Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases
Abstract
:1. Introduction
2. Characterization of H19 and Its Participation in Epigenetic Regulation
2.1. LncRNAs
2.2. H19
2.3. Regulation of H19 Expression by Epigenetic Mechanisms
2.4. H19 Regulates Gene Expression by Epigenetic Mechanisms
3. Characterization of H19 and Its Participation in Epigenetic Regulation
3.1. The Roles of H19 in Liver Development
3.2. The Roles of H19 in the Regulation of Xenobiotic Metabolism and Transport
4. The Roles of H19 in the Progression of Liver Diseases
4.1. The Roles of H19 in the Development of Steatosis, Fibrosis, and Cirrhosis
4.2. The Roles of H19 in the Regulation of Diabetes
4.3. The Roles of H19 in Hepatocellular Carcinoma
4.4. The Roles of H19 in the Epithelial-to-Mesenchymal Transition
5. Targeting H19 for Development of Therapeutic Approaches for Liver Diseases
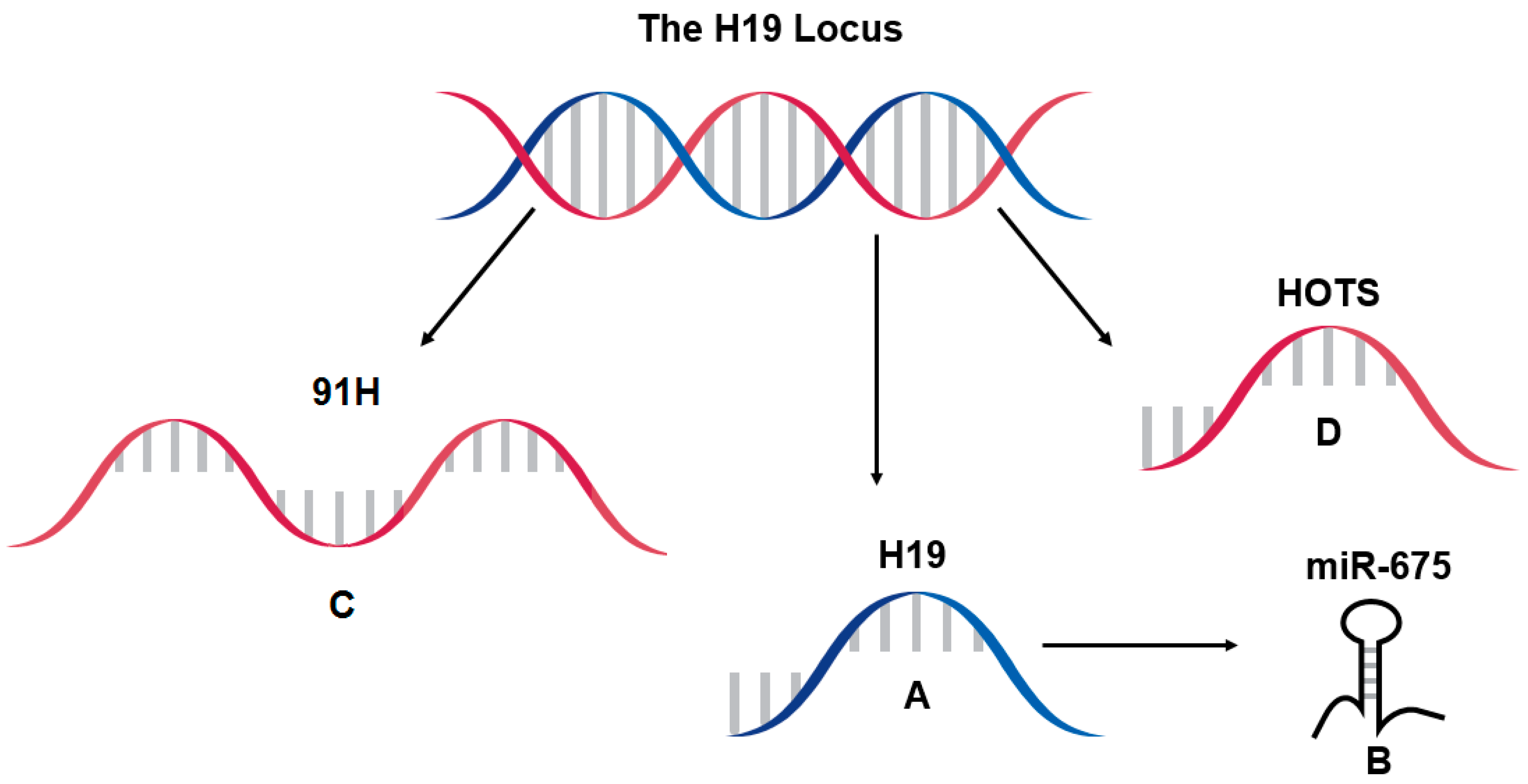
6. Further Considerations at the H19 Locus, 91H and HOTS
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BCL2 | B-cell lymphoma 2 |
| CTCF | CCCTC-binding factor |
| CDKN1C | Cyclin-dependent kinase inhibitor 1C |
| DCN | Decorin |
| DLK1 | Delta-like non-canonical notch ligand 1 |
| DM | Diabetes mellitus |
| DMR | Differentially-methylated region |
| EGR1 | Early growth response protein 1 |
| EMT | Epithelial-mesenchymal transition |
| FXR | Farnesoid X receptor |
| EZH2 | Enhancer of zeste homolog 2 |
| GSTP1 | Glutathione S-transferase pi 1 |
| GNAS | GNAS complex locus |
| H3K9 | Histone H3 at lysine 9 |
| H3K27 | Histone H3 at lysine 27 |
| HCC | Hepatocellular carcinoma |
| HDAC | Histone deacetylase |
| HnRNP U | Heterogeneous nuclear ribonucleoprotein U |
| HOTS | H19 opposite tumor suppressor |
| ICR | Imprinting control region |
| IGF2 | Insulin-like growth factor 2 |
| IGF1R | Insulin-like growth factor 1 receptor |
| IGN | Imprinted gene network |
| IVF | In vitro fertilization |
| KMT | Lysine methyltransferase |
| LncRNA | Long non-coding RNA |
| MBD1 | Methyl-CpG-binding domain protein 1 |
| MDR1 | Multidrug resistance protein 1 |
| MeCP2 | Methyl-CpG binding protein 2 |
| MEG3 | Maternally-expressed 3 |
| NAFLD | Nonalcoholic fatty liver disease |
| NASH | Nonalcoholic steatohepatitis |
| NTCP | Na+-taurocholate cotransporting polypeptide |
| P450 | Cytochrome P450 |
| PEG1 | Paternally-expressed gene 1 |
| PCAF | P300/CBP-associated factor |
| PKM2 | Pyruvate kinase isozyme M2 |
| PRC2 | Polycomb repressive complex 2 |
| SETDB1 | SET domain bifurcated 1 |
| SHP | Small heterodimer partner |
| SLC38A4 | Solute carrier family 38 member 4 |
| SUV39H1 | Suppressor of variegation 3–9 homolog 1 |
| TERT | Telomerase reverse transcriptase |
| VEGF | Vascular endothelial growth factor |
| ZEB1 | Zinc finger E-box-binding homeobox 1 |
| ZEB2 | Zinc finger E-box-binding homeobox 2 |
| ZHX2 | Zinc fingers and homeoboxes 2 |
References
