Camel Milk Resistome in Kuwait: Genotypic and Phenotypic Characterization
Abstract
:1. Introduction
2. Results
2.1. Genotypic Resistance Profile of Camel Milk
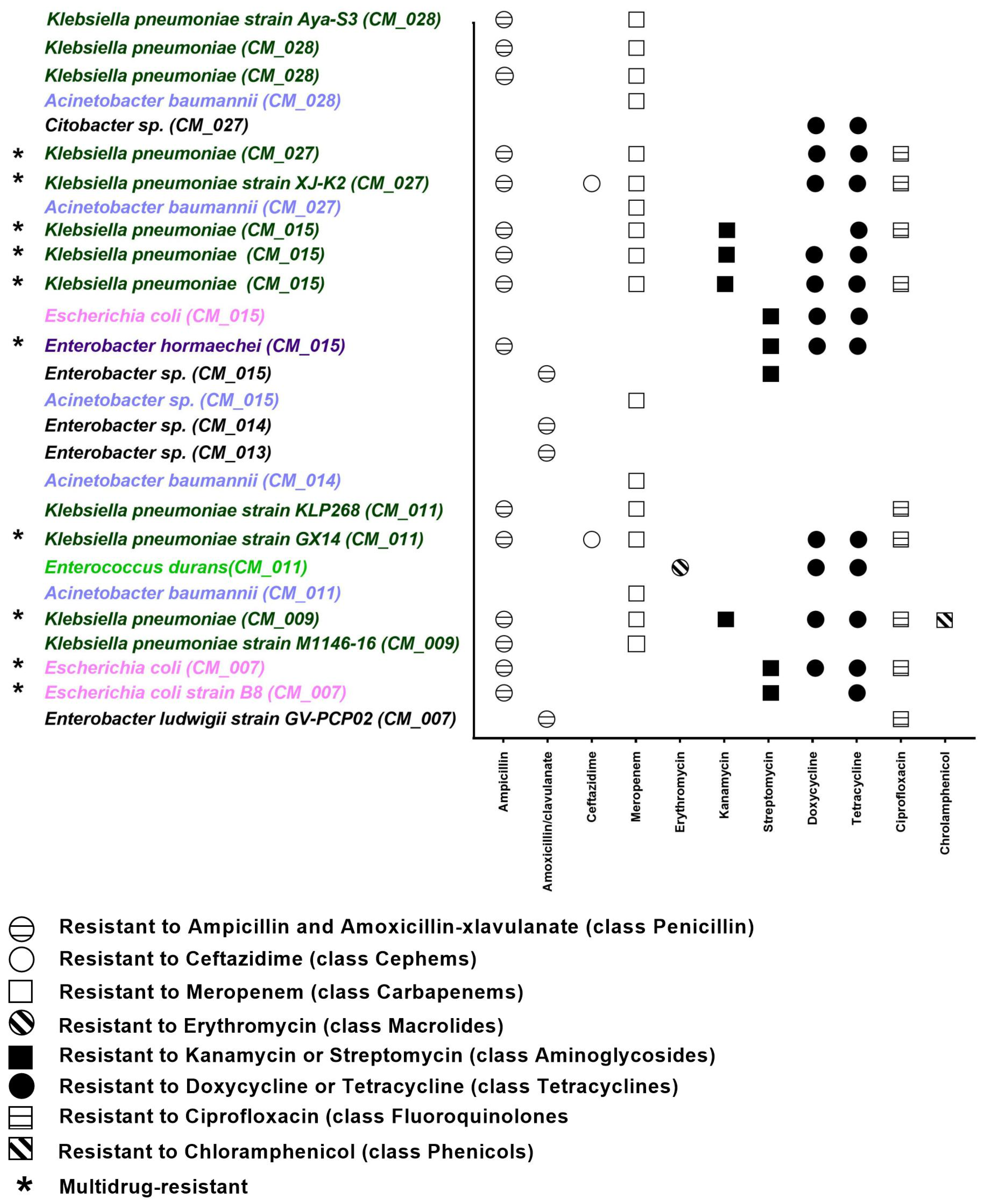
2.2. Phenotypic Resistance Profile of Camel Milk
3. Discussion
4. Material and Methods
4.1. Sample Collection and Genomic DNA Extraction
4.2. Library Preparation and Sequencing
4.3. Bioinformatics Analysis
4.4. Bacterial Isolation and Identification
4.5. Resistance Profile of Bacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pokharel, S.; Shrestha, P.; Adhikari, B. Antimicrobial use in food animals and human health: Time to implement ‘One Health’ approach. Antimicrob. Resist. Infect. Control. 2020, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.G.; Csabai, I.; Krikó, E.; Tőzsér, D.; Maróti, G.; Patai, V.; Makrai, L.; Szita, G.; Solymosi, N. Antimicrobial resistance genes in raw milk for human consumption. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Rubiola, S.; Chiesa, F.; Dalmasso, A.; Di Ciccio, P.; Civera, T. Detection of Antimicrobial Resistance Genes in the Milk Production Environment: Impact of Host DNA and Sequencing Depth. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef] [PubMed]
- Nwobodo, D.C.; Ugwu, M.C.; Anie, C.O.; Al-Ouqaili, M.T.S.; Ikem, J.C.; Chigozie, U.V.; Saki, M. Antibiotic resistance: The challenges and some emerging strategies for tackling a global menace. J. Clin. Lab. Anal. 2022, 36, e24655. [Google Scholar] [CrossRef] [PubMed]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, F.; Taban, B.M. A State-of-Art Review on Multi-Drug Resistant Pathogens in Foods of Animal Origin: Risk Factors and Mitigation Strategies. Front. Microbiol. 2019, 10, 2091. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Jay-Russell, M.; Lemay, D.G.; Mills, D.A. Reservoirs of antimicrobial resistance genes in retail raw milk. Microbiome 2020, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Godziszewska, J.; Pogorzelska-Nowicka, E.; Brodowska, M.; Jagura-Burdzy, G.; Wierzbicka, A. Detection in raw cow’s milk of coliform bacteria - reservoir of antibiotic resistance. LWT 2018, 93, 634–640. [Google Scholar] [CrossRef]
- Oselu, S.; Ebere, R.; Arimi, J.M. Camels, Camel Milk, and Camel Milk Product Situation in Kenya in Relation to the World. Int. J. Food Sci. 2022, 2022, 1237423. [Google Scholar] [CrossRef]
