A Dual Long Short-Term Memory Model in Forecasting the Number of COVID-19 Infections
Abstract
:1. Introduction
2. The DULSTMGA in Forecasting Infectious Cases of COVID-19
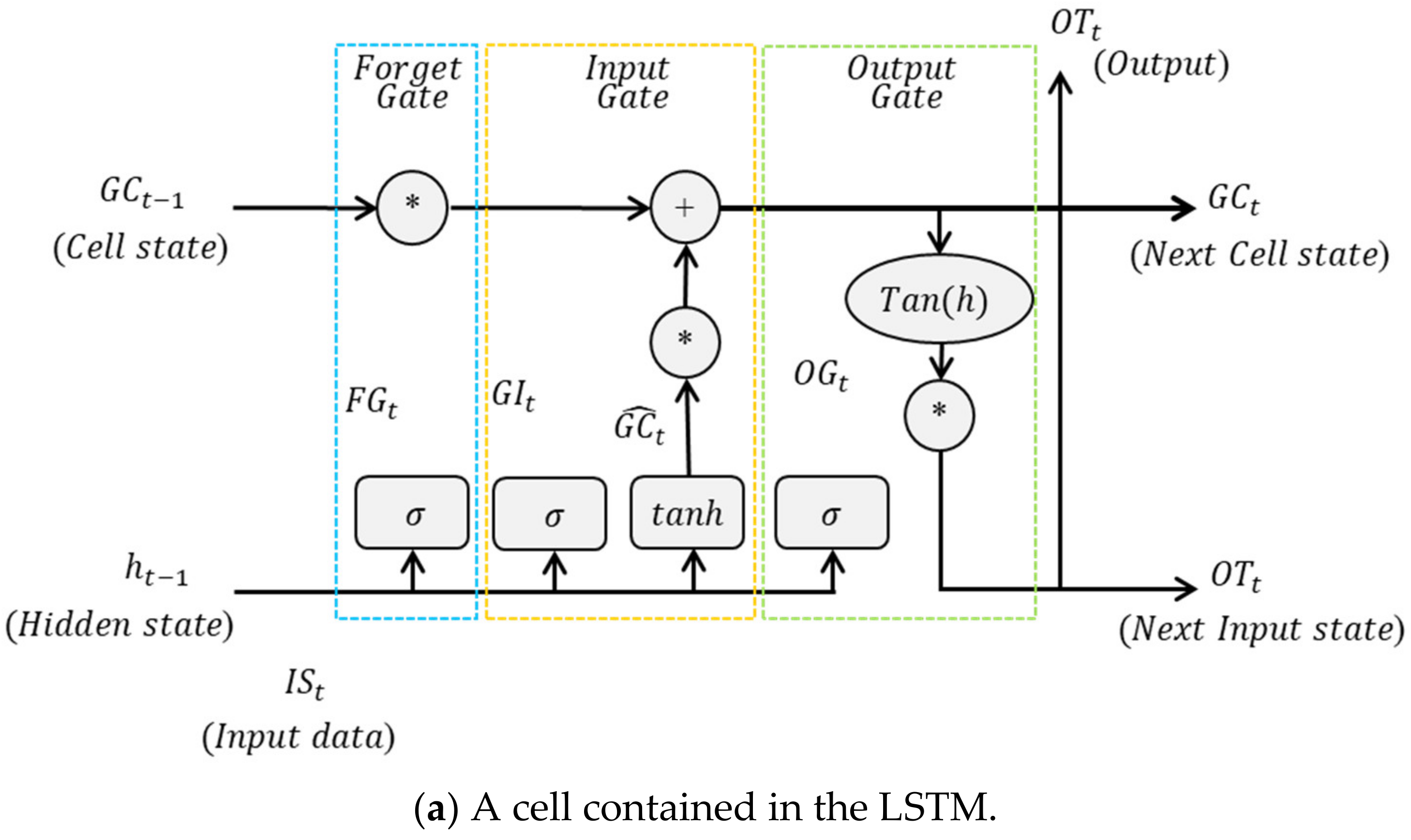
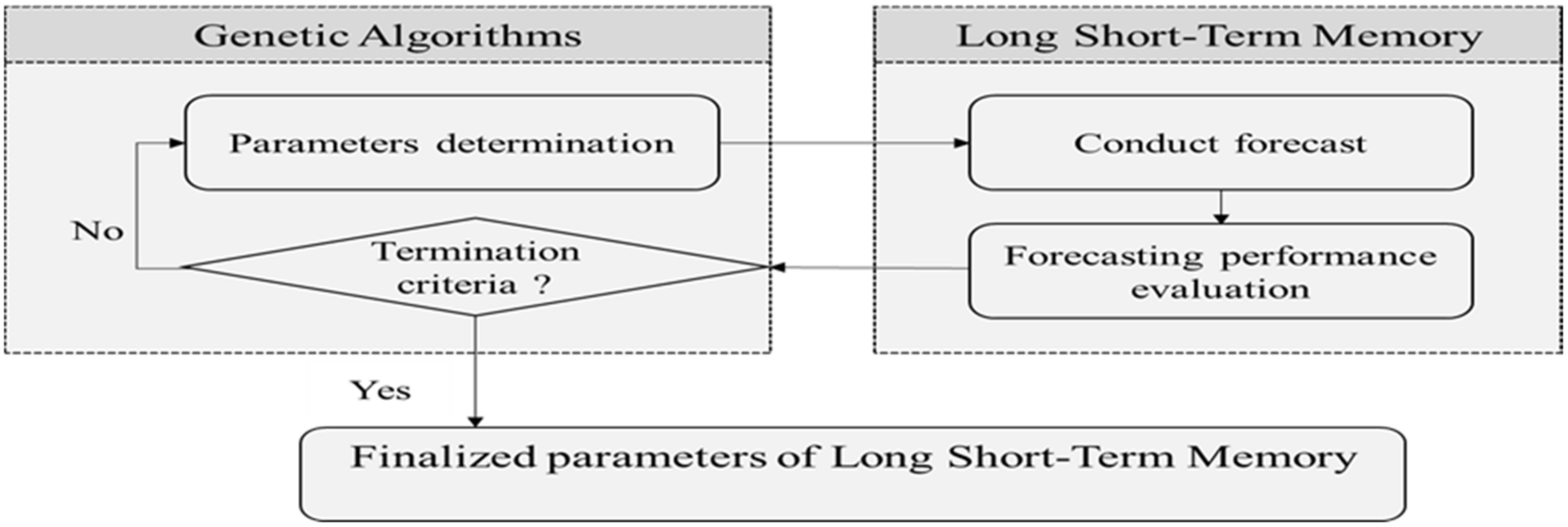
2.1. Long Short-Term Memory with Genetic Algorithms
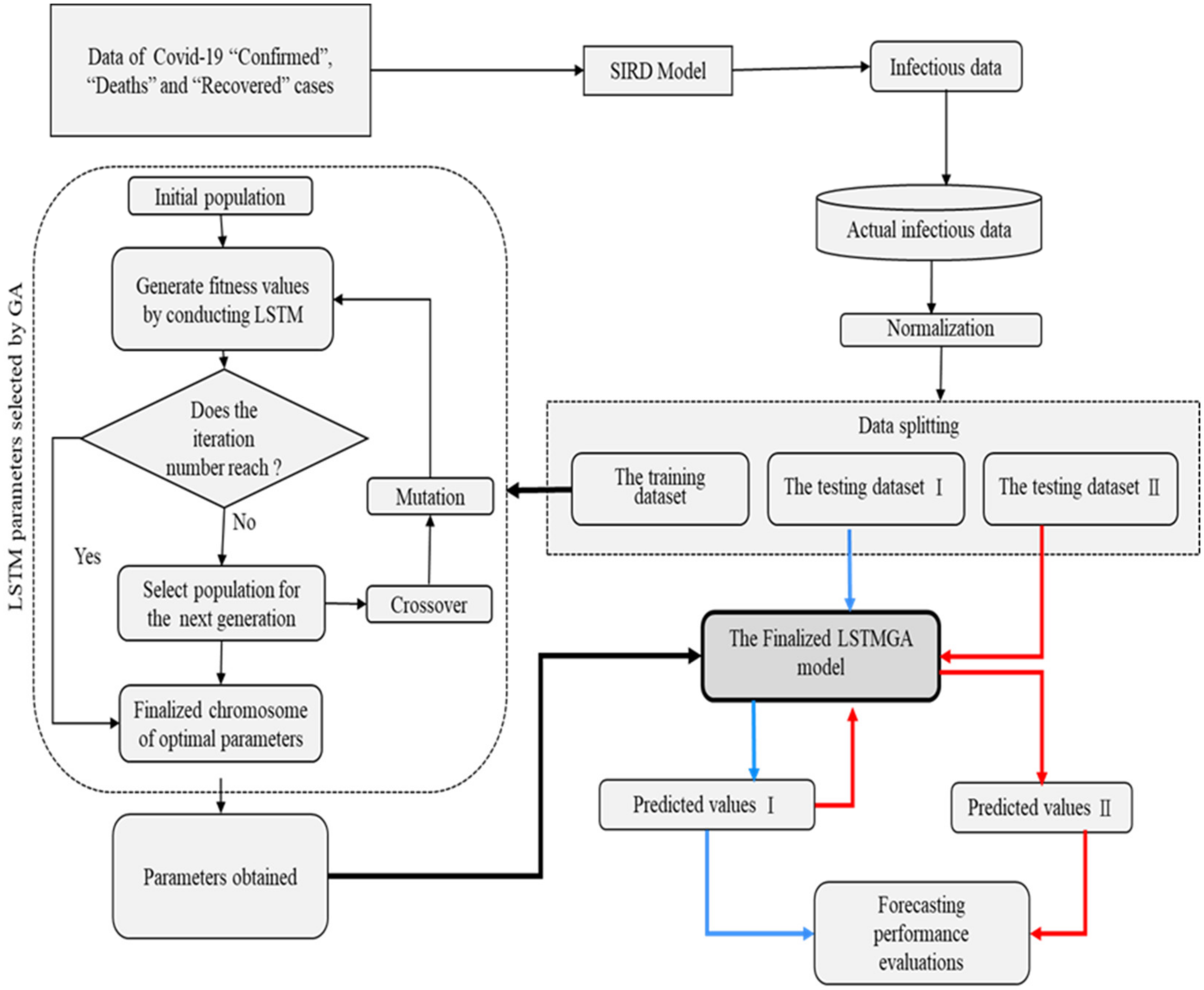
2.2. The Proposed Dual Long Short-Term Memory with Genetic Algorithms (DULSTMGA) Model
3. Numerical Examples
3.1. Forecasting of Infectious COVID-19 Cases
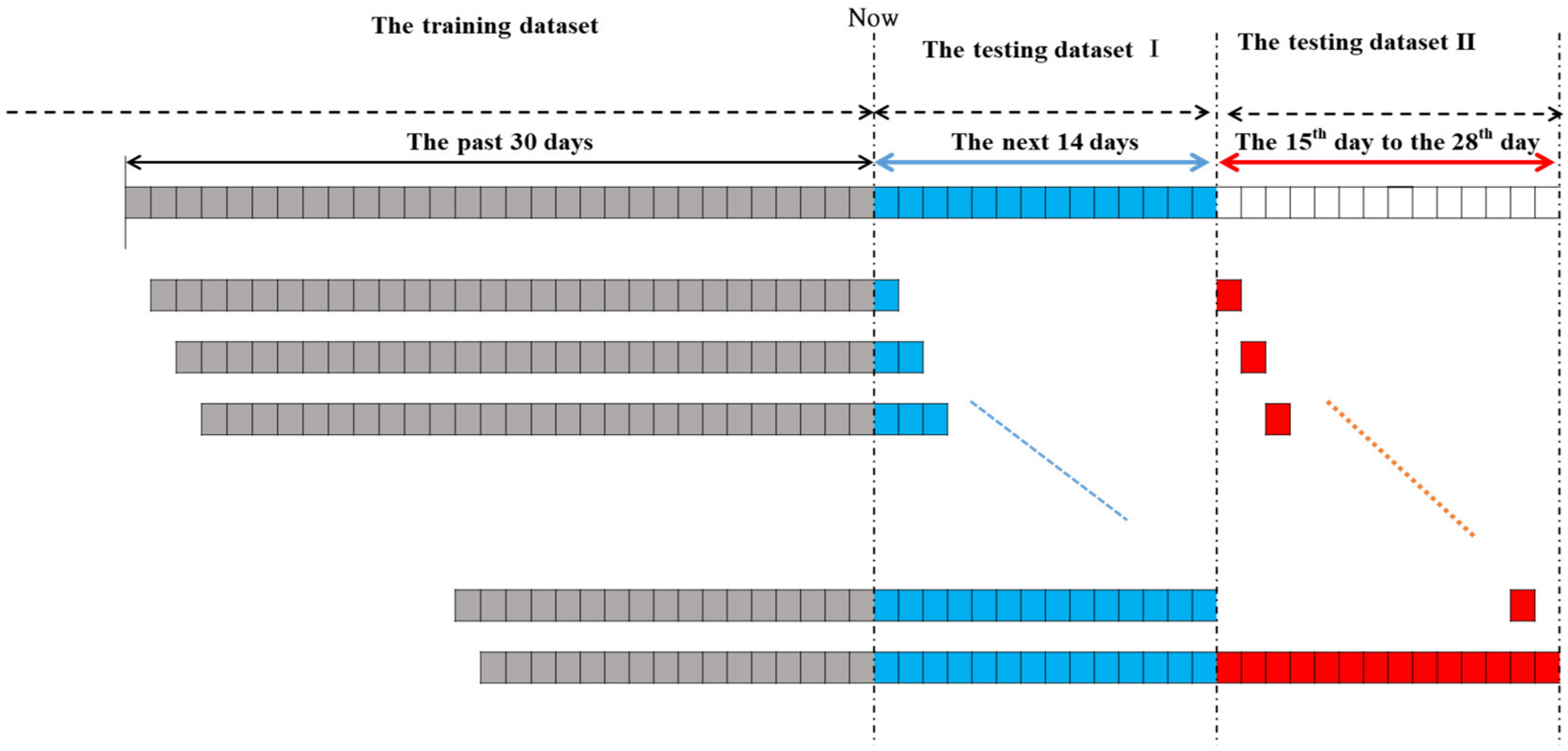
3.2. Datasets
3.3. Numerical Results
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Daley, D.J.; Gani, J. Epidemic Modelling: An Introduction; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Kermack, W.O.; McKendrick, A.G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. Ser. A Contain. Pap. A Math. Phys. Character 1927, 115, 700–721. [Google Scholar]
- Harko, T.; Lobo, F.S.; Mak, M. Exact analytical solutions of the susceptible-infected-recovered (sir) epidemic model and of the sir model with equal death and birth rates. Appl. Math. Comput. 2014, 236, 184–194. [Google Scholar] [CrossRef]
- Cruz-Mendoza, I.; Quevedo-Pulido, J.; Adanaque-Infante, L. Lstm Perfomance Analysis for Predictive Models Based on COVID-19 Dataset. In Proceedings of the 2020 IEEE XXVII International Conference on Electronics, Electrical Engineering and Computing (INTERCON), Lima, Peru, 3–5 September 2020; IEEE: New York, NY, USA, 2020; pp. 1–4. [Google Scholar]
- Shastri, S.; Singh, K.; Kumar, S.; Kour, P.; Mansotra, V. Time series forecasting of COVID-19 using deep learning models: India-USA comparative case study. Chaos Solitons Fractals 2020, 140, 110227. [Google Scholar] [CrossRef]
- Verma, H.; Mandal, S.; Gupta, A. Temporal deep learning architecture for prediction of COVID-19 cases in India. Expert Syst. Appl. 2022, 195, 116611. [Google Scholar] [CrossRef]