- Pachnis, V.; Belayew, A.; Tilghman, S.M. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. USA 1984, 81, 5523–5527. [Google Scholar] [CrossRef] [PubMed]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Nordin, M.; Bergman, D.; Halje, M.; Engstrom, W.; Ward, A. Epigenetic regulation of the Igf2/H19 gene cluster. Cell Prolif. 2014, 47, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Gabory, A.; Ripoche, M.A.; Yoshimizu, T.; Dandolo, L. The H19 gene: Regulation and function of a non-coding RNA. Cytogenet. Genome Res. 2006, 113, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Smallwood, A.; Lamond, S.; Campbell, S.; Nargund, G. Igf2/H19 imprinting control region (ICR): An insulator or a position-dependent silencer? Sci. World J. 2001, 1, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Ishihara, K.; Kato, R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. 2000, 127, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Zhu, M.; Zhang, X.W.; Pan, Z.Y.; Gao, S.S.; Zhou, H.; Qiu, S.L.; Liang, C.Z.; Tu, J.C. The significance of long noncoding RNA H19 in predicting progression and metastasis of cancers: A meta-analysis. BioMed Res. Int. 2016, 2016, 5902678. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; Halle, D.; Raveh, E.; Gilon, M.; Sorin, V.; Hochberg, A. The role of the oncofetal H19 lncRNA in tumor metastasis: Orchestrating the emt-met decision. Oncotarget 2016, 7, 3748–3765. [Google Scholar] [PubMed]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 long non-coding RNA in cancer initiation, progression and metastasis—a proposed unifying theory. Mol. Cancer 2015, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, L.; Wei, W.; Zhang, X.; Ying, R. Long noncoding RNA H19 in digestive system cancers: A meta-analysis of its association with pathological features. BioMed Res. Int. 2016, 2016, 4863609. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; Halle, D.; Gilon, M.; Hochberg, A. The non-coding RNAs of the H19-Igf2 imprinted loci: A focus on biological roles and therapeutic potential in lung cancer. J. Transl. Med. 2015, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Wake, N.; Arima, T.; Matsuda, T. Involvement of Igf2 and H19 imprinting in choriocarcinoma development. Int. J. Gynaecol. Obstet. Off. Org. Int. Fed. Gynaecol. Obstet. 1998, 60 (Suppl. S1), S1–S8. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The gencode v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at ucsc. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Leighton, P.A.; Saam, J.R.; Ingram, R.S.; Stewart, C.L.; Tilghman, S.M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995, 9, 2079–2089. [Google Scholar] [CrossRef] [PubMed]
- Drewell, R.A.; Goddard, C.J.; Thomas, J.O.; Surani, M.A. Methylation-dependent silencing at the H19 imprinting control region by mecp2. Nucleic Acids Res. 2002, 30, 1139–1144. [Google Scholar] [CrossRef] [PubMed]
- Brunkow, M.E.; Tilghman, S.M. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 1991, 5, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Nishikawa, Y.; Tokairin, T.; Omori, Y.; Enomoto, K. Increased expression of H19 non-coding mRNA follows hepatocyte proliferation in the rat and mouse. J. Hepatol. 2004, 40, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Pfeifer, K.; Dutta, A. The H19 long noncoding RNA gives rise to microRNAs miR-675–3p and miR-675–5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014, 28, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Hibi, K.; Nakamura, H.; Hirai, A.; Fujikake, Y.; Kasai, Y.; Akiyama, S.; Ito, K.; Takagi, H. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996, 56, 480–482. [Google Scholar] [PubMed]
