Plant Growth Promotion and Stress Tolerance Enhancement through Inoculation with Bacillus proteolyticus OSUB18
Abstract
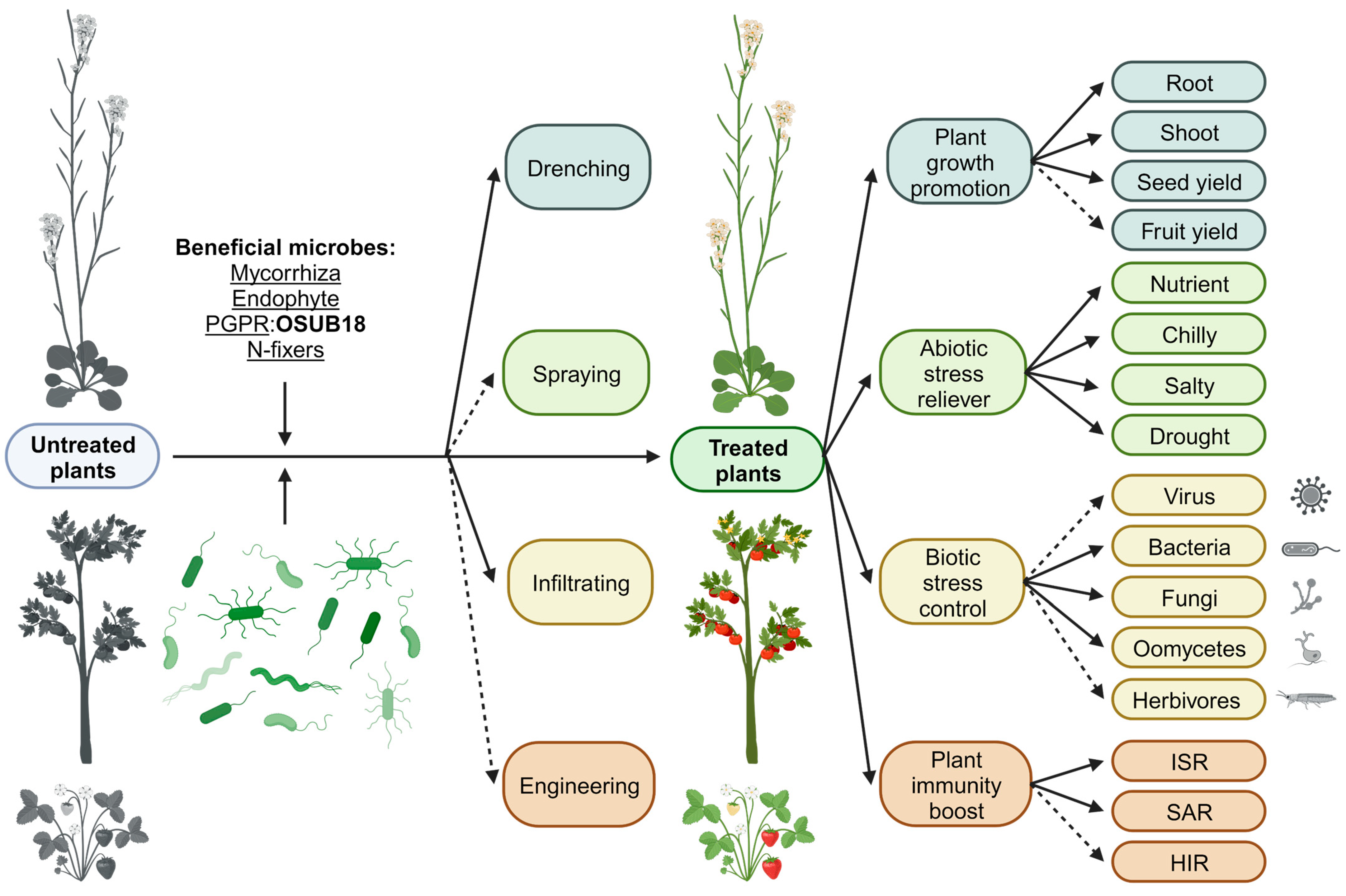
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Plant Growth Analysis
2.3. RNA-Seq and Bioinformatics Analysis
2.4. Pathogen Inoculations, Phloem Exudate Collection, and SAR Assay
2.5. Microbial Materials and Antagonistic Assays
2.6. Statistical Analysis
3. Results
3.1. Alterations in Root Architecture and the Promotion of Plant Growth under Various Abiotic Stresses in Arabidopsis Are Discerned through the Impact of OSUB18
3.2. RNA-Seq Analysis Reveals Qualitative and Quantitative Differences in Arabidopsis Gene Expressions Due to OSUB18-Drenching Treatment
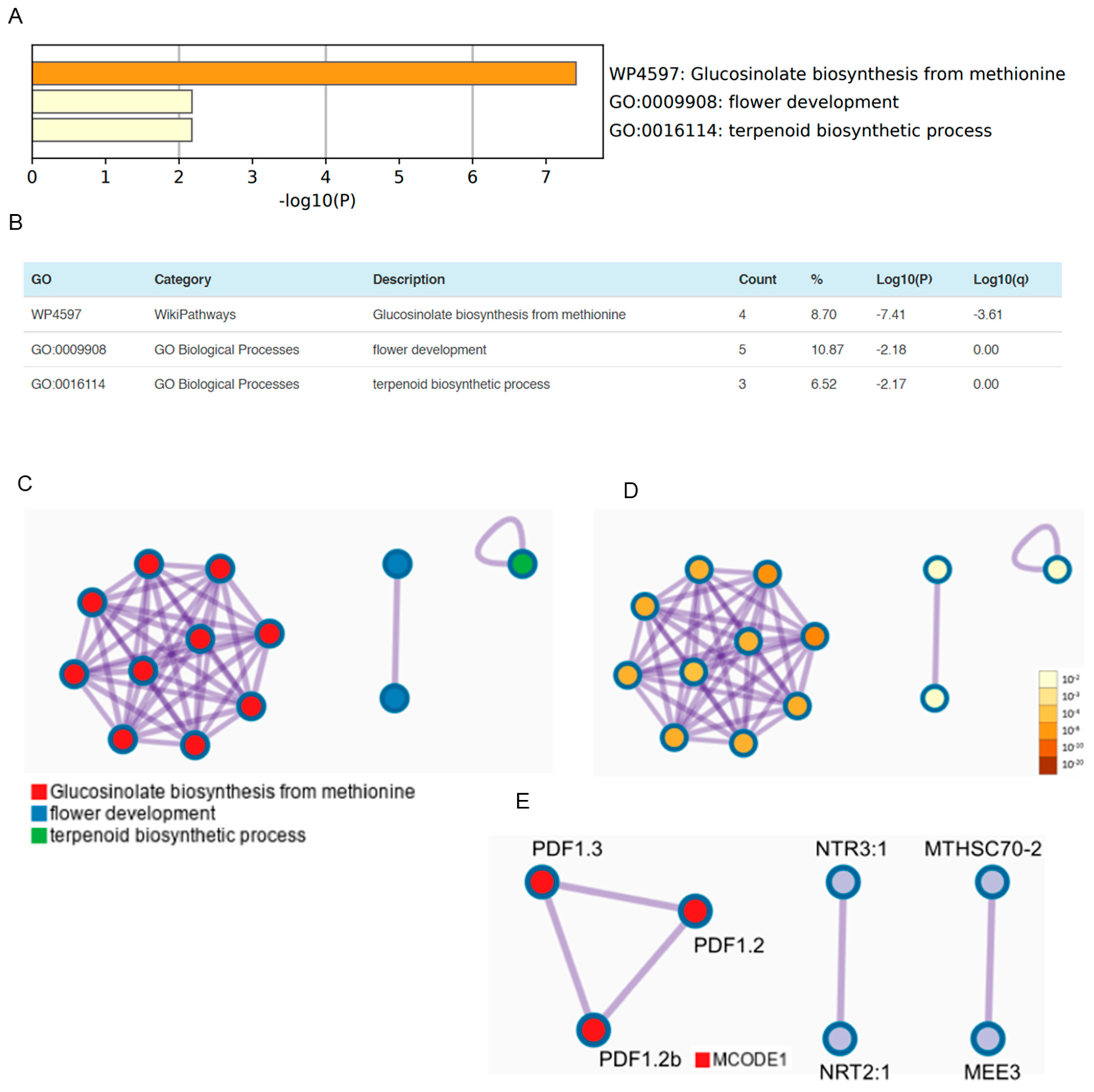
3.3. OSUB18 Elicits the Upregulation of Glucosinolate Biosynthesis and Defense-Related Genes in Arabidopsis
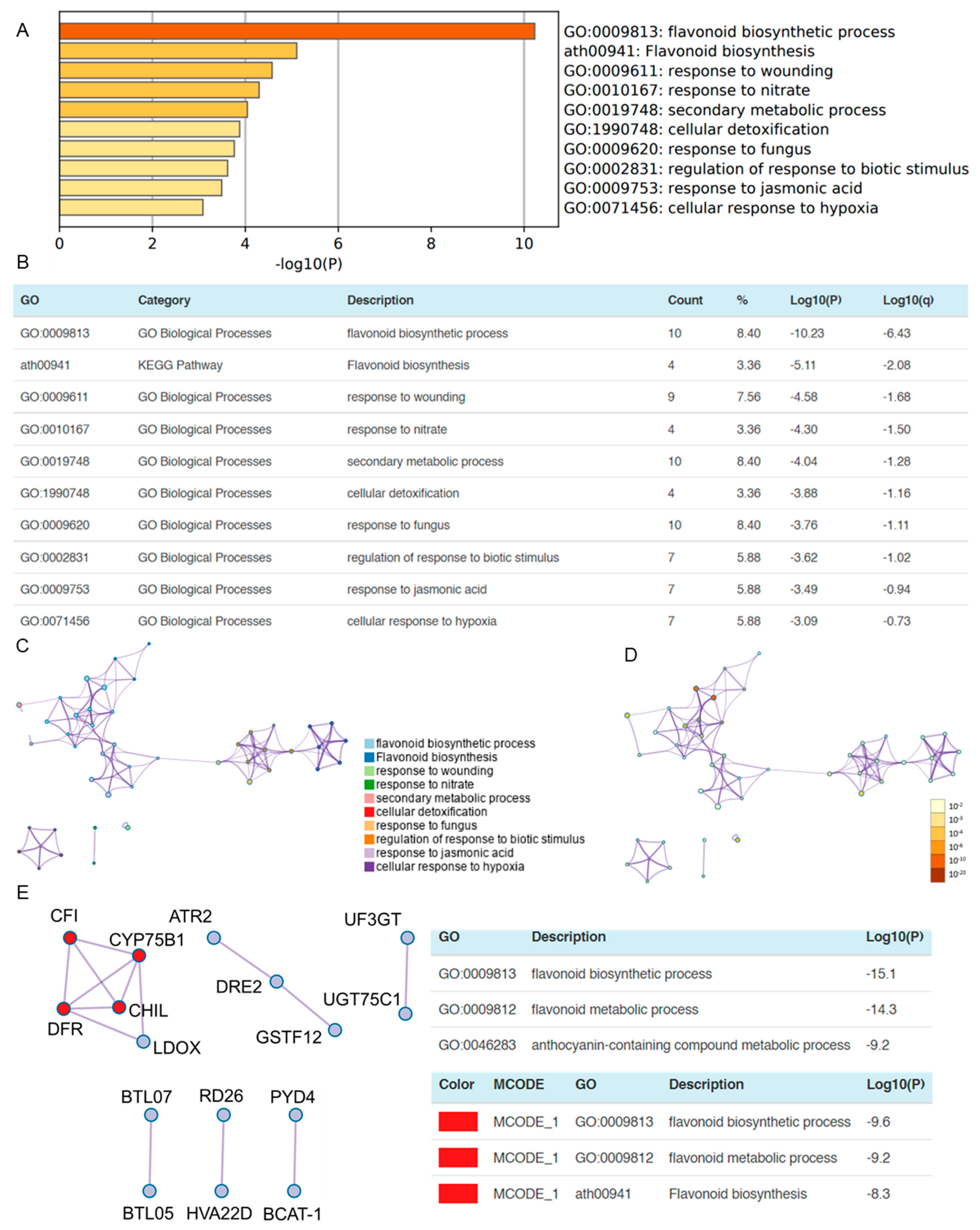
3.4. OSUB18 Exerts the Downregulation of Plant Flavonoid Biosynthetic and Wound Responsive Genes in Arabidopsis
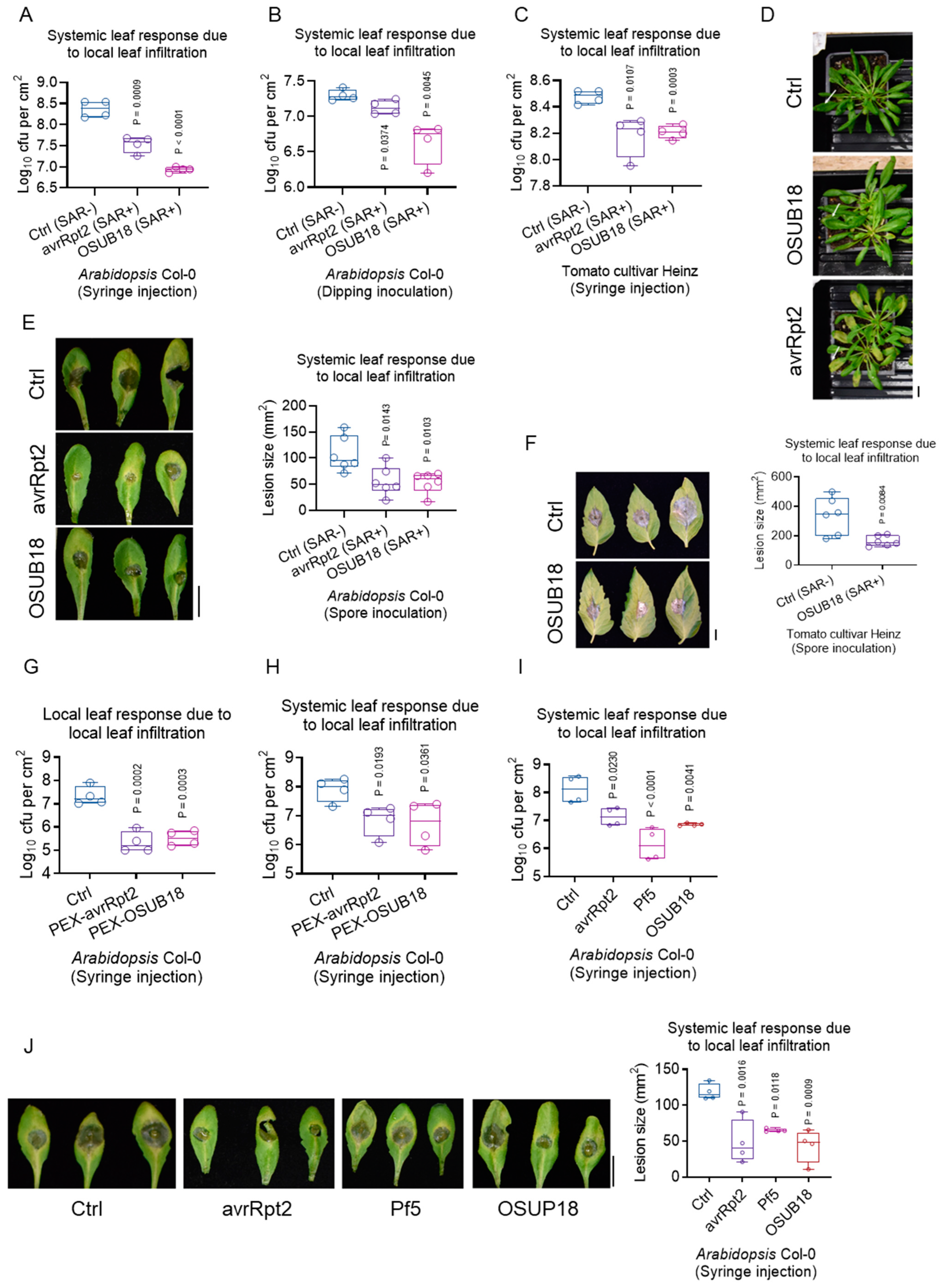
3.5. OSUB18 Elicits Systemic Resistance in Host Plants against Bacterial and Fungal Pathogens
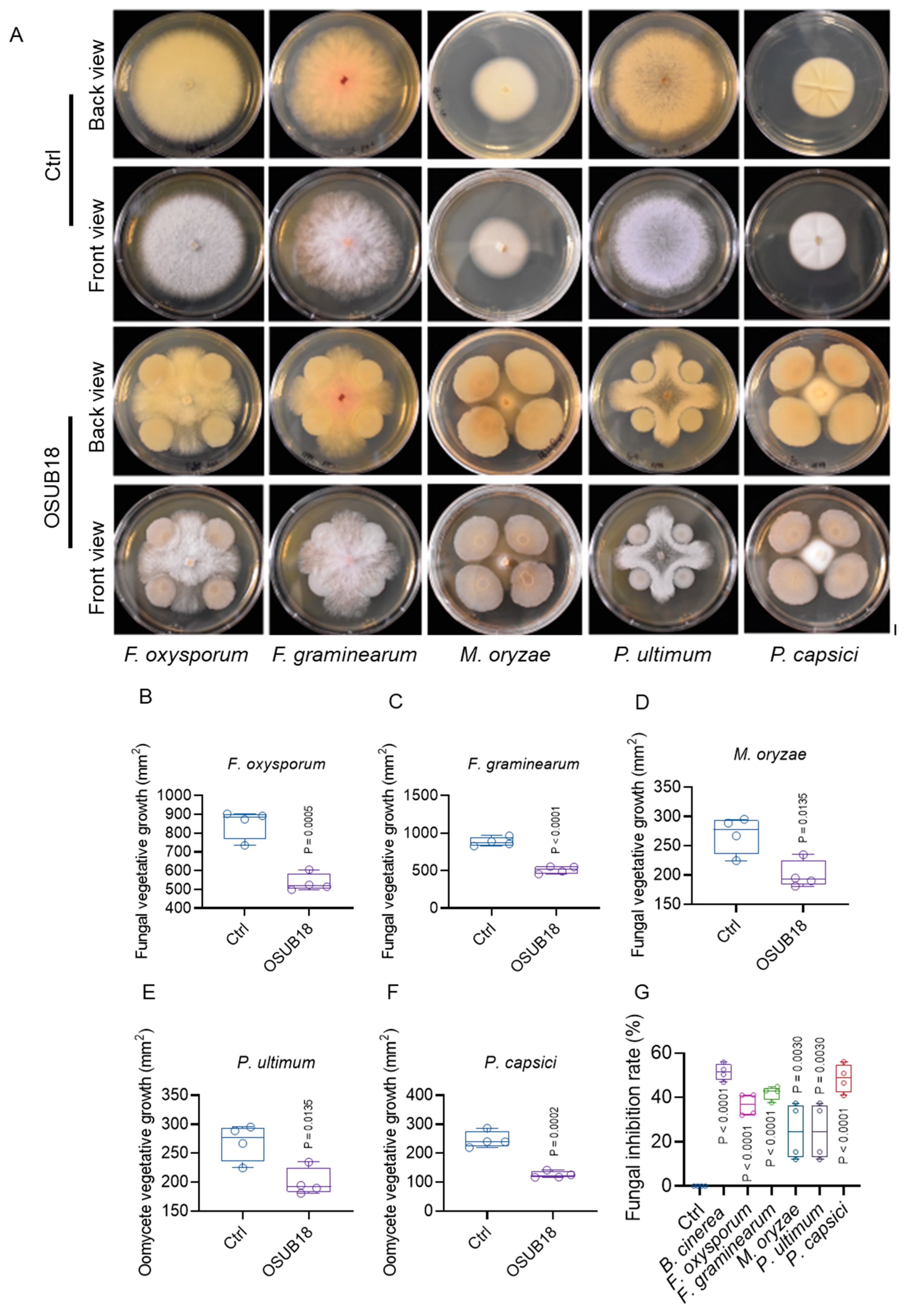
3.6. OSUB18 Exhibits Antagonism towards Phytopathogenic Bacteria, Fungi, and Oomycetes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olaniyan, O.T.; Adetunji, C.O. Biochemical Role of Beneficial Microorganisms: An Overview on Recent Development in Environmental and Agro Science. In Microbial Rejuvenation of Polluted Environment; Adetunji, C.O., Panpatte, D.G., Jhala, Y.K., Eds.; Microorganisms for Sustainability; Springer: Singapore, 2021; Volume 3, pp. 21–33. ISBN 9789811574597. [Google Scholar]