- Prasanth, S.; Singh, U.; Kumar, A.; Tikkiwal, V.A.; Chong, P.H. Forecasting spread of COVID-19 using google trends: A hybrid gwo-deep learning approach. Chaos Solitons Fractals 2021, 142, 110336. [Google Scholar] [CrossRef]
- Dairi, A.; Harrou, F.; Zeroual, A.; Hittawe, M.M.; Sun, Y. Comparative study of machine learning methods for COVID-19 transmission forecasting. J. Biomed. Inform. 2021, 118, 103791. [Google Scholar] [CrossRef]
- Nabi, K.N.; Tahmid, M.T.; Rafi, A.; Kader, M.E.; Haider, M.A. Forecasting COVID-19 cases: A comparative analysis between recurrent and convolutional neural networks. Results Phys. 2021, 24, 104137. [Google Scholar] [CrossRef]
- Rguibi, M.A.; Moussa, N.; Madani, A.; Aaroud, A.; Zine-Dine, K. Forecasting COVID-19 transmission with arima and lstm techniques in morocco. SN Comput. Sci. 2022, 3, 1–14. [Google Scholar] [CrossRef]
- Ketu, S.; Mishra, P.K. India perspective: Cnn-lstm hybrid deep learning model-based COVID-19 prediction and current status of medical resource availability. Soft Comput. 2022, 26, 645–664. [Google Scholar] [CrossRef]
- Jiao, F.; Huang, L.; Song, R.; Huang, H. An improved stl-lstm model for daily bus passenger flow prediction during the COVID-19 pandemic. Sensors 2021, 21, 5950. [Google Scholar] [CrossRef]
- Tuli, S.; Tuli, S.; Verma, R.; Tuli, R. Modelling for prediction of the spread and severity of COVID-19 and its association with socioeconomic factors and virus types. Biomed. Res. Clin. Rev. 2020. [Google Scholar] [CrossRef]
- Cao, R.-M.; Liu, X.F.; Xu, X.-K. Why cannot long-term cascade be predicted? Exploring temporal dynamics in information diffusion processes. R. Soc. Open Sci. 2021, 8, 202245. [Google Scholar] [CrossRef]