- Barsyte-Lovejoy, D.; Lau, S.K.; Boutros, P.C.; Khosravi, F.; Jurisica, I.; Andrulis, I.L.; Tsao, M.S.; Penn, L.Z. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006, 66, 5330–5337. [Google Scholar] [CrossRef] [PubMed]
- Yoshimizu, T.; Miroglio, A.; Ripoche, M.A.; Gabory, A.; Vernucci, M.; Riccio, A.; Colnot, S.; Godard, C.; Terris, B.; Jammes, H.; et al. The H19 locus acts in vivo as a tumor suppressor. Proc. Natl. Acad. Sci. USA 2008, 105, 12417–12422. [Google Scholar] [CrossRef] [PubMed]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. BioEssays News Rev. Mol. Cell. Dev. Biol. 2010, 32, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Onyango, P.; Feinberg, A.P. A nucleolar protein, H19 opposite tumor suppressor (hots), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc. Natl. Acad. Sci. USA 2011, 108, 16759–16764. [Google Scholar] [CrossRef] [PubMed]
- Berteaux, N.; Aptel, N.; Cathala, G.; Genton, C.; Coll, J.; Daccache, A.; Spruyt, N.; Hondermarck, H.; Dugimont, T.; Curgy, J.J.; et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal Igf2 expression. Mol. Cell. Biol. 2008, 28, 6731–6745. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, H.; Babcock, K.; Pitot, H.C. Changes in the DNA methylation profile of the rat H19 gene upstream region during development and transgenic hepatocarcinogenesis and its role in the imprinted transcriptional regulation of the H19 gene. Mol. Carcinog. 2004, 41, 1–16. [Google Scholar] [CrossRef] [PubMed]
- DeChiara, T.M.; Robertson, E.J.; Efstratiadis, A. Parental imprinting of the mouse insulin-like growth factor ii gene. Cell 1991, 64, 849–859. [Google Scholar] [CrossRef]
- Ripoche, M.A.; Kress, C.; Poirier, F.; Dandolo, L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997, 11, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Kurukuti, S.; Tiwari, V.K.; Tavoosidana, G.; Pugacheva, E.; Murrell, A.; Zhao, Z.; Lobanenkov, V.; Reik, W.; Ohlsson, R. Ctcf binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA 2006, 103, 10684–10689. [Google Scholar] [CrossRef] [PubMed]
- Murrell, A.; Heeson, S.; Reik, W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004, 36, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Ramani, K.; Mavila, N.; Ko, K.S.; Mato, J.M.; Lu, S.C. Prohibitin 1 regulates the H19-Igf2 axis and proliferation in hepatocytes. J. Biol. Chem. 2016, 291, 24148–24159. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, J.F.; Qiu, X.; Ling, J.; Chen, H.; Wang, S.; Hou, A.; Vu, T.H.; Hoffman, A.R. Ctcf regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 2008, 28, 6473–6482. [Google Scholar] [CrossRef] [PubMed]