- Gupta, V.V. Beneficial Microorganisms for Sustainable Agriculture. Microbiol. Aust. 2012, 33, 113–115. [Google Scholar] [CrossRef]
- Zeng, Z.; He, X.; Li, F.; Zhang, Y.; Huang, Z.; Wang, Y.; Li, K.; Bao, Y.; Iqbal, M.; Fakhar-e-Alam Kulyar, M.; et al. Probiotic Properties of Bacillus proteolyticus Isolated from Tibetan Yaks, China. Front. Microbiol. 2021, 12, 649207. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zhao, Z.; Fan, J.; Liang, Y.; Bernier, M.C.; Gao, Y.; Zhao, L.; Opiyo, S.O.; Xia, Y. Bacillus proteolyticus OSUB18 Triggers Induced Systemic Resistance against Bacterial and Fungal Pathogens in Arabidopsis. Front. Plant Sci. 2023, 14, 1078100. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Fan, J.; Yang, P.; Wang, Z.; Stephen, O.; Mackey, D.; Ye, X. Involvement of Arabidopsis Acyl Carrier Protein 1 in PAMP-Triggered Immunity. Mol. Plant-Microbe Interact. 2022, 35, 681–693. [Google Scholar] [CrossRef] [PubMed]
- The Biological Resources of Model Organisms; Jarret, R.L.; McCluskey, K. (Eds.) CRC Press: Boca Raton, FL, USA, 2019; ISBN 978-1-138-29461-5. [Google Scholar]
- Zhao, L.; Yang, P.; Li, W.; Zhao, Z.; Xia, Y. First Report of Trichoderma crassum Causing Leaf Spot on Tomato (Solanum lycopersicum) in Ohio. Plant Disease 2023, 107, 582. [Google Scholar] [CrossRef]
- Zhao, L.; Yang, P.; Zhao, Z.; Xia, Y. First Report of Cephaliophora Tropica Causing Leaf Spot on Tomato (Solanum lycopersicum cv. Picus) in U.S.A. Crop Prot. 2023, 172, 106324. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Cheng, C.-Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A Complete Reannotation of the Arabidopsis Thaliana Reference Genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Haynes, W. Benjamini–Hochberg Method. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; p. 78. ISBN 978-1-4419-9863-7. [Google Scholar]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved Gene Annotation and New Tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Zhao, Z.; Fan, J.; Gao, Y.G.; Wang, Z.; Yang, P.; Liang, Y.; Opiyo, S.; Xia, Y. Arabidopsis Plasma Membrane ATPase AHA5 Is Negatively Involved in PAMP-Triggered Immunity. Int. J. Mol. Sci. 2022, 23, 3857. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis Thaliana-Pseudomonas Syringae Interaction. In Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2002. [Google Scholar] [CrossRef]
- Hoffmann-Benning, S. Collection and Analysis of Phloem Lipids. In Plant Lipids: Methods and Protocols; Bartels, D., Dörmann, P., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; pp. 351–361. ISBN 978-1-07-161362-7. [Google Scholar]
- Wang, C.; Huang, X.; Li, Q.; Zhang, Y.; Li, J.-L.; Mou, Z. Extracellular Pyridine Nucleotides Trigger Plant Systemic Immunity through a Lectin Receptor Kinase/BAK1 Complex. Nat. Commun. 2019, 10, 4810. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, M.; Chhajed, S.; Yu, F.; Chen, S.; Zhang, Y.; Mou, Z. N-Hydroxypipecolic Acid Triggers Systemic Acquired Resistance through Extracellular NAD(P). Nat. Commun. 2023, 14, 6848. [Google Scholar] [CrossRef]