- Rahmeh, R.; Akbar, A.; Alomirah, H.; Kishk, M.; Al-Ateeqi, A.; Al-Milhm, S.; Shajan, A.; Akbar, B.; Al-Merri, S.; Alotaibi, M.; et al. Camel milk microbiota: A culture-independent assessment. Food Res. Int. 2022, 159, 111629. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Lundborg, C.S.; Sun, X.; Hu, X.; Dong, H. Economic burden of antibiotic resistance in ESKAPE organisms: A systematic review. Antimicrob. Resist. Infect. Control. 2019, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Huang, W.; Yang, J.; Zhao, Y.; Zhao, M.; Xu, H.; Zhang, M. The Antibiotic Resistome and Its Association with Bacterial Communities in Raw Camel Milk from Altay Xinjiang. Foods 2023, 12, 3928. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Weese, J.S. Methods and basic concepts for microbiota assessment. Veter. J. 2019, 249, 10–15. [Google Scholar] [CrossRef]
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-Based Methods and Resources to Study Antimicrobial Resistance. Nat. Rev. Genet. 2019, 20, 356–370. [Google Scholar] [CrossRef]
- Zaheer, R.; Noyes, N.; Polo, R.O.; Cook, S.R.; Marinier, E.; Van Domselaar, G.; Belk, K.E.; Morley, P.S.; McAllister, T.A. Impact of sequencing depth on the characterization of the microbiome and resistome. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Yee, R.; Bard, J.D.; Simner, P.J. The Genotype-to-Phenotype Dilemma: How Should Laboratories Approach Discordant Susceptibility Results? J. Clin. Microbiol. 2021, 59, e00138-20. [Google Scholar] [CrossRef] [PubMed]
- McLain, J.E.; Cytryn, E.; Durso, L.M.; Young, S. Culture-based Methods for Detection of Antibiotic Resistance in Agroecosystems: Advantages, Challenges, and Gaps in Knowledge. J. Environ. Qual. 2016, 45, 432–440. [Google Scholar] [CrossRef]
- Hoque, M.N.; Istiaq, A.; Clement, R.A.; Gibson, K.M.; Saha, O.; Islam, O.K.; Abir, R.A.; Sultana, M.; Siddiki, A.M.A.M.Z.; Crandall, K.A.; et al. Insights into the Resistome of Bovine Clinical Mastitis Microbiome, a Key Factor in Disease Complication. Front. Microbiol. 2020, 11, 860. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; A Wlodarski, M.; Edalatmand, A.; Petkau, A.; A Syed, S.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2022, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- von Meijenfeldt, F.A.B.; Arkhipova, K.; Cambuy, D.D.; Coutinho, F.H.; Dutilh, B.E. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Barth, P.; Falgenhauer, L.; Hain, T.; Chakraborty, T.; Goesmann, A. Platon: Identification and Characterization of Bacterial Plasmid Contigs in Short-Read Draft Assemblies Exploiting Protein Sequence-Based Replicon Distribution Scores. Microb. Genom. 2020, 6, e000398. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Narváez, S.; Giguère, S.; Anastasi, E.; Hearn, J.; Scortti, M.; Vázquez-Boland, J.A. Clonal Confinement of a Highly Mobile Resistance Element Driven by Combination Therapy in Rhodococcus equi. mBio 2019, 10, e02260-19. [Google Scholar] [CrossRef] [PubMed]
- M100-Ed33; Performance Standards for Antimicrobial Susceptibility Testing. CLSI: Wayne, PA, USA, 2023.
