Antibacterial and Antibiofilm Activity of Endophytic Alternaria sp. Isolated from Eremophila longifolia
Abstract
:1. Introduction
2. Results
2.1. Molecular Identification of Endophytic Fungi
2.2. Antibacterial Activity of Fungal Extracts
2.3. Antibiofilm Activity of Fungal Extracts
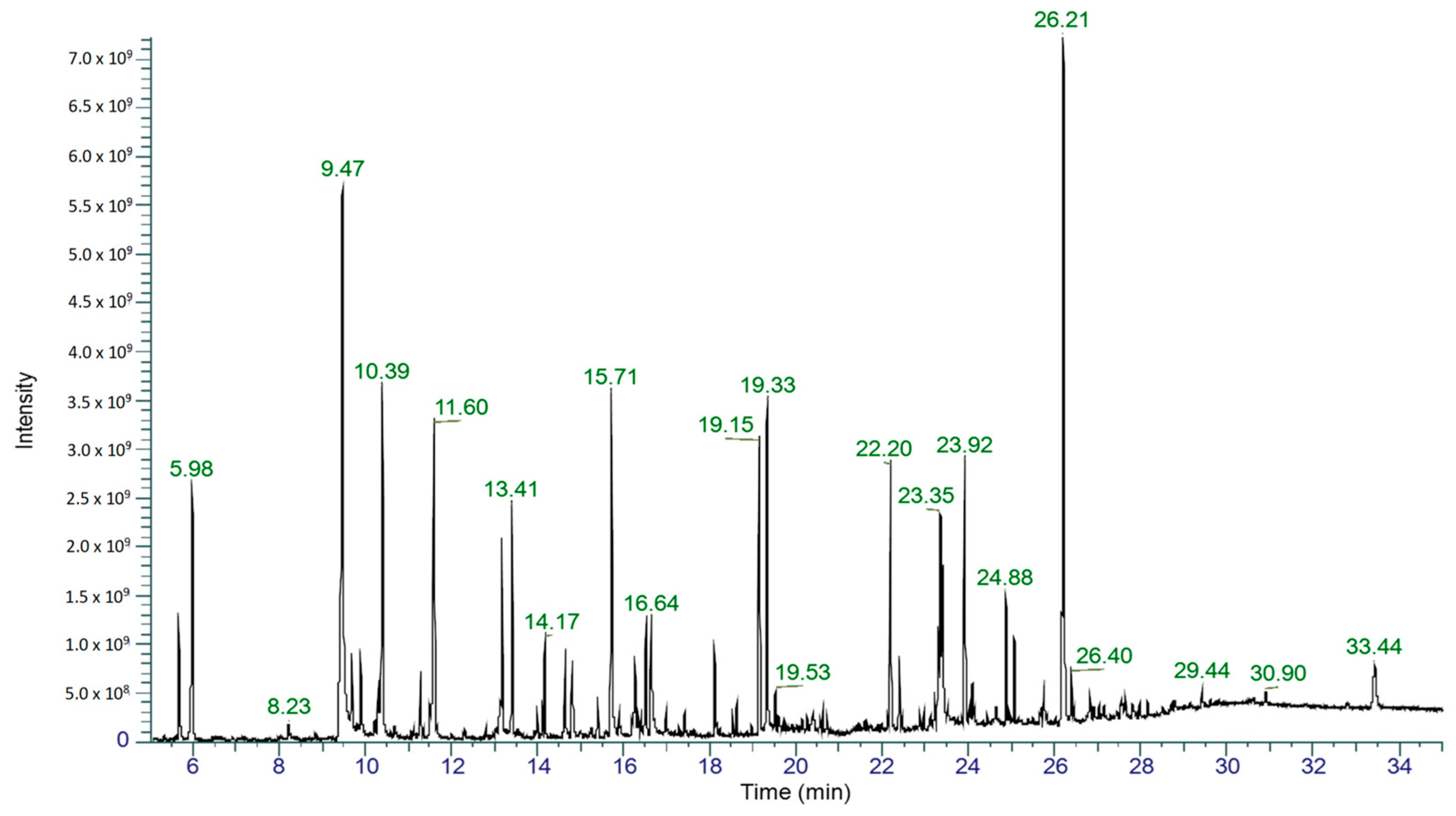
2.4. Gas Chromatography–Mass Spectrometry Analysis
3. Discussion
4. Materials and Methods
4.1. Molecular Identification of Endophytic Fungi
4.2. Preparation of Fungal Extracts
4.3. Bacterial Cultures
4.4. Disk Diffusion Assay
4.5. Determination of MIC and MBC
4.6. Antibiofilm Activity
4.6.1. Inhibition of Cell Attachment
4.6.2. Eradication of Established Biofilms
4.7. GC-MS Analysis of the EL 24 EtOAc Extract
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. 2016. Available online: https://iica.int/en/press/news/tackling-drug-resistant-infections-globally-final-report-and-recommendations (accessed on 6 August 2023).
