Chem. 2018, 160, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.G.; Parker, J.B.; Sagar, V.; Truica, M.I.; Soni, P.N.; Han, H.; Schiltz, G.E.; Abdulkadir, S.A.; Chakravarti, D. A MYC Inhibitor Selectively Alters the MYC and MAX Cistromes and Modulates the Epigenomic Landscape to Regulate Target Gene Expression. Sci. Adv. 2022, 8, eabh3635. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, G.; Supino, R.; Zuco, V.; Lanzi, C.; Scovassi, A.I.; Semple, S.C.; Zunino, F. Role of C-Myc Protein in Hormone Refractory Prostate Carcinoma: Cellular Response to Paclitaxel. Biochem. Pharmacol. 2004, 68, 923–931. [Google Scholar] [CrossRef]
- Kim, J.; Roh, M.; Abdulkadir, S.A. Pim1 Promotes Human Prostate Cancer Cell Tumorigenicity and C-MYC Transcriptional Activity. BMC Cancer 2010, 10, 248. [Google Scholar] [CrossRef]
- Manzanares, D.; Pérez-Carrión, M.D.; Blanco, J.L.J.; Mellet, C.O.; Fernández, J.M.G.; Ceña, V. Cyclodextrin-Based Nanostructure Efficiently Delivers Sirna to Glioblastoma Cells Preferentially via Macropinocytosis. Int. J. Mol. Sci. 2020, 21, 9306. [Google Scholar] [CrossRef] [PubMed]
- Bechara, C.; Sagan, S. Cell-penetrating Peptides: 20 Years Later, Where Do We Stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.Z.; Ariyama, H.; Alam, J.M.; Yamazaki, M. Entry of Cell-Penetrating Peptide Transportan 10 into a Single Vesicle by Translocating across Lipid Membrane and Its Induced Pores. Biochemistry 2014, 53, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; García, A.E. Free Energy of Translocating an Arginine-Rich Cell-Penetrating Peptide across a Lipid Bilayer Suggests Pore Formation. Biophys. J. 2013, 104, 412–420. [Google Scholar] [CrossRef] [PubMed]





| IC | Sequence |
|---|---|
| CME Allevato F | 5′-CCG GCC ACG TGC ACG TGT TAA TAG CTC AGA CTA CTG TGT CGA CG-3′ |
| CME Allevato R | 5′-CGT CGA CAC AGT AGT CTG AGC TAT TAA CAC GTG CAC GTG GCC GG-3′ |
| CME F | 5′-AGA TCT CGA GCT GCA TGC TGT ACA CGT GAT GTC GTA CGT CGA GCT CTA GT-3′ |
| CME R | 5′-ACT AGA GCT CGA CGT ACG ACA TCA CGT GTA CAG CAT GCA GCT CGA GAT CT-3′ |
| NE F | 5′-AGA TCT CGA GCT GCA TGC TGT AAA CGT TAT GTC GTA CGT CGA GCT CTA GT-3′ |
| NE R | 5′-ACT AGA GCT CGA CGT ACG ACA TAA CGT TTA CAG CAT GCA GCT CGA GAT CT-3′ |
| CTRL F | 5′-AGA TCT CGA GCT GCA TGC TGT ATT AGC AAT GTC GTT ATC AGA GCT CTA GT-3′ |
| CTRL R | 5′-ACT AGA GCT CTG ATA ACG ACA TTG CTA ATA CAG CAT GCA GCT CGA GAT CT-3′ |
| Cell Line | Myc mRNA (nRPM) | Max mRNA (nRPM) | Myc Relative Protein Expression (a) | Max Relative Protein Expression (a) | Myc/Max Relative Protein Expression (a) |
|---|---|---|---|---|---|
| PC-3 | 256.91 | 502.38 | 0.63 | 1.51 | 0.42 |
| DU145 | NA | NA | 0.00 | 0.08 | 0.00 |
| LNCaP Clone FGC | 1548.99 | 315.14 | 0.96 | 0.33 | 2.95 |
| Vero CCL-81 | NA | NA | NA | NA | NA |
| Tm (°C) | ΔGr 1 (kcal/mol) | |
|---|---|---|
| pMyc | 80.1 | −8.4 |
| pMax | 81.4 | −6 |
| pMyc:pMax dimer | 76.6 | −4.2 |
| Native Myc | 66.4 | −5.1 |
| Native Max | 69.2 | −13 |
| Omomyc | 70.4 | −5.1 |
| Diameter of AuNPs Core (nm) | Hydrodynamic Diameter of Nanocomplexes(nm) | PDI | Zeta Potential | Peptide Concentration (nM) 1 | |
|---|---|---|---|---|---|
| AuNPs | 5.68 ± 0.84 | 243.03 ± 12.83 | 0.257 ± 0.04 | −9.05 ± 6.09 | 0.00 |
| pMyc:AuNPs | 3.28 ± 1.16 | 264.97 ± 4.39 | 0.177 ± 0.02 | −5.05 ± 3.42 | 163.60 |
| pMax:AuNPs | 5.60 ± 1.19 | 262. 57 ± 23.40 | 0.168 ± 0.02 | −7.67 ± 5.71 | 200.86 |
| pMyc:pMax:AuNPs | 5.78 ± 1.41 | 225.03 ± 17.65 | 0.083 ± 0.04 | −10.09 ± 4.04 | 223.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Longoria-García, S.; Sánchez-Domínguez, C.N.; Sánchez-Domínguez, M.; Delgado-Balderas, J.R.; Islas-Cisneros, J.F.; Vidal-Gutiérrez, O.; Gallardo-Blanco, H.L. Design and Characterization of pMyc/pMax Peptide-Coupled Gold Nanosystems for Targeting Myc in Prostate Cancer Cell Lines. Nanomaterials 2023, 13, 2802. https://doi.org/10.3390/nano13202802
Longoria-García S, Sánchez-Domínguez CN, Sánchez-Domínguez M, Delgado-Balderas JR, Islas-Cisneros JF, Vidal-Gutiérrez O, Gallardo-Blanco HL. Design and Characterization of pMyc/pMax Peptide-Coupled Gold Nanosystems for Targeting Myc in Prostate Cancer Cell Lines. Nanomaterials. 2023; 13(20):2802. https://doi.org/10.3390/nano13202802
Chicago/Turabian StyleLongoria-García, Samuel, Celia N. Sánchez-Domínguez, Margarita Sánchez-Domínguez, Jesús R. Delgado-Balderas, José F. Islas-Cisneros, Oscar Vidal-Gutiérrez, and Hugo L. Gallardo-Blanco. 2023. "Design and Characterization of pMyc/pMax Peptide-Coupled Gold Nanosystems for Targeting Myc in Prostate Cancer Cell Lines" Nanomaterials 13, no. 20: 2802. https://doi.org/10.3390/nano13202802