- Terreni, M.; Taccani, M.; Pregnolato, M. New Antibiotics for Multidrug-Resistant Bacterial Strains: Latest Research Developments and Future Perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.A.; Mir, R.A.; Qadri, H.; Dhiman, R.; Almilaibary, A.; Alkhanani, M.; Mir, M.A. Integrons in the development of antimicrobial resistance: Critical review and perspectives. Front. Microbiol. 2023, 14, 1231938. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Njage, P.; Jans, C.; Wangoh, J.; Lacroix, C. Detection, isolation and molecular characterisation of Shigatoxigenic O157 and non-O157 Escherichia coli in raw and fermented camel milk. Afr. J. Microbiol. Res. 2012, 6, 6031–6038. [Google Scholar] [CrossRef]
- Ntuli, V.; Njage, P.; Buys, E. Characterization of Escherichia coli and other Enterobacteriaceae in producer-distributor bulk milk. J. Dairy Sci. 2016, 99, 9534–9549. [Google Scholar] [CrossRef]
- WHO Publishes the WHO Medically Important Antimicrobials List for Human Medicine. Available online: https://www.who.int/samoa/news/detail-global/08-02-2024-who-medically-important-antimicrobial-list-2024 (accessed on 17 April 2024).
- O’connell, A.; Ruegg, P.; Jordan, K.; O’brien, B.; Gleeson, D. The effect of storage temperature and duration on the microbial quality of bulk tank milk. J. Dairy Sci. 2016, 99, 3367–3374. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2018. [Google Scholar]
- Zaharia, M.; Bolosky, W.J.; Curtis, K.; Fox, A.; Patterson, D.; Shenker, S. Faster and More Accurate Sequence Alignment with SNAP 2011. arXiv 2011, arXiv:1111.5572. [Google Scholar]
- Bushnell, B. BBTools; JGI: Berkeley, CA, USA, 2019. [Google Scholar]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K. MEGAHIT v1.0: A Fast and Scalable Metagenome Assembler Driven by Advanced Methodologies and Community Practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Bradnam, K.R.; Fass, J.N.; Alexandrov, A.; Baranay, P.; Bechner, M.; Birol, I.; Boisvert, S.; Chapman, J.A.; Chapuis, G.; Chikhi, R.; et al. Assemblathon 2: Evaluating de novo methods of genome assembly in three vertebrate species. GigaScience 2013, 2, 10. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- R Core Team. _R: A Language and Environment for Statistical Computing_. R Foundation for Statistical Computing, Vienna, Austria. 2023. Available online: https://www.R-project.org/ (accessed on 21 September 2023).
- Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Stetsenko, P.; Short, T.; Lianoglou, S.; Antonyan, E.; Bonsch, M.; Parsonage, H.; et al. Data.Table: Extension of “Data.Frame”. 2019. Available online: https://CRAN.R-project.org/package=data.table (accessed on 21 September 2023).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Kalvari, I.; Nawrocki, E.P.; Argasinska, J.; Quinones-Olvera, N.; Finn, R.D.; Bateman, A.; Petrov, A.I. Non-Coding RNA Analysis Using the Rfam Database. Curr. Protoc. Bioinform. 2018, 62, e51. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; LoCascio, P.F.; Hauser, L.J.; Uberbacher, E.C. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 2012, 28, 2223–2230. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2005, 33, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2013, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Pérochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 2006, 34, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.V.; Lund, O.; Villa, L. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Redondo-Salvo, S.; Vielva, L.; de la Cruz, F. MOBscan: Automated Annotation of MOB Relaxases. In Horizontal Gene Transfer. Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2075, pp. 295–308. [Google Scholar]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Federhen, S. The NCBI Taxonomy Database. Nucleic Acids Res. 2012, 40, 136–143. [Google Scholar] [CrossRef]
- Gao, J.; Barkema, H.W.; Zhang, L.; Liu, G.; Deng, Z.; Cai, L.; Shan, R.; Zhang, S.; Zou, J.; Kastelic, J.P.; et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J. Dairy Sci. 2017, 100, 4797–4806. [Google Scholar] [CrossRef]


| Drug Class | Mean | SD |
|---|---|---|
| fluoroquinolone antibiotic | 12.48 | 1.68 |
| disinfecting agents and antiseptics | 9.02 | 1.85 |
| cephalosporin | 8.42 | 1.26 |
| penam | 8.07 | 1.23 |
| tetracycline antibiotic | 8.06 | 1.20 |
| peptide antibiotic | 5.92 | 1.84 |
| glycopeptide antibiotic | 5.62 | 5.21 |
| rifamycin antibiotic | 5.29 | 0.84 |
| aminoglycoside antibiotic | 5.27 | 2.34 |
| macrolide antibiotic | 4.84 | 1.01 |
| phenicol antibiotic | 4.25 | 0.59 |
| cephamycin | 3.87 | 1.18 |
| glycylcycline | 3.83 | 0.67 |
| carbapenem | 3.35 | 0.76 |
| penem | 3.21 | 0.60 |
| monobactam | 2.01 | 0.55 |
| phosphonic acid antibiotic | 1.92 | 0.52 |
| aminocoumarin antibiotic | 1.18 | 0.24 |
| nucleoside antibiotic | 1.13 | 0.27 |
| diaminopyrimidine antibiotic | 0.84 | 0.17 |
| nitroimidazole antibiotic | 0.47 | 0.19 |
| lincosamide antibiotic | 0.29 | 0.47 |
| nitrofuran antibiotic | 0.23 | 0.29 |
| streptogramin antibiotic | 0.14 | 0.22 |
| sulfonamide antibiotic | 0.12 | 0.05 |
| pleuromutilin antibiotic | 0.10 | 0.17 |
| streptogramin A antibiotic | 0.03 | 0.05 |
| streptogramin B antibiotic | 0.03 | 0.05 |
| Genes of Resistance | Drug Class | Resistance Mechanism | AMR Gene Family | Genus | CM 007 | CM 009 | CM 011 | CM 013 | CM 014 | CM 015 | CM 027 | CM 028 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vanY gene in vanB cluster | glycopeptide antibiotic | antibiotic target alteration | vanY | Enterococcus | 0.29 | 1.53 | 0.08 | 0.09 | 0.09 | 0.15 | 0.09 | 0.22 |
| vanT gene in vanG cluster | glycopeptide antibiotic | antibiotic target alteration | vanT | Enterococcus | 0.18 | 0.98 | 0.05 | 0.05 | 0.04 | 0.12 | 0.05 | 0.15 |