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Rossolini, G.M.; Arena, F.; Pecile, P.; Pollini, S. Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Founou, R.C.; Founou, L.L.; Essack, S.Y. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0189621. [Google Scholar] [CrossRef]
- Siddiqui, A.H.; Koirala, J. Methicillin Resistant Staphylococcus aureus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Masimen, M.A.A.; Harun, N.A.; Maulidiani, M.; Ismail, W.I.W. Overcoming Methicillin-Resistance Staphylococcus aureus (MRSA) Using Antimicrobial Peptides-Silver Nanoparticles. Antibiotics 2022, 11, 951. [Google Scholar] [CrossRef]
- Cameron, D.R.; Jiang, J.-H.; Kostoulias, X.; Foxwell, D.J.; Peleg, A.Y. Vancomycin susceptibility in methicillin-resistant Staphylococcus aureus is mediated by YycHI activation of the WalRK essential two-component regulatory system. Sci. Rep. 2016, 6, 30823. [Google Scholar] [CrossRef]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.-S.; Patra, J.K. Endophytes: A Treasure House of Bioactive Compounds of Medicinal Importance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef]
- Hashem, A.H.; Attia, M.S.; Kandil, E.K.; Fawzi, M.M.; Abdelrahman, A.S.; Khader, M.S.; Khodaira, M.A.; Emam, A.E.; Goma, M.A.; Abdelaziz, A.M. Bioactive compounds and biomedical applications of endophytic fungi: A recent review. Microb. Cell Fact. 2023, 22, 107. [Google Scholar] [CrossRef]
- Kaul, S.; Gupta, S.; Ahmed, M.; Dhar, M.K. Endophytic fungi from medicinal plants: A treasure hunt for bioactive metabolites. Phytochem. Rev. 2012, 11, 487–505. [Google Scholar] [CrossRef]
- Jia, M.; Chen, L.; Xin, H.-L.; Zheng, C.-J.; Rahman, K.; Han, T.; Qin, L.-P. A Friendly Relationship between Endophytic Fungi and Medicinal Plants: A Systematic Review. Front. Microbiol. 2016, 7, 906. [Google Scholar] [CrossRef]
- Gupta, A.; Meshram, V.; Gupta, M.; Goyal, S.; Qureshi, K.A.; Jaremko, M.; Shukla, K.K. Fungal Endophytes: Microfactories of Novel Bioactive Compounds with Therapeutic Interventions; A Comprehensive Review on the Biotechnological Developments in the Field of Fungal Endophytic Biology over the Last Decade. Biomolecules 2023, 13, 1038. [Google Scholar] [CrossRef] [PubMed]
- Stierle, A.; Strobel, G.; Stierle, D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 1993, 260, 214–216. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Zühlke, S.; Spiteller, M. An Endophytic Fungus from Camptotheca acuminata That Produces Camptothecin and Analogues. J. Nat. Prod. 2009, 72, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Manganyi, M.C.; Ateba, C.N. Untapped Potentials of Endophytic Fungi: A Review of Novel Bioactive Compounds with Biological Applications. Microorganisms 2020, 8, 1934. [Google Scholar] [CrossRef]
- Caruso, D.J.; Palombo, E.A.; Moulton, S.E.; Zaferanloo, B. Exploring the promise of endophytic fungi: A review of novel antimicrobial compounds. Microorganisms 2022, 10, 1990. [Google Scholar] [CrossRef] [PubMed]
- Richmond, G. A review of the use of Eremophila (Myoporaceae) by Australian Aborigines. J. Adel. Bot. Gard. 1993, 15, 101–107. [Google Scholar]
- Zaferanloo, B.; Virkar, A.; Mahon, P.J.; Palombo, E.A. Endophytes from an Australian native plant are a promising source of industrially useful enzymes. World J. Microbiol. Biotechnol. 2013, 29, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Zaferanloo, B.; Pepper, S.A.; Coulthard, S.A.; Redfern, C.P.F.; Palombo, E.A. Metabolites of endophytic fungi from Australian native plants as potential anticancer agents. FEMS Microbiol. Lett. 2018, 365, fny078. [Google Scholar] [CrossRef]