- Lai, J.-P. Intervention and Prediction of the Spread of COVID-19 Infectious Disease. Ph.D. Thesis, National Chi Nan University, Taiwan, China, 2021, (unpublished doctoral dissertation). Available online: https://hdl.handle.net/11296/z3gr7k (accessed on 30 May 2021).
- Okamura, H.; Osada, Y.; Nishijima, S.; Eguchi, S. Novel robust time series analysis for long-term and short-term prediction. Sci. Rep. 2021, 11, 11938. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Hua, Y.; Zhao, Z.; Li, R.; Chen, X.; Liu, Z.; Zhang, H. Deep learning with long short-term memory for time series prediction. IEEE Commun. Mag. 2019, 57, 114–119. [Google Scholar] [CrossRef]
- Zeroual, A.; Harrou, F.; Dairi, A.; Sun, Y. Deep learning methods for forecasting COVID-19 time-series data: A comparative study. Chaos Solitons Fractals 2020, 140, 110121. [Google Scholar] [CrossRef]
- Chandra, R.; Jain, A.; Singh Chauhan, D. Deep learning via lstm models for COVID-19 infection forecasting in india. PloS ONE 2022, 17, e0262708. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.H. Adaptation in Natural and Artificial Systems; The University of Michigan Press: Ann Arbor, MI, USA, 1975. [Google Scholar]
- Holland, J.H. Genetic algorithms. Sci. Am. 1992, 267, 66–73. [Google Scholar] [CrossRef]
- Armano, G.; Marchesi, M.; Murru, A. A hybrid genetic-neural architecture for stock indexes forecasting. Inf. Sci. 2005, 170, 3–33. [Google Scholar] [CrossRef]
- Kostiantis, S. Supervised machine learning: A review of classification techniques. Informatica 2007, 31, 249–268. [Google Scholar]
- Bhasin, H.; Bhatia, S. Application of genetic algorithms in machine learning. IJCSIT 2011, 2, 2412–2415. [Google Scholar]
- Bouktif, S.; Fiaz, A.; Ouni, A.; Serhani, M.A. Multi-sequence lstm-rnn deep learning and metaheuristics for electric load forecasting. Energies 2020, 13, 391. [Google Scholar] [CrossRef]
- Stajkowski, S.; Kumar, D.; Samui, P.; Bonakdari, H.; Gharabaghi, B. Genetic-algorithm-optimized sequential model for water temperature prediction. Sustainability 2020, 12, 5374. [Google Scholar] [CrossRef]