| vanW gene in vanG cluster | glycopeptide antibiotic | antibiotic target alteration | vanW | Streptococcus | 0.42 | 0.20 | 0.00 | 0.00 | 0.03 | 0.02 | 0.01 | 0.06 |
| patB | fluoroquinolone antibiotic | antibiotic efflux | ATP-binding cassette (ABC) antibiotic efflux pump | Streptococcus | 0.40 | 0.18 | 0.00 | 0.00 | 0.00 | 0.02 | 0.01 | 0.06 |
| vanY gene in vanF cluster | glycopeptide antibiotic | antibiotic target alteration | vanY | Streptococcus | 0.35 | 0.16 | 0.00 | 0.00 | 0.02 | 0.02 | 0.00 | 0.05 |
| vanY gene in vanM cluster | glycopeptide antibiotic | antibiotic target alteration | vanY | Streptococcus | 0.35 | 0.16 | 0.00 | 0.00 | 0.02 | 0.02 | 0.00 | 0.05 |
| qacJ | disinfecting agents and antiseptics | antibiotic efflux | small multidrug resistance (SMR) antibiotic efflux pump | Streptococcus | 0.14 | 0.06 | 0.00 | 0.00 | 0.01 | 0.01 | 0.00 | 0.02 |
| mreA | macrolide antibiotic | antibiotic efflux | major facilitator superfamily (MFS) antibiotic efflux pump | Streptococcus | 0.09 | 0.05 | 0.00 | 0.00 | 0.02 | 0.01 | 0.00 | 0.02 |
| Streptococcus agalactiae mprF | peptide antibiotic | antibiotic target alteration | defensin resistant mprF | Streptococcus | 0.00 | 0.00 | 0.01 | 0.01 | 0.06 | 0.00 | 0.00 | 0.01 |
| lmrD | lincosamide antibiotic | antibiotic efflux | ATP-binding cassette (ABC) antibiotic efflux pump | Lactococcus | 0.01 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 |
| vanY gene in vanG cluster | glycopeptide antibiotic | antibiotic target alteration | vanY | Lactococcus | 0.02 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 |
| CrcB | aminoglycoside antibiotic | antibiotic efflux | multidrug and toxic compound extrusion (MATE) transporter | Klebsiella | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.06 | 0.00 | 0.00 |
| adeC | glycylcycline | antibiotic efflux | resistance-nodulation-cell division (RND) antibiotic efflux pump | Acinetobacter | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | 0.01 |
| adeC | tetracycline antibiotic | antibiotic efflux | resistance-nodulation-cell division (RND) antibiotic efflux pump | Acinetobacter | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | 0.01 |
| vanY gene in vanB cluster | glycopeptide antibiotic | antibiotic target alteration | vanY | Paenibacillus | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 |
| tet(36) | tetracycline antibiotic | antibiotic target protection | tetracycline-resistant ribosomal protection protein | Paenibacillus | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 |
| vanW gene in vanI cluster | glycopeptide antibiotic | antibiotic target alteration | vanW | Paenibacillus | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 |
| Contig | On Plasmid | CDS ID | Target Distance | ISfinder Description | CARD Description | CARD Drug Class | CARD Resistance Mechanism |
|---|---|---|---|---|---|---|---|
| 1450 | 1 | 1450_5 | 7, −1, 1 | lnuB | lincosamide | antibiotic inactivation | |
| 1450 | 1 | 1450_6 | 6, −2, 0 | ISRe46_aa4 | lsaE | lincosamide; streptogramin; pleuromutilin, | antibiotic target protection |
| 1450 | 1 | 1450_6 | 6, −2, 0 | ISRe46_aa4 | lsaE | lincosamide; streptogramin; pleuromutilin, | antibiotic target protection |
| 2967 | 1 | 2967_3 | −2, −1 | tet(A) | tetracycline | antibiotic efflux | |
| 3207 | 1 | 3207_1 | 4 | EreA | macrolide | antibiotic inactivation | |