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA controls gene expression of the imprinted gene network by recruiting mbd1. Proc. Natl. Acad. Sci. USA 2013, 110, 20693–20698. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Li, Z.; Wang, W.; Zeng, Y.; Liu, Z.; Qiu, J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting e-cadherin expression. Cancer Lett. 2013, 333, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Au, S.L.; Wong, C.C.; Lee, J.M.; Fan, D.N.; Tsang, F.H.; Ng, I.O.; Wong, C.M. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology 2012, 56, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, F.; Yuan, J.H.; Yuan, S.X.; Zhou, W.P.; Huo, X.S.; Xu, D.; Bi, H.S.; Wang, F.; Sun, S.H. Epigenetic activation of the miR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013, 34, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Tsang, W.P.; Kwok, T.T. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene 2007, 26, 4877–4881. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, Z. Cooperation of liver cells in health and disease. Adv. Anat. Embryol. Cell Biol. 2001, 161, 1–151. [Google Scholar]
- VanPutte, C.L.; Regan, J.L.; Seeley, R.R.; Russo, A. Seeley’s Anatomy and Physiology; McGraw-Hill Education: New York, NY, USA, 2013. [Google Scholar]
- Georgiades, C.S.; Neyman, E.G.; Francis, I.R.; Sneider, M.B.; Fishman, E.K. Typical and atypical presentations of extramedullary hemopoiesis. AJR. Am. J. Roentgenol. 2002, 179, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Paulson, A.; Li, H.; Piekos, S.; He, X.; Li, L.; Zhong, X.B. Developmental programming of long non-coding RNAs during postnatal liver maturation in mice. PLoS ONE 2014, 9, e114917. [Google Scholar] [CrossRef] [PubMed]
- Perincheri, S.; Dingle, R.W.; Peterson, M.L.; Spear, B.T. Hereditary persistence of alpha-fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene. Proc. Natl. Acad. Sci. USA 2005, 102, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Lui, J.C.; Finkielstain, G.P.; Barnes, K.M.; Baron, J. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R189–R196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Lodish, H.F. Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells. Blood 2004, 103, 2513–2521. [Google Scholar] [CrossRef] [PubMed]
- Sakajiri, S.; O’Kelly, J.; Yin, D.; Miller, C.W.; Hofmann, W.K.; Oshimi, K.; Shih, L.Y.; Kim, K.H.; Sul, H.S.; Jensen, C.H.; et al. Dlk1 in normal and abnormal hematopoiesis. Leukemia 2005, 19, 1404–1410. [Google Scholar] [CrossRef] [PubMed]
- Apte, U.; Zeng, G.; Thompson, M.D.; Muller, P.; Micsenyi, A.; Cieply, B.; Kaestner, K.H.; Monga, S.P. Beta-catenin is critical for early postnatal liver growth. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G1578–G1585. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, X.; Liu, Y.; Yuan, J.; Yang, F.; Huang, J.; Meng, Q.; Zhou, C.; Liu, F.; Ma, J.; et al. Long noncoding RNA H19 inhibits the proliferation of fetal liver cells and the wnt signaling pathway. FEBS Lett. 2016, 590, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gray, S.G.; Flam, F.; Pietsch, T.; Ekstrom, T.J. Developmental-dependent DNA methylation of the Igf2 and H19 promoters is correlated to the promoter activities in human liver development. Int. J. Dev. Biol. 1998, 42, 687–693. [Google Scholar] [PubMed]