- Yu, T.; Zhu, H. Hyper-parameter optimization: A review of algorithms and applications. arXiv 2020, arXiv:2003.05689. [Google Scholar]
- Hassanat, A.; Almohammadi, K.; Alkafaween, E.a.; Abunawas, E.; Hammouri, A.; Prasath, V.S. Choosing mutation and crossover ratios for genetic algorithms—A review with a new dynamic approach. Information 2019, 10, 390. [Google Scholar] [CrossRef]
- Rudolph, G. Convergence analysis of canonical genetic algorithms. IEEE Trans. Neural Netw. 1994, 5, 96–101. [Google Scholar] [CrossRef]
- Katoch, S.; Chauhan, S.S.; Kumar, V. A review on genetic algorithm: Past, present, and future. Multimed. Tools Appl. 2021, 80, 8091–8126. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. In Proceedings of the ICLR 2015-Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Alqahtani, F.; Abotaleb, M.; Kadi, A.; Makarovskikh, T.; Potoroko, I.; Alakkari, K.; Badr, A. Hybrid deep learning algorithm for forecasting sars-cov-2 daily infections and death cases. Axioms 2022, 11, 620. [Google Scholar] [CrossRef]
- Hamed, H.; Helmy, A.; Mohammed, A. Holy quran-italian seq2seq machine translation with attention mechanism. In Proceedings of the 2022 2nd International Mobile, Intelligent, and Ubiquitous Computing Conference (MIUCC), Cairo, Egypt, 8–9 May 2022; IEEE: New York, NY, USA, 2022; pp. 11–20. [Google Scholar]
- Chen, W.; Wu, H.; Ren, S. Cm-lstm based spectrum sensing. Sensors 2022, 22, 2286. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.; Chen, Z. Dynamic graph conv-lstm model with dynamic positional encoding for the large-scale traveling salesman problem. Math. Biosci. Eng. 2022, 19, 9730–9748. [Google Scholar] [CrossRef]
- 2019. Available online: https://data.humdata.org/dataset/novel-coronavirus-2019-ncov-cases (accessed on 15 May 2021).