- Maldonado, A.M.; Doerner, P.; Dixon, R.A.; Lamb, C.J.; Cameron, R.K. A Putative Lipid Transfer Protein Involved in Systemic Resistance Signalling in Arabidopsis. Nature 2002, 419, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.-F.; He, S.Y. Pseudomonas Syringae Pv. Tomato Dc3000: A Model Pathogen for Probing Disease Susceptibility and Hormone Signaling in Plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Press, C.M.; Ravel, J.; Kobayashi, D.Y.; Myers, G.S.A.; Mavrodi, D.V.; DeBoy, R.T.; Seshadri, R.; Ren, Q.; Madupu, R.; et al. Complete Genome Sequence of the Plant Commensal Pseudomonas Fluorescens Pf-5. Nat. Biotechnol. 2005, 23, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Barba, F.J.; Nikmaram, N.; Roohinejad, S.; Khelfa, A.; Zhu, Z.; Koubaa, M. Bioavailability of Glucosinolates and Their Breakdown Products: Impact of Processing. Front. Nutr. 2016, 3, 24. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Benítez, M.; Corvera-Poiré, A.; Chaos Cador, Á.; de Folter, S.; Gamboa de Buen, A.; Garay-Arroyo, A.; García-Ponce, B.; Jaimes-Miranda, F.; Pérez-Ruiz, R.V.; et al. Flower Development. Arab. Book 2010, 8, e0127. [Google Scholar] [CrossRef]
- Cheng, A.-X.; Lou, Y.-G.; Mao, Y.-B.; Lu, S.; Wang, L.-J.; Chen, X.-Y. Plant Terpenoids: Biosynthesis and Ecological Functions. J. Integr. Plant Biol. 2007, 49, 179–186. [Google Scholar] [CrossRef]
- Brown, R.L.; Kazan, K.; McGrath, K.C.; Maclean, D.J.; Manners, J.M. A Role for the GCC-Box in Jasmonate-Mediated Activation of the PDF1.2 Gene of Arabidopsis. Plant Physiol. 2003, 132, 1020–1032. [Google Scholar] [CrossRef]
- Mathesius, U. Flavonoid Functions in Plants and Their Interactions with Other Organisms. Plants 2018, 7, 30. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- De Kesel, J.; Conrath, U.; Flors, V.; Luna, E.; Mageroy, M.H.; Mauch-Mani, B.; Pastor, V.; Pozo, M.J.; Pieterse, C.M.J.; Ton, J.; et al. The Induced Resistance Lexicon: Do’s and Don’ts. Trends Plant Sci. 2021, 26, 685–691. [Google Scholar] [CrossRef]