- Arnaud, P. Genomic imprinting in germ cells: Imprints are under control. Reproduction 2010, 140, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Le, F.; Wang, L.Y.; Wang, N.; Li, L.; Li le, J.; Zheng, Y.M.; Lou, H.Y.; Liu, X.Z.; Xu, X.R.; Sheng, J.Z.; et al. In vitro fertilization alters growth and expression of Igf2/H19 and their epigenetic mechanisms in the liver and skeletal muscle of newborn and elder mice. Biol. Reprod. 2013, 88, 75. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes p450: A success story. Genome Biol. 2000, 1, REVIEWS3003. [Google Scholar] [CrossRef] [PubMed]
- Jancova, P.; Anzenbacher, P.; Anzenbacherova, E. Phase ii drug metabolizing enzymes. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czechoslov. 2010, 154, 103–116. [Google Scholar] [CrossRef]
- Klaassen, C.D. Xenobiotic transporters: Another protective mechanism for chemicals. Int. J. Toxicol. 2002, 21, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Bonin, S.; Pascolo, L.; Croce, L.S.; Stanta, G.; Tiribelli, C. Gene expression of abc proteins in hepatocellular carcinoma, perineoplastic tissue, and liver diseases. Mol. Med. 2002, 8, 318–325. [Google Scholar] [PubMed]
- Zheng, Z.G.; Xu, H.; Suo, S.S.; Xu, X.L.; Ni, M.W.; Gu, L.H.; Chen, W.; Wang, L.Y.; Zhao, Y.; Tian, B.; et al. The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci. Rep. 2016, 6, 26093. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.N.; Cui, Y.; Klaassen, C.D.; Zhong, X.B. Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metab. Dispos. Biol. Fate Chem. 2009, 37, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Townsend Creasy, K.; Jiang, J.; Ren, H.; Peterson, M.L.; Spear, B.T. Zinc fingers and homeoboxes 2 (Zhx2) regulates sexually dimorphic Cyp gene expression in the adult mouse liver. Gene Expr. 2016, 17, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, C.; Barbier, O.; Smalling, R.; Tsuchiya, H.; Lee, S.; Delker, D.; Zou, A.; Hagedorn, C.H.; Wang, L. Bcl2 is a critical regulator of bile acid homeostasis by dictating shp and lncRNA H19 function. Sci. Rep. 2016, 6, 20559. [Google Scholar] [CrossRef] [PubMed]
- Zollner, G.; Trauner, M. Nuclear receptors as therapeutic targets in cholestatic liver diseases. Br. J. Pharmacol. 2009, 156, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, X.; Yi, Z.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L.; Huang, F. Genome-wide analysis of long noncoding RNA expression profiles in patients with non-alcoholic fatty liver disease. IUBMB Life 2015, 67, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Boyle, S.; Varela, G.M.; Caron, E.; Yin, X.; Dhir, R.; Dhir, R.; Graham, M.J.; Ahima, R.S. Effects of perilipin 2 antisense oligonucleotide treatment on hepatic lipid metabolism and gene expression. Physiol. Genom. 2012, 44, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Tybl, E.; Shi, F.D.; Kessler, S.M.; Tierling, S.; Walter, J.; Bohle, R.M.; Wieland, S.; Zhang, J.; Tan, E.M.; Kiemer, A.K. Overexpression of the Igf2-mRNA binding protein p62 in transgenic mice induces a steatotic phenotype. J. Hepatol. 2011, 54, 994–1001. [Google Scholar] [CrossRef] [PubMed]