- Lewis, C.D. Industrial and Business Forecasting Methods: A Practical Guide to Exponential Smoothing and Curve Fitting; Butterworth-Heinemann: Oxford, UK, 1982. [Google Scholar]
- Shen, M.; Yang, J.; Li, S.; Zhang, A.; Bai, Q. Nonlinear hyperparameter optimization of a neural network in image processing for micromachines. Micromachines 2021, 12, 1504. [Google Scholar] [CrossRef] [PubMed]

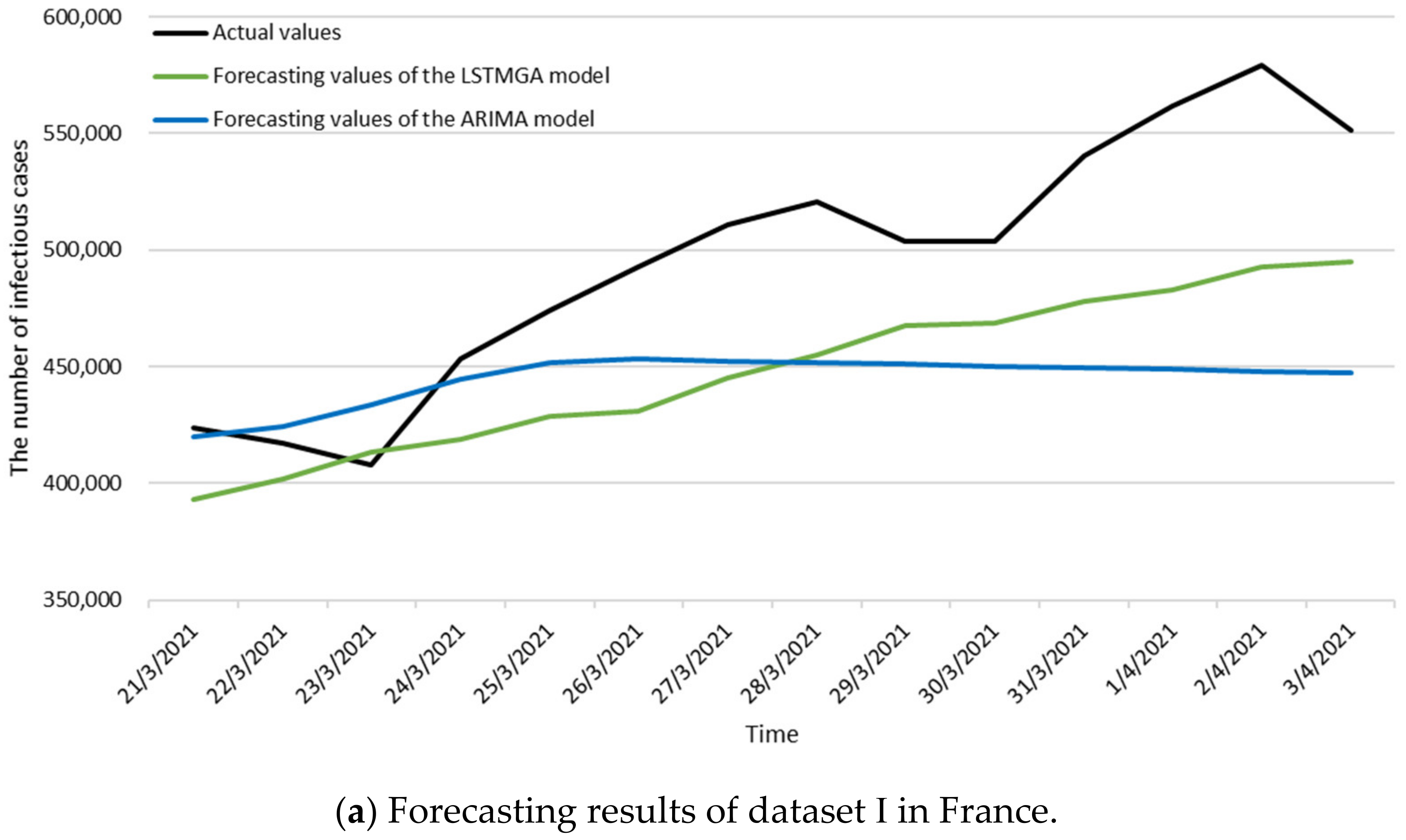
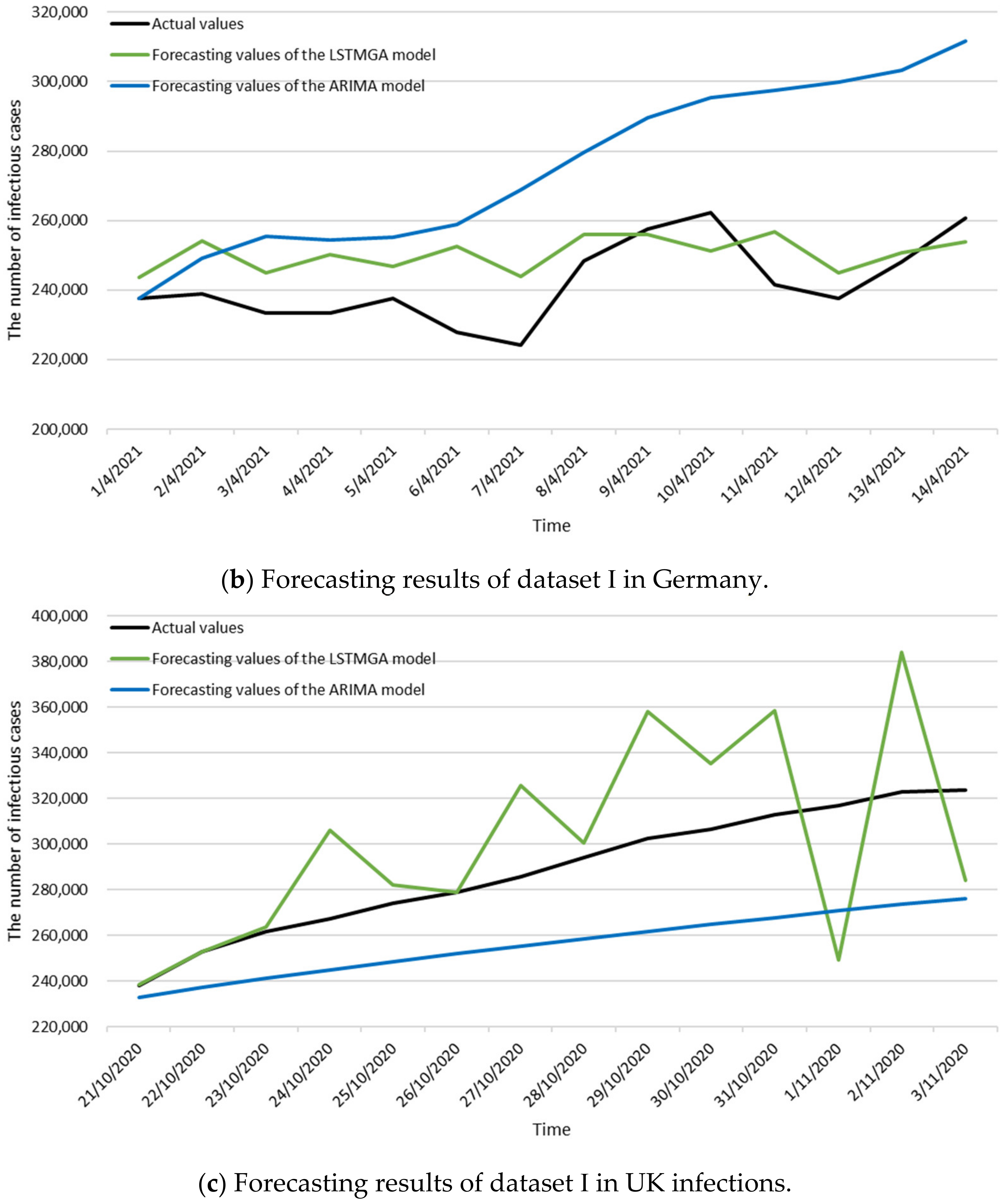
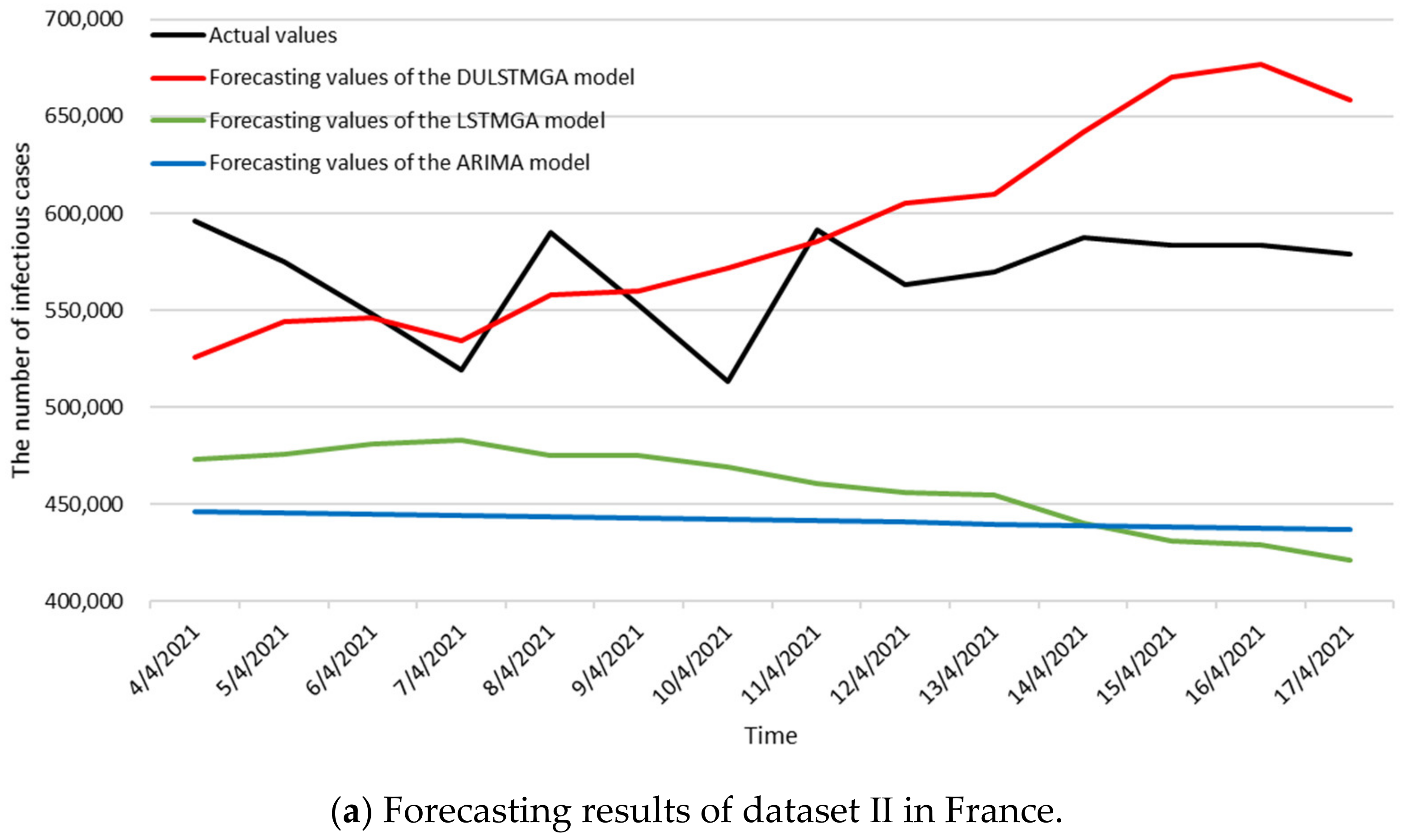
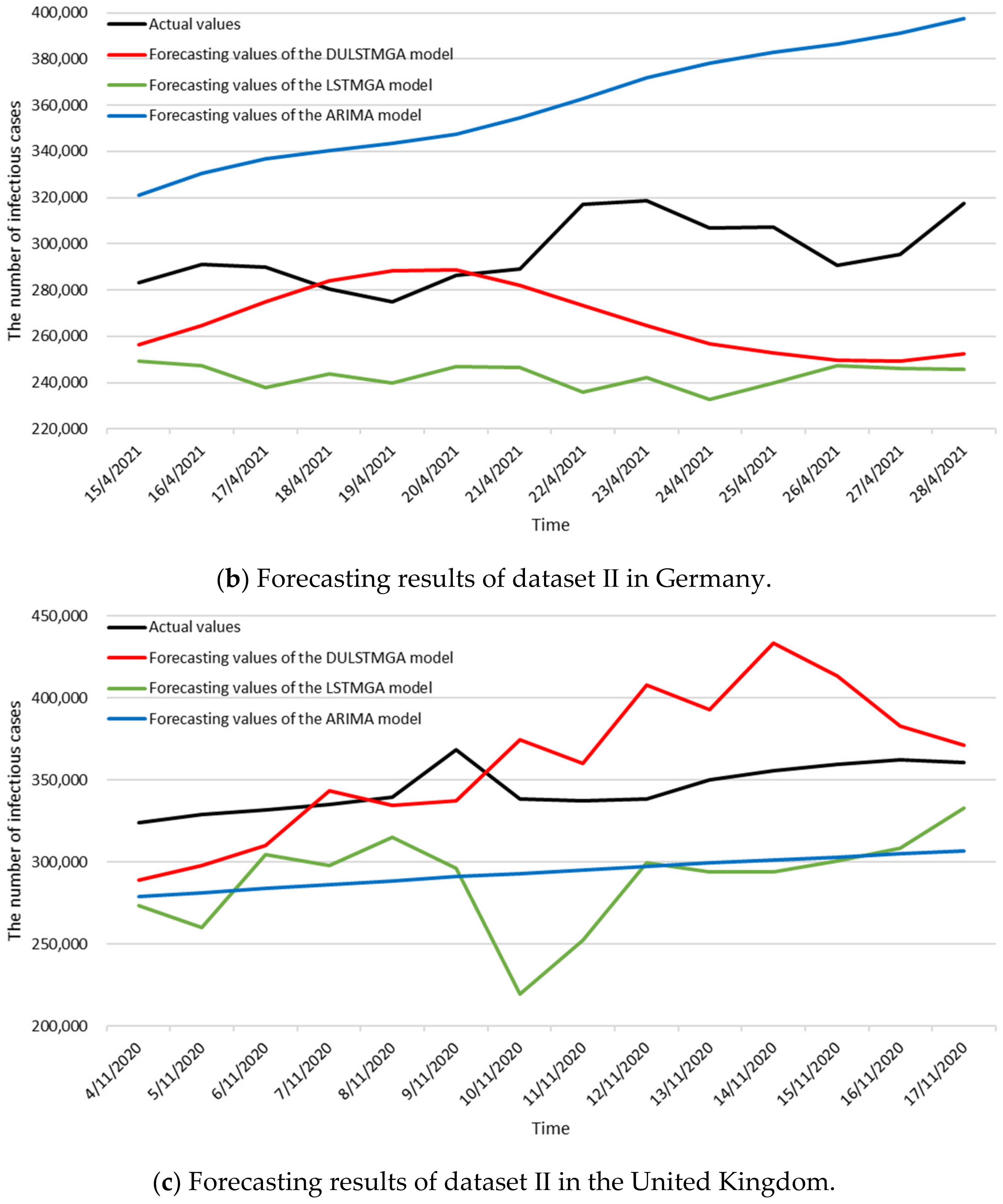
| References | Scenarios | Time Intervals of Data | MAPE |
|---|---|---|---|
| [6] | Predicting COVID-19 cases in India | 30 January 2020 to 10 July 2021 | 17.01% (confirmed cases) |
| [7] | Forecasting spread of COVID-19 | 24 February 2020 to 20 May 2020 | 8.808% (total cumulative cases) |
| [8] | Comparative study for COVID-19 transmission forecasting | 22 January to 6 September 2020 | 20.394% (averaged for confirmed and recovered cases) |
| [5] | Prediction and analysis of COVID-19 cases | 7 February 2020 to 7 July 2020 (USA, confirmed cases); 26 February 2020 to 7 July 2020 (USA, deaths) | 10.00% (confirmed cases) |
| [9] | Forecasting COVID-19 cases | 2 February 2020 to 18 November 2020 | 8.00% (deaths) |