- DeMers, M. Alternaria alternata as endophyte and pathogen. Microbiology 2022, 168, 001153. [Google Scholar] [CrossRef]
- Tao, J.; Bai, X.; Zeng, M.; Li, M.; Hu, Z.; Hua, Y.; Zhang, H. Whole-Genome Sequence Analysis of an Endophytic Fungus Alternaria sp. SPS-2 and Its Biosynthetic Potential of Bioactive Secondary Metabolites. Microorganisms 2022, 10, 1789. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Shi, R.; Li, X.; Yu, Z.; Hu, L.; Hu, H.; Zhang, M.; Chang, J.; Li, C. Friend or Foe? The Endophytic Fungus Alternaria tenuissima Might Be a Major Latent Pathogen Involved in Ginkgo Leaf Blight. Forests 2023, 14, 1452. [Google Scholar] [CrossRef]
- Sudharshana, T.N.; Venkatesh, H.N.; Nayana, B.; Manjunath, K.; Mohana, D.C. Anti-microbial and anti-mycotoxigenic activities of endophytic Alternaria alternata isolated from Catharanthus roseus (L.) G. Don.: Molecular characterisation and bioactive compound isolation. Mycology 2019, 10, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Khazaal, H.T.; Khazaal, M.T.; Abdel-Razek, A.S.; Hamed, A.A.; Ebrahim, H.Y.; Ibrahim, R.R.; Bishr, M.; Mansour, Y.E.; El Dib, R.A.; Soliman, H.S.M. Antimicrobial, antiproliferative activities and molecular docking of metabolites from Alternaria alternata. AMB Express 2023, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Hamed, A.A.; Soldatou, S.; Qader, M.M.; Arjunan, S.; Miranda, K.J.; Casolari, F.; Pavesi, C.; Diyaolu, O.A.; Thissera, B.; Eshelli, M.; et al. Screening Fungal Endophytes Derived from Under-Explored Egyptian Marine Habitats for Antimicrobial and Antioxidant Properties in Factionalised Textiles. Microorganisms 2020, 8, 1617. [Google Scholar] [CrossRef]
- Mousa, A.A.A.; Mohamed, H.; Hassane, A.M.A.; Abo-Dahab, N.F. Antimicrobial and cytotoxic potential of an endophytic fungus Alternaria tenuissima AUMC14342 isolated from Artemisia judaica L. growing in Saudi Arabia. J. King Saud Univ. Sci. 2021, 33, 101462. [Google Scholar] [CrossRef]
- Arivudainambi, U.S.E.; Kanugula, A.K.; Kotamraju, S.; Karunakaran, C.; Rajendran, A. Antibacterial effect of an extract of the endophytic fungus and its cytotoxic activity on MCF-7 and MDA MB-231 tumour cell lines. Biol. Lett. 2014, 51, 7–17. [Google Scholar] [CrossRef]
- Cai, S.; King, J.B.; Du, L.; Powell, D.R.; Cichewicz, R.H. Bioactive Sulfur-Containing Sulochrin Dimers and Other Metabolites from an Alternaria sp. Isolate from a Hawaiian Soil Sample. J. Nat. Prod. 2014, 77, 2280–2287. [Google Scholar] [CrossRef]
- Techaoei, S.; Jarmkom, K.; Dumrongphuttidecha, T.; Khobjai, W. Evaluation of the stability and antibacterial activity of crude extracts of hydro-endophytic fungi. J. Adv. Pharm. Technol. Res. 2021, 12, 61–66. [Google Scholar] [CrossRef]
- Khiralla, A.; Mohamed, I.E.; Tzanova, T.; Schohn, H.; Slezack-Deschaumes, S.; Hehn, A.; André, P.; Carre, G.; Spina, R.; Lobstein, A.; et al. Endophytic fungi associated with Sudanese medicinal plants show cytotoxic and antibiotic potential. FEMS Microbiol. Lett. 2016, 363, fnw089. [Google Scholar] [CrossRef]
- EUCAST. Reading Guide for Broth Microdilution ver. 4.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2022_manuals/Reading_guide_BMD_v_4.0_2022.pdf (accessed on 17 July 2023).