- Laggai, S.; Simon, Y.; Ranssweiler, T.; Kiemer, A.K.; Kessler, S.M. Rapid chromatographic method to decipher distinct alterations in lipid classes in nafld/nash. World J. Hepatol. 2013, 5, 558–567. [Google Scholar] [PubMed]
- Simon, Y.; Kessler, S.M.; Gemperlein, K.; Bohle, R.M.; Muller, R.; Haybaeck, J.; Kiemer, A.K. Elevated free cholesterol in a p62 overexpression model of non-alcoholic steatohepatitis. World J. Gastroenterol. 2014, 20, 17839–17850. [Google Scholar] [PubMed]
- Mortality, G.B.D.; Causes of Death, C. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the global burden of disease study 2013. Lancet 2015, 385, 117–171. [Google Scholar]
- Su, Z.; Zhi, X.; Zhang, Q.; Yang, L.; Xu, H.; Xu, Z. LncRNA H19 functions as a competing endogenous RNA to regulate aqp3 expression by sponging miR-874 in the intestinal barrier. FEBS Lett. 2016, 590, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Kitano, M.; Bloomston, P.M. Hepatic stellate cells and microRNAs in pathogenesis of liver fibrosis. J. Clin. Med. 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Esposti, D.D.; Hernandez-Vargas, H.; Voegele, C.; Fernandez-Jimenez, N.; Forey, N.; Bancel, B.; Le Calvez-Kelm, F.; McKay, J.; Merle, P.; Herceg, Z. Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget 2016, 7, 31862–31877. [Google Scholar] [CrossRef] [PubMed]
- Roder, P.V.; Wu, B.; Liu, Y.; Han, W. Pancreatic regulation of glucose homeostasis. Exp. Mol. Med. 2016, 48, e219. [Google Scholar] [CrossRef] [PubMed]
- Han, D.K.; Khaing, Z.Z.; Pollock, R.A.; Haudenschild, C.C.; Liau, G. H19, a marker of developmental transition, is reexpressed in human atherosclerotic plaques and is regulated by the insulin family of growth factors in cultured rabbit smooth muscle cells. J. Clin. Investig. 1996, 97, 1276–1285. [Google Scholar] [CrossRef] [PubMed]
- Leighton, P.A.; Ingram, R.S.; Eggenschwiler, J.; Efstratiadis, A.; Tilghman, S.M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 1995, 375, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.G.; Forsen, T.J.; Osmond, C.; Barker, D.J. Pathways of infant and childhood growth that lead to type 2 diabetes. Diabetes Care 2003, 26, 3006–3010. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.; Matte, A.; Perfilyev, A.; de Mello, V.D.; Kakela, P.; Pihlajamaki, J.; Ling, C. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J. Clin. Endocrinol. Metab. 2015, 100, E1491–E1501. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.; Ibanez, L.; Hattersley, A.; Tost, J. Igf2/H19 hypomethylation in a patient with very low birthweight, preocious pubarche and insulin resistance. BMC Med. Genet. 2012, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, F.; Zhou, J.; Yan, L.; Jurczak, M.J.; Lee, H.Y.; Yang, L.; Mueller, M.; Zhou, X.B.; Dandolo, L.; et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014, 42, 13799–13811. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.F.; Liu, Y.; Fu, W.M.; Xu, J.; Wang, B.; Sun, Y.X.; Wu, T.Y.; Xu, L.L.; Chan, K.M.; Zhang, J.F.; et al. Long noncoding RNA H19 accelerates tenogenic differentiation and promotes tendon healing through targeting miR-29b-3p and activating tgf-beta1 signaling. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 31, 954–964. [Google Scholar]