| [10] | Forecasting COVID-19 transmission | 22 January 2020 to 22 November 2020 | 13.33% (confirmed cases) |
| [11] | COVID-19 prediction and current status of medical resource availability | 30 January to 10 June 2020 | 40.98% (confirmed cases) |
| [12] | Prediction during COVID-19 pandemic | 22 January 2020 to 21 January 2021 | 35.94% (deaths) |
| [13] | Modeling for prediction of spread and severity of COVID-19 | 7 March 2020 to 22 August 2020 | 15.7 % (confirmed cases) |
| Datasets/Countries | Forecasting Models | France | Germany | United Kingdom |
|---|---|---|---|---|
| Training data | LSTMGA DULATMGA | From 22 January 2020 to 20 March 2021 | From 22 January 2020 to 31 March 2021 | 22 January 2020 to 20 October 2020 |
| Testing dataset I (1st to 14th day) | LSTMGA | From 21 March 2021 to 3 April 2021 | From 1 April 2021 to 14 April 2021 | From 21 October 2020 to 3 November 2020 |
| Testing dataset II (15th to 28th day) | LSTMGA DULATMGA | From 4 April 2021 to 17 April 2021 | From 15 April 2021 to 28 April 2021 | From 4 November 2020 to 17 November 2020 |
| France | Germany | United Kingdom | |
|---|---|---|---|
| MAPE (%) | 9.449 | 4.692 | 9.261 |
| RMSE | 53,590 | 12,815 | 36,961 |