- Yang, P.; Zhao, L.; Gao, Y.G.; Xia, Y. Detection, Diagnosis, and Preventive Management of the Bacterial Plant Pathogen Pseudomonas Syringae. Plants 2023, 12, 1765. [Google Scholar] [CrossRef]
- Cheng, W.; Li, H.-P.; Zhang, J.-B.; Du, H.-J.; Wei, Q.-Y.; Huang, T.; Yang, P.; Kong, X.-W.; Liao, Y.-C. Tissue-Specific and Pathogen-Inducible Expression of a Fusion Protein Containing a Fusarium-Specific Antibody and a Fungal Chitinase Protects Wheat against Fusarium Pathogens and Mycotoxins. Plant Biotechnol. J. 2015, 13, 664–674. [Google Scholar] [CrossRef]
- Yun, Y.; Zhou, X.; Yang, S.; Wen, Y.; You, H.; Zheng, Y.; Norvienyeku, J.; Shim, W.-B.; Wang, Z. Fusarium oxysporum f. Sp. Lycopersici C2H2 Transcription Factor FolCzf1 Is Required for Conidiation, Fusaric Acid Production, and Early Host Infection. Curr. Genet. 2019, 65, 773–783. [Google Scholar] [CrossRef]
- Cheng, W.; Song, X.-S.; Li, H.-P.; Cao, L.-H.; Sun, K.; Qiu, X.-L.; Xu, Y.-B.; Yang, P.; Huang, T.; Zhang, J.-B.; et al. Host-Induced Gene Silencing of an Essential Chitin Synthase Gene Confers Durable Resistance to Fusarium Head Blight and Seedling Blight in Wheat. Plant Biotechnol. J. 2015, 13, 1335–1345. [Google Scholar] [CrossRef]
- Yang, P.; Chen, Y.; Wu, H.; Fang, W.; Liang, Q.; Zheng, Y.; Olsson, S.; Zhang, D.; Zhou, J.; Wang, Z.; et al. The 5-Oxoprolinase Is Required for Conidiation, Sexual Reproduction, Virulence and Deoxynivalenol Production of Fusarium Graminearum. Curr. Genet. 2018, 64, 285–301. [Google Scholar] [CrossRef]