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Lade, H.; Park, J.H.; Chung, S.H.; Kim, I.H.; Kim, J.M.; Joo, H.S.; Kim, J.S. Biofilm Formation by Staphylococcus aureus Clinical Isolates is Differentially Affected by Glucose and Sodium Chloride Supplemented Culture Media. J. Clin. Med. 2019, 8, 1853. [Google Scholar] [CrossRef] [PubMed]
- Hulankova, R. The Influence of Liquid Medium Choice in Determination of Minimum Inhibitory Concentration of Essential Oils against Pathogenic Bacteria. Antibiotics 2022, 11, 150. [Google Scholar] [CrossRef]
- Lee, B.L.; Sachdeva, M.; Chambers, H.F. Effect of protein binding of daptomycin on MIC and antibacterial activity. Antimicrob. Agents Chemother. 1991, 35, 2505–2508. [Google Scholar] [CrossRef]
- Juven, B.J.; Kanner, J.; Schved, F.; Weisslowicz, H. Factors that interact with the antibacterial action of thyme essential oil and its active constituents. J. Appl. Bacteriol. 1994, 76, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Devlieghere, F.; Vermeulen, A.; Debevere, J. Chitosan: Antimicrobial activity, interactions with food components and applicability as a coating on fruit and vegetables. Food Microbiol. 2004, 21, 703–714. [Google Scholar] [CrossRef]
- Gutierrez, J.; Barry-Ryan, C.; Bourke, P. The antimicrobial efficacy of plant essential oil combinations and interactions with food ingredients. Int. J. Food Microbiol. 2008, 124, 91–97. [Google Scholar] [CrossRef]
- Imani Rad, H.; Arzanlou, M.; Ranjbar Omid, M.; Ravaji, S.; Peeri Doghaheh, H. Effect of culture media on chemical stability and antibacterial activity of allicin. J. Funct. Foods 2017, 28, 321–325. [Google Scholar] [CrossRef]
- Oh, D.-H.; Marshall, D.L. Effect of pH on the Minimum Inhibitory Concentration of Monolaurin against Listeria monocytogenes. J. Food Protect. 1992, 55, 449–450. [Google Scholar] [CrossRef]
- Rewak-Soroczynska, J.; Dorotkiewicz-Jach, A.; Drulis-Kawa, Z.; Wiglusz, R.J. Culture Media Composition Influences the Antibacterial Effect of Silver, Cupric, and Zinc Ions against Pseudomonas aeruginosa. Biomolecules 2022, 12, 963. [Google Scholar] [CrossRef]
- Buyck, J.; Plesiat, P.; Traore, H.; Vanderbist, F.; Tulkens, P.; Van Bambeke, F. Increased Susceptibility of Pseudomonas aeruginosa to Macrolides and Ketolides in Eukaryotic Cell Culture Media and Biological Fluids Due to Decreased Expression of oprM and Increased Outer-Membrane Permeability. Clin. Infect. Dis. 2012, 55, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Trimble, M.J.; Cheng, J.T.; Vallé, Q.; Hancock, R.E.W. Critical Assessment of Methods to Quantify Biofilm Growth and Evaluate Antibiofilm Activity of Host Defence Peptides. Biomolecules 2018, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Trimble, M.J.; Hancock, R.E.W. Microtiter plate assays to assess antibiofilm activity against bacteria. Nat. Protoc. 2021, 16, 2615–2632. [Google Scholar] [CrossRef]