- Kwon, D.N.; Chang, B.S.; Kim, J.H. MicroRNA dysregulation in liver and pancreas of cmp-neu5ac hydroxylase null mice disrupts insulin/pi3k-akt signaling. BioMed Res. Int. 2014, 2014, 236385. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.L.; Wang, F.F.; Shu, J.; Tian, S.; Jiang, Y.; Zhang, D.; Wang, N.; Luo, Q.; Zhang, Y.; Jin, F.; et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 2012, 61, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA A Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Ahmed, I.; Lobo, D.N. Malignant tumours of the liver. Surgery (Oxford) 2009, 27, 30–37. [Google Scholar] [CrossRef]
- Tanaka, M.; Katayama, F.; Kato, H.; Tanaka, H.; Wang, J.; Qiao, Y.L.; Inoue, M. Hepatitis b and c virus infection and hepatocellular carcinoma in china: A review of epidemiology and control measures. J. Epidemiol. Jpn. Epidemiol. Assoc. 2011, 21, 401–416. [Google Scholar] [CrossRef]
- Vitale, A.; Gringeri, E.; Valmasoni, M.; D’Amico, F.; Carraro, A.; Pauletto, A.; D’Amico, F.J.; Polacco, M.; D’Amico, D.F.; Cillo, U. Long-term results of liver transplantation for hepatocellular carcinoma: An update of the university of padova experience. Transplant. Proc. 2007, 39, 1892–1894. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Ang, S.F.; Ng, E.S.; Li, H.; Ong, Y.H.; Choo, S.P.; Ngeow, J.; Toh, H.C.; Lim, K.H.; Yap, H.Y.; Tan, C.K.; et al. Correction: The singapore liver cancer recurrence (slicer) score for relapse prediction in patients with surgically resected hepatocellular carcinoma. PLoS ONE 2015, 10, e0128058. [Google Scholar] [CrossRef] [PubMed]
- Dugimont, T.; Montpellier, C.; Adriaenssens, E.; Lottin, S.; Dumont, L.; Iotsova, V.; Lagrou, C.; Stehelin, D.; Coll, J.; Curgy, J.J. The H19 tata-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene 1998, 16, 2395–2401. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Sohn, B.H.; Choo, J.H.; Joe, C.O.; Seong, J.K.; Lee, Y.I.; Chung, J.H. Deregulation of DNA methyltransferases and loss of parental methylation at the insulin-like growth factor ii (Igf2)/H19 loci in p53 knockout mice prior to tumor development. J. Cell. Biochem. 2005, 94, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Ariel, I.; Ayesh, S.; Perlman, E.J.; Pizov, G.; Tanos, V.; Schneider, T.; Erdmann, V.A.; Podeh, D.; Komitowski, D.; Quasem, A.S.; et al. The product of the imprinted H19 gene is an oncofetal RNA. Mol. Pathol. MP 1997, 50, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Cariani, E.; Lasserre, C.; Seurin, D.; Hamelin, B.; Kemeny, F.; Franco, D.; Czech, M.P.; Ullrich, A.; Brechot, C. Differential expression of insulin-like growth factor ii mRNA in human primary liver cancers, benign liver tumors, and liver cirrhosis. Cancer Res. 1988, 48, 6844–6849. [Google Scholar] [PubMed]
- Kim, K.S.; Lee, Y.I. Biallelic expression of the H19 and Igf2 genes in hepatocellular carcinoma. Cancer Lett. 1997, 119, 143–148. [Google Scholar] [CrossRef]