- Chen, X.; Pan, S.; Bai, H.; Fan, J.; Batool, W.; Shabbir, A.; Han, Y.; Zheng, H.; Lu, G.; Lin, L.; et al. A Nonclassically Secreted Effector of Magnaporthe Oryzae Targets Host Nuclei and Plays Important Roles in Fungal Growth and Plant Infection. Mol. Plant Pathol. 2023, 24, 1093–1106. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Selvaraj, P.; Lin, L.; Fang, W.; Wu, C.; Yang, P.; Zhang, J.; Abubakar, Y.S.; Yang, F.; Lu, G.; et al. Rab7/Retromer-Based Endolysosomal Trafficking Is Essential for Proper Host Invasion in Rice Blast. New Phytol. 2023, 239, 1384–1403. [Google Scholar] [CrossRef]
- Lévesque, C.A.; Brouwer, H.; Cano, L.; Hamilton, J.P.; Holt, C.; Huitema, E.; Raffaele, S.; Robideau, G.P.; Thines, M.; Win, J.; et al. Genome Sequence of the Necrotrophic Plant Pathogen Pythium Ultimum Reveals Original Pathogenicity Mechanisms and Effector Repertoire. Genome Biol. 2010, 11, R73. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, S.; Furzer, O.; Jones, J.D.G.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.D.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The Top 10 Oomycete Pathogens in Molecular Plant Pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Jiang, Y.; Peng, J.; Guo, J.; Lin, M.; Jin, C.; Huang, J.; Tang, W.; Guan, D.; He, S. The Transcriptional Reprograming and Functional Identification of WRKY Family Members in Pepper’s Response to Phytophthora Capsici Infection. BMC Plant Biol. 2020, 20, 256. [Google Scholar] [CrossRef]
- Cheng, W.; Lin, M.; Qiu, M.; Kong, L.; Xu, Y.; Li, Y.; Wang, Y.; Ye, W.; Dong, S.; He, S.; et al. Chitin Synthase Is Involved in Vegetative Growth, Asexual Reproduction and Pathogenesis of Phytophthora Capsici and Phytophthora Sojae. Environ. Microbiol. 2019, 21, 4537–4547. [Google Scholar] [CrossRef]