| MAE | 48,545 | 11,130 | 28,186 |
| Parameters of LSTMGA | |||
| Dropouts | 0.17 | 0.06 | 0.25 |
| Learning rates | 0.0082 | 0.0159 | 0.02784 |
| Batch sizes | 27 | 28 | 23 |
| French | Germany | United Kingdom | |
|---|---|---|---|
| ARIMA(p,d,q) | ARIMA(1,0,6) | ARIMA(6,2,6) | ARIMA(4,1,0) |
| MAPE (%) | 10.571 | 13.764 | 10.858 |
| RMSE | 68,543.62 | 37,880.53 | 34,821.33 |
| MAE | 55,773.42 | 33,412.59 | 32,285.87 |
| Ranges of MAPE Values (%) | Ranks |
|---|---|
| Less than 10 | Highly accurate prediction |
| From 10 to 20 | Good prediction |
| From 20 to 50 | Reasonable prediction |
| Larger than 50 | Inaccurate prediction |
| Performance Measurements | ARIMA | LSTMGA | DULSTMGA |
|---|---|---|---|
| MAPE (%) | 22.06 | 18.950 | 7.711 |
| RMSE | 128,907.91 | 115,652 | 53,090 |
| MAE | 126,251.57 | 109,020 | 44,162 |
| ARIMA(p,d,q) | ARIMA(1,0,6) | ||
| Parameters of DULSTMGA | |||
| Dropouts | 0.17 | ||
| Learning rates | 0.0082 | ||
| Batch sizes | 27 | ||
| Performance Measurements | ARIMA | LSTMGA | DULSTMGA |
|---|---|---|---|
| MAPE (%) | 21.63 | 17.797 | 10.572 |
| RMSE | 66,508.56 | 55,813 | 37,987 |
| MAE | 64,025.78 | 53,346 | 32,048 |
| ARIMA(p,d,q) | ARIMA(6,2,6) | ||
| Parameters of DULSTMGA | |||