| 3207 | 1 | 3207_2 | 3 | qacEdelta1 | disinfecting agents and antiseptics | antibiotic efflux | |
| 3207 | 1 | 3207_3 | 2 | sul1 | sulfonamide | antibiotic target replacement | |
| 3238 | 1 | 3238_1 | 0, 1 | Tn2_aa1 | TEM-1 | monobactam; cephalosporin; penam; penem | antibiotic inactivation |
| 3348 | 1 | 3348_1 | 1, 5 | qacL | disinfecting agents and antiseptics | antibiotic efflux | |
| 3348 | 1 | 3348_3 | −1, 3 | sul3 | sulfonamide | antibiotic target replacement | |
| 3369 | 1 | 3369_1 | 0 | ISArsp14_aa11 | tet(45) | tetracycline | antibiotic efflux |
| 4993 | 1 | 4993_2 | 0, 1 | Tn2_aa1 | CTX-M-15 | cephalosporin; penam | antibiotic inactivation |
| 6435 | 1 | 6435_1 | 0 | MICBce1_aa1 | DHA-1 | cephalosporin; cephamycin | antibiotic inactivation |
| 7063 | 1 | 7063_1 | 0 | Tn2_aa1 | LAP-2 | cephalosporin; penam; penem | antibiotic inactivation |
| 9281 | 1 | 9281_1 | 0 | ISAba61_aa1 | cmlA1 | phenicol | antibiotic efflux |
| 10144 | 1 | 10144_2 | −1 | QnrS1 | fluoroquinolone | antibiotic target protection | |
| 13504 | 1 | 13504_1 | 0 | ISBce8_aa2 | AAC(6′)-Ib-cr6 | fluoroquinolone; aminoglycoside | antibiotic inactivation |
| 13534 | 1 | 13534_1 | 0 | ISCco2_aa4 | APH(3′)-IIIa | aminoglycoside | antibiotic inactivation |
| 14655 | 1 | 14655_1 | 0 | ISSsu9_aa2 | dfrA12 | diaminopyrimidine | antibiotic target replacement |
| 16142 | 1 | 16142_1 | 0 | ISRe46_aa6 | ErmB | macrolide; lincosamid; streptogramin; streptogramin A; streptogramin B | antibiotic target alteration |
| Genus | Contig | On Plasmid | CDS ID | Target Distance | ISfinder Description | CARD Description | CARD Drug Class | CARD Resistance Mechanism |
|---|---|---|---|---|---|---|---|---|
| Paenibacillus | 1 | 0 | 1_943 | 4 | vanT gene in vanG cluster | glycopeptide antibiotic | antibiotic target alteration | |
| Paenibacillus | 3 | 0 | 3_46 | −1 | vanW gene in vanI cluster | glycopeptide antibiotic | antibiotic target alteration | |
| Paenibacillus | 3 | 0 | 3_546 | −8, 7, 8, 9 | tet(36) | tetracycline antibiotic | antibiotic target protection | |
| Paenibacillus | 4 | 0 | 4_279 | −5, 4, 9, 10 | vanY gene in vanB cluster | glycopeptide antibiotic | antibiotic target alteration | |
| Paenibacillus | 9 | 0 | 9_284 | −8, −2, 4 | LlmA 23S ribosomal RNA methyltransferase | lincosamide antibiotic | antibiotic target alteration | |
| Paenibacillus | 9 | 0 | 9_367 | 0, 1, 9 | ISArsp14_aa11 | tet(45) | tetracycline antibiotic | antibiotic efflux |
| Paenibacillus | 20 | 0 | 20_197 | 8 | vanH gene in vanO cluster | glycopeptide antibiotic | antibiotic target alteration | |
| Enterococcus | 39 | 0 | 39_47 | 1 | vanY gene in vanB cluster | glycopeptide antibiotic | antibiotic target alteration | |
| Enterococcus | 111 | 0 | 111_30 | −6 | vanT gene in vanG cluster | glycopeptide antibiotic | antibiotic target alteration | |
| Enterococcus | 111 | 0 | 111_30 | −6 | vanT gene in vanG cluster | glycopeptide antibiotic | antibiotic target alteration | |
| Streptococcus | 228 | 0 | 228_29 | 7 | vanY gene in vanM cluster | glycopeptide antibiotic | antibiotic target alteration | |
| Streptococcus | 242 | 0 | 242_12 | 0, 1 | ISArsp14_aa5 | patB | fluoroquinolone antibiotic | antibiotic efflux |
| Streptococcus | 679 | 0 | 679_11 | −2 | Streptococcus agalactiae mprF | peptide antibiotic | antibiotic target alteration | |