- Sarkar, D.; Rakshit, A. Safeguarding the Fragile Rice–Wheat Ecosystem of the Indo-Gangetic Plains through Bio-Priming and Bioaugmentation Interventions. FEMS Microbiol. Ecol. 2020, 96, fiaa221. [Google Scholar] [CrossRef]
- Nie, P.; Li, X.; Wang, S.; Guo, J.; Zhao, H.; Niu, D. Induced Systemic Resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-Dependent Signaling Pathway and Activates PAMP-Triggered Immunity in Arabidopsis. Front. Plant Sci. 2017, 8, 238. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, Y.; Gu, C.; He, C.; Yang, M.; Zhang, X.; Guo, J.; Zhao, H.; Niu, D. Bacillus cereus AR156 Activates Defense Responses to Pseudomonas Syringae Pv. Tomato in Arabidopsis Thaliana Similarly to Flg22. Mol. Plant-Microbe Interact. 2017, 31, 311–322. [Google Scholar] [CrossRef]
- Ferdous, U.T.; Shishir, M.A.; Khan, S.N.; Hoq, M.M. Bacillus spp.: Attractive Sources of Anti-Cancer and Anti-Proliferative Biomolecules. Microb. Bioact. 2018, 1, E033–E045. [Google Scholar] [CrossRef]
- Scholthof, K.-B.G. The Disease Triangle: Pathogens, the Environment and Society. Nat. Rev. Microbiol. 2007, 5, 152–156. [Google Scholar] [CrossRef]
- Leppla, N.; LeBeck, L.M. Guidelines for Purchasing and Using Commercial Natural Enemies and Biopesticides in North America. EDIS 2021, 2021, 14. [Google Scholar] [CrossRef]