| Dropouts | 0.06 | ||
| Learning rates | 0.0159 | ||
| Batch sizes | 28 | ||
| Performance Measurements | ARIMA | LSTMGA | DULSTMGA |
|---|---|---|---|
| MAPE (%) | 14.82 | 16.178 | 9.628 |
| RMSE | 52,054.21 | 60,994 | 39,332 |
| MAE | 51,324.39 | 55,768 | 33,297 |
| ARIMA(p,d,q) | ARIMA(4,1,0) | ||
| Parameters of DULSTMGA | |||
| Dropouts | 0.25 | ||
| Learning rates | 0.02784 | ||
| Batch sizes | 23 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, J.-P.; Pai, P.-F. A Dual Long Short-Term Memory Model in Forecasting the Number of COVID-19 Infections. Electronics 2023, 12, 759. https://doi.org/10.3390/electronics12030759
Lai J-P, Pai P-F. A Dual Long Short-Term Memory Model in Forecasting the Number of COVID-19 Infections. Electronics. 2023; 12(3):759. https://doi.org/10.3390/electronics12030759
Chicago/Turabian StyleLai, Jung-Pin, and Ping-Feng Pai. 2023. "A Dual Long Short-Term Memory Model in Forecasting the Number of COVID-19 Infections" Electronics 12, no. 3: 759. https://doi.org/10.3390/electronics12030759
APA StyleLai, J.-P., & Pai, P.-F. (2023). A Dual Long Short-Term Memory Model in Forecasting the Number of COVID-19 Infections. Electronics, 12(3), 759. https://doi.org/10.3390/electronics12030759