- Fernández, L.; Breidenstein, E.B.M.; Hancock, R.E.W. Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Update 2011, 14, 1–21. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 121, 1–58. [Google Scholar] [CrossRef] [PubMed]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Abdelgawad, M.A.; Hamed, A.A.; Nayl, A.A.; Badawy, M.; Ghoneim, M.M.; Sayed, A.M.; Hassan, H.M.; Gamaleldin, N.M. The Chemical Profiling, Docking Study, and Antimicrobial and Antibiofilm Activities of the Endophytic fungi Aspergillus sp. AP5. Molecules 2022, 27, 1704. [Google Scholar] [CrossRef]
- Fathallah, N.; Raafat, M.M.; Issa, M.Y.; Abdel-Aziz, M.M.; Bishr, M.; Abdelkawy, M.A.; Salama, O. Bio-Guided Fractionation of Prenylated Benzaldehyde Derivatives as Potent Antimicrobial and Antibiofilm from Ammi majus L. Fruits-Associated Aspergillus amstelodami. Molecules 2019, 24, 4118. [Google Scholar] [CrossRef]
- Jalil, M.T.M.; Ibrahim, D. Fungal Extract of Lasiodiplodia pseudotheobromae IBRL OS-64 Inhibits the Growth of Skin Pathogenic Bacterium and Attenuates Biofilms of Methicillin-Resistant Staphylococcus aureus. Malays. J. Med. Sci. 2021, 28, 24–36. [Google Scholar] [CrossRef]
- Malhadas, C.; Malheiro, R.; Pereira, J.A.; de Pinho, P.G.; Baptista, P. Antimicrobial activity of endophytic fungi from olive tree leaves. World J. Microbiol. Biotechnol. 2017, 33, 46. [Google Scholar] [CrossRef]
- Corre, J.; Lucchini, J.J.; Mercier, G.M.; Cremieux, A. Antibacterial activity of phenethyl alcohol and resulting membrane alterations. Res. Microbiol. 1990, 141, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Cheng, Y.; Yang, M.; Liu, Y.; Chen, K.; Long, C.-a.; Deng, X. Mechanisms of action for 2-phenylethanol isolated from Kloeckera apiculata in control of Penicillium molds of citrus fruits. BMC Microbiol. 2014, 14, 242. [Google Scholar] [CrossRef] [PubMed]
- Kleinwächter, I.S.; Pannwitt, S.; Centi, A.; Hellmann, N.; Thines, E.; Bereau, T.; Schneider, D. The Bacteriostatic Activity of 2-Phenylethanol Derivatives Correlates with Membrane Binding Affinity. Membranes 2021, 11, 254. [Google Scholar] [CrossRef] [PubMed]