- Sahib, M.R.; Yang, P.; Bokros, N.; Shapiro, N.; Woyke, T.; Kyrpides, N.C.; Xia, Y.; DeBolt, S. Improved Draft Genome Sequence of Microbacterium Sp. Strain LKL04, a Bacterial Endophyte Associated with Switchgrass Plants. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Bokros, N.; DeBolt, S.; Yang, P.; Xia, Y. Genome Sequence Resource of Bacillus sp. RRD69, a Beneficial Bacterial Endophyte Isolated from Switchgrass Plants. Mol. Plant-Microbe Interact. 2021, 34, 1320–1323. [Google Scholar] [CrossRef]
- Yang, P.; Bokros, N.; Debolt, S.; Zhao, Z.; Xia, Y. Genome Sequence Source of Bacillus Amyloliquefaciens Strain GD4a, a Bacterial Endophyte Associated with Switchgrass Plants. Phytobiomes J. 2022, 6, 354–357. [Google Scholar] [CrossRef]
- Yang, P.; Zhao, Z.; Virag, A.; Becker, T.; Zhao, L.; Liu, W.; Xia, Y. Botrytis cinerea In-Vivo Inoculation Assays for Early-, Middle- and Late-Stage Strawberries. Bio-Protocol 2023, 13, e4859. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, P.; Liu, W.; Yuan, P.; Zhao, Z.; Zhang, C.; Opiyo, S.O.; Adhikari, A.; Zhao, L.; Harsh, G.; Xia, Y. Plant Growth Promotion and Stress Tolerance Enhancement through Inoculation with Bacillus proteolyticus OSUB18. Biology 2023, 12, 1495. https://doi.org/10.3390/biology12121495
Yang P, Liu W, Yuan P, Zhao Z, Zhang C, Opiyo SO, Adhikari A, Zhao L, Harsh G, Xia Y. Plant Growth Promotion and Stress Tolerance Enhancement through Inoculation with Bacillus proteolyticus OSUB18. Biology. 2023; 12(12):1495. https://doi.org/10.3390/biology12121495
Chicago/Turabian StyleYang, Piao, Wenshan Liu, Pu Yuan, Zhenzhen Zhao, Chunquan Zhang, Stephen Obol Opiyo, Ashna Adhikari, Lijing Zhao, Garrett Harsh, and Ye Xia. 2023. "Plant Growth Promotion and Stress Tolerance Enhancement through Inoculation with Bacillus proteolyticus OSUB18" Biology 12, no. 12: 1495. https://doi.org/10.3390/biology12121495