| Streptococcus | 758 | 0 | 758_6 | 7, 8 | mreA | macrolide antibiotic | antibiotic efflux | |
| Streptococcus | 854 | 0 | 854_16 | −6, −5, −8 | qacJ | disinfecting agents and antiseptics | antibiotic efflux | |
| Lactococcus | 1373 | 0 | 1373_2 | −1, 9, 0 | ISArsp14_aa5 | lmrD | lincosamide antibiotic | antibiotic efflux |
| Antibiotic | Antibiotic Class | Disk Content | Interpretive Categories and Zone Diameter Breakpoints (Nearest Whole mm) | Isolates | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Klebsiella pneumonia | Escherichia coli | Enterobacter sp. | |||||||||||||||||||||
| No. of Isolates | (%) | No. of Isolates | (%) | No. of Isolates | (%) | ||||||||||||||||||
| S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | |||
| Ampicillin | Penicillin | 10 µg | ≥17 | 14–16 | ≤13 | 0 | 0 | 12 | 0 | 0 | 100 | 3 | 0 | 2 | 60 | 0 | 40 | 3 | 0 | 2 | 20 | 60 | 20 |
| AMC | Penicillin | 30 µg | ≥18 | 14–17 | ≤13 | 11 | 1 | 0 | 92 | 8 | 0 | 5 | 0 | 0 | 100 | 0 | 0 | 5 | 0 | 0 | 20 | 0 | 80 |
| Ceftazidime | Cephems | 30 µg | ≥21 | 18–20 | ≤17 | 8 | 2 | 2 | 67 | 17 | 17 | 5 | 0 | 0 | 100 | 0 | 0 | 5 | 0 | 0 | 100 | 0 | 0 |
| Meropenem | Carbapenems | 10 µg | ≥23 | 20–22 | ≤19 | 0 | 0 | 12 | 0 | 0 | 100 | 5 | 0 | 0 | 100 | 0 | 0 | 5 | 0 | 0 | 100 | 0 | 0 |
| Imipenem | Carbapenems | 10 µg | ≥23 | 20–22 | ≤19 | 12 | 0 | 0 | 100 | 0 | 0 | 4 | 1 | 0 | 80 | 20 | 0 | 4 | 1 | 0 | 100 | 0 | 0 |
| Gentamicin | Aminoglycosides | 10 µg | ≥18 | 15–17 | ≤14 | 11 | 1 | 0 | 92 | 8 | 0 | 5 | 0 | 0 | 100 | 0 | 0 | 5 | 0 | 0 | 100 | 0 | 0 |
| Kanamycin | Aminoglycosides | 30 µg | ≥18 | 14–17 | ≤13 | 8 | 0 | 4 | 67 | 0 | 33 | 5 | 0 | 0 | 100 | 0 | 0 | 5 | 0 | 0 | 100 | 0 | 0 |
| Streptomycin | Aminoglycosides | 10 µg | ≥15 | 12–14 | ≤11 | - | - | - | - | - | - | 2 | 0 | 3 | 40 | 0 | 60 | 2 | 0 | 3 | 60 | 0 | 40 |
| Tetracycline | Tetracyclines | 30 µg | ≥15 | 12–14 | ≤11 | 5 | 0 | 7 | 42 | 0 | 58 | 2 | 0 | 3 | 40 | 0 | 60 | 2 | 0 | 3 | 60 | 20 | 20 |
| Doxycycline | Tetracyclines | 30 µg | ≥14 | 11–13 | ≤10 | 6 | 0 | 6 | 50 | 0 | 50 | 3 | 0 | 2 | 60 | 0 | 40 | 3 | 0 | 2 | 60 | 20 | 20 |
| Ciprofloxacin | Fluoroquinolones | 5 µg | ≥26 | 22–25 | ≤21 | 2 | 7 | 3 | 17 | 58 | 25 | 3 | 1 | 1 | 60 | 20 | 20 | 3 | 1 | 1 | 20 | 60 | 20 |
| Chloramphenicol | Phenicols | 30 µg | ≥18 | 13–17 | ≤12 | 11 | 0 | 1 | 92 | 0 | 8 | 5 | 0 | 0 | 100 | 0 | 0 | 5 | 0 | 0 | 100 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahmeh, R.; Akbar, A.; Almutairi, B.; Kishk, M.; Jordamovic, N.B.; Al-Ateeqi, A.; Shajan, A.; Al-Sherif, H.; Esposito, A.; Al-Momin, S.; et al. Camel Milk Resistome in Kuwait: Genotypic and Phenotypic Characterization. Antibiotics 2024, 13, 380. https://doi.org/10.3390/antibiotics13050380
Rahmeh R, Akbar A, Almutairi B, Kishk M, Jordamovic NB, Al-Ateeqi A, Shajan A, Al-Sherif H, Esposito A, Al-Momin S, et al. Camel Milk Resistome in Kuwait: Genotypic and Phenotypic Characterization. Antibiotics. 2024; 13(5):380. https://doi.org/10.3390/antibiotics13050380
Chicago/Turabian StyleRahmeh, Rita, Abrar Akbar, Batlah Almutairi, Mohamed Kishk, Naida Babic Jordamovic, Abdulaziz Al-Ateeqi, Anisha Shajan, Heba Al-Sherif, Alfonso Esposito, Sabah Al-Momin, and et al. 2024. "Camel Milk Resistome in Kuwait: Genotypic and Phenotypic Characterization" Antibiotics 13, no. 5: 380. https://doi.org/10.3390/antibiotics13050380