- Saraswathi, M.; Meshram, S.H.; Siva, B.; Misra, S.; Suresh Babu, K. Isolation, purification and structural elucidation of Mellein from endophytic fungus Lasiodiplodia theobromae strain (SJF-1) and its broad-spectrum antimicrobial and pharmacological properties. Lett. Appl. Microbiol. 2022, 75, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.; Niu, Q.; Kong, J.; Li, L.; Liang, X.; Cao, Y.; Zhou, X.; Sun, X.; Ma, H.; Wang, M.; et al. Genomic and Transcriptomic Characterization of Alternaria alternata during Infection. Agronomy 2023, 13, 809. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.-X.; Chen, Y.-C.; Sun, Z.-H.; Li, H.-H.; Li, S.-N.; Yan, M.-L.; Zhang, W.-M. Two New Metabolites from the Endophytic Fungus Alternaria sp. A744 Derived from Morinda officinalis. Molecules 2017, 22, 765. [Google Scholar] [CrossRef]
- Hussein, M.E.; Mohamed, O.G.; El-Fishawy, A.M.; El-Askary, H.I.; El-Senousy, A.S.; El-Beih, A.A.; Nossier, E.S.; Naglah, A.M.; Almehizia, A.A.; Tripathi, A.; et al. Identification of Antibacterial Metabolites from Endophytic Fungus Aspergillus fumigatus, Isolated from Albizia lucidior Leaves (Fabaceae), Utilizing Metabolomic and Molecular Docking Techniques. Molecules 2022, 27, 1117. [Google Scholar] [CrossRef]
- Leyte-Lugo, M.; Richomme, P.; Peña-Rodriguez, L.M. Diketopiperazines from Alternaria dauci. J. Mex. Chem. Soc. 2020, 64, 283–290. [Google Scholar] [CrossRef]
- Gowrishankar, S.; Poornima, B.; Pandian, S.K. Inhibitory efficacy of cyclo(l-leucyl-l-prolyl) from mangrove rhizosphere bacterium–Bacillus amyloliquefaciens (MMS-50) toward cariogenic properties of Streptococcus mutans. Res. Microbiol. 2014, 165, 278–289. [Google Scholar] [CrossRef]
- Gowrishankar, S.; Sivaranjani, M.; Kamaladevi, A.; Ravi, A.V.; Balamurugan, K.; Karutha Pandian, S. Cyclic dipeptide cyclo(l-leucyl-l-prolyl) from marine Bacillus amyloliquefaciens mitigates biofilm formation and virulence in Listeria monocytogenes. Pathog. Dis. 2016, 74, ftw017. [Google Scholar] [CrossRef]
- Graz, M.; Hunt, A.; Jamie, H.; Grant, G.; Milne, P. Antimicrobial activity of selected cyclic dipeptides. Pharmazie 1999, 54, 772–775. [Google Scholar]
- Viswapriya, V.; Saravana Kumari, P. Synergistic Activity of Cyclo(phe-pro) Antibiotic from Streptomyces violascens VS in Reducing the Drug Resistance Burden of Clinically Significant Pathogens. J. Biol. Act. Prod. Nat. 2022, 12, 450–460. [Google Scholar] [CrossRef]
- Li, H.; Li, C.; Ye, Y.; Cui, H.; Lin, L. Inhibition mechanism of cyclo (L-Phe-L-Pro) on early stage Staphylococcus aureus biofilm and its application on food contact surface. Food Biosci. 2022, 49, 101968. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.; Shah, R.; Bhave, M.; Palombo, E.A. Inhibitory activity of yarrow essential oil on Listeria planktonic cells and biofilms. Food Control 2013, 29, 125–130. [Google Scholar] [CrossRef]
- Skogman, M.E.; Vuorela, P.M.; Fallarero, A. A Platform of Anti-biofilm Assays Suited to the Exploration of Natural Compound Libraries. J. Vis. Exp. 2016, 118, 54829. [Google Scholar] [CrossRef]

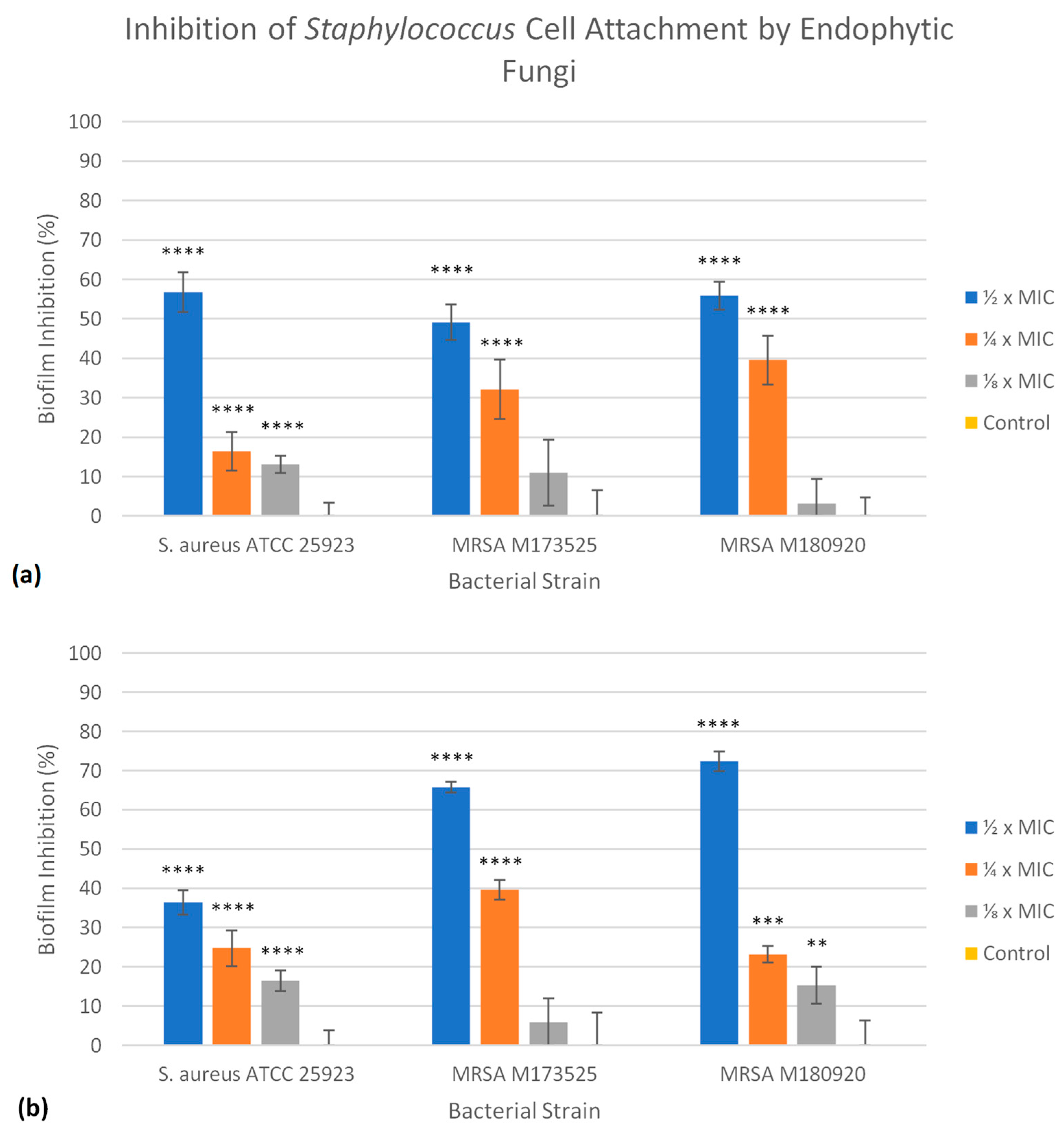


| Sample | ATCC 25923 | M173525 | M180920 | |||
|---|---|---|---|---|---|---|
| MHB | TSBG | MHB | TSBG | MHB | TSBG | |
| EL 24 | 78 | 0.8 | 50 | 0.8 | 50 | 0.4 |
| EL 35 | 156 | 4.9 | 312 | 2.4 | 156 | 1.2 |
| Chloramphenicol | 6.2 | 1.6 | 6.2 | 0.8 | 6.2 | 0.4 |
| Sample | ATCC 25923 | M173525 | M180920 | |||
|---|---|---|---|---|---|---|
| MHB | TSBG | MHB | TSBG | MHB | TSBG | |
| EL 24 | 156 | 6.2 | 200 | 1.6 | 100 | 1.6 |
| EL 35 | 156 | 9.8 | 625 | 2.4 | 312 | 1.2 |
| Chloramphenicol | >100 | 12.5 | >100 | 1.6 | >100 | 0.8 |
| RT (min) | Chemical Formula | Common Name (IUPAC Name) | Peak Area (%) | Molecular Weight | Retention Index | Match Factor (/1000) |
|---|---|---|---|---|---|---|
| 9.47 | C8H10O | Phenylethyl alcohol (2-phenylethanol) | 12 | 122.16 | 1100 | 927 |
| 15.71 | C10H10O3 | Mellein (8-hydroxy-3-methyl-3,4-dihydro-1H-isochromen-1-one) | 4.4 | 178.18 | 1570 | 844 |
| 16.64 | C10H10O3 | Isosclerone (4,8-dihydroxy-3,4-dihydro-1(2H)-naphthalenone) | 1.8 | 178.18 | 1640 | 888 |
| 19.15 | C11H18N2O2 | Cyclo(leucylprolyl) (3-isobutylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione) | 3.8 | 210.27 | 1820 | 809 |
| 23.40 | C14H16N2O2 | Cyclo(phenylalanylprolyl) (3-benzylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione) | 2.4 | 244.29 | 2140 | 883 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caruso, D.J.; Palombo, E.A.; Moulton, S.E.; Duggan, P.J.; Zaferanloo, B. Antibacterial and Antibiofilm Activity of Endophytic Alternaria sp. Isolated from Eremophila longifolia. Antibiotics 2023, 12, 1459. https://doi.org/10.3390/antibiotics12091459
Caruso DJ, Palombo EA, Moulton SE, Duggan PJ, Zaferanloo B. Antibacterial and Antibiofilm Activity of Endophytic Alternaria sp. Isolated from Eremophila longifolia. Antibiotics. 2023; 12(9):1459. https://doi.org/10.3390/antibiotics12091459
Chicago/Turabian StyleCaruso, Daniel J., Enzo A. Palombo, Simon E. Moulton, Peter J. Duggan, and Bita Zaferanloo. 2023. "Antibacterial and Antibiofilm Activity of Endophytic Alternaria sp. Isolated from Eremophila longifolia" Antibiotics 12, no. 9: 1459. https://doi.org/10.3390/antibiotics12091459