- Lv, Z.; Zhang, M.; Bi, J.; Xu, F.; Hu, S.; Wen, J. Promoter hypermethylation of a novel gene, zhx2, in hepatocellular carcinoma. Am. J. Clin. Pathol. 2006, 125, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; DeGroot, N.; Mezan, S.; Ayesh, S.; Abu-lail, R.; Hochberg, A.; Galun, E. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2007, 2, e845. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Elahi, A.; Clark, C.W.; Wang, J.; Humphries, L.A.; Centeno, B.; Bloom, G.; Fuchs, B.C.; Yeatman, T.; Shibata, D. MiR-675 mediates downregulation of twist1 and rb in afp-secreting hepatocellular carcinoma. Ann. Surg. Oncol. 2013, 20 (Suppl. S3), S625–S635. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Jia, S.; Wu, M.; An, J.; Zheng, Q.; Zhang, W.; Lu, D. Mir675 upregulates long noncoding RNA H19 through activating egr1 in human liver cancer. Oncotarget 2015, 6, 31958–31984. [Google Scholar] [PubMed]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. Cd90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- Gotink, K.J.; Verheul, H.M. Anti-angiogenic tyrosine kinase inhibitors: What is their mechanism of action? Angiogenesis 2010, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pu, H.; Zheng, Q.; Li, H.; Wu, M.; An, J.; Gui, X.; Li, T.; Lu, D. Cudr promotes liver cancer stem cell growth through upregulating tert and C-Myc. Oncotarget 2015, 6, 40775–40798. [Google Scholar] [PubMed]
- Hung, C.S.; Liu, H.H.; Liu, J.J.; Yeh, C.T.; Chang, T.C.; Wu, C.H.; Ho, Y.S.; Wei, P.L.; Chang, Y.J. MicroRNA-200a and -200b mediated hepatocellular carcinoma cell migration through the epithelial to mesenchymal transition markers. Ann. Surg. Oncol. 2013, 20 (Suppl. S3), S360–S368. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of e-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910–14914. [Google Scholar] [CrossRef] [PubMed]
- Ariel, I.; Miao, H.Q.; Ji, X.R.; Schneider, T.; Roll, D.; de Groot, N.; Hochberg, A.; Ayesh, S. Imprinted H19 oncofetal RNA is a candidate tumour marker for hepatocellular carcinoma. Mol. Pathol. MP 1998, 51, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Sorin, V.; Ohana, P.; Mizrahi, A.; Matouk, I.; Birman, T.; Hochberg, A.; Czerniak, A. Regional therapy with dta-H19 vector suppresses growth of colon adenocarcinoma metastases in the rat liver. Int. J. Oncol. 2011, 39, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; He, B.; Pan, Y.; Xu, Y.; Li, R.; Deng, Q.; Sun, H.; Wang, S. Long non-coding RNA 91H contributes to the occurrence and progression of esophageal squamous cell carcinoma by inhibiting Igf2 expression. Mol. Carcinog. 2015, 54, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; He, B.; Gao, T.; Pan, Y.; Sun, H.; Xu, Y.; Li, R.; Ying, H.; Wang, F.; Liu, X.; et al. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS ONE 2014, 9, e103022. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.K.; Lin, Q.F.; Shen, D.; Liu, Z.L.; Su, J.; Mao, W.D. Clinical implication of long noncoding RNA 91H expression profile in osteosarcoma patients. OncoTargets Ther. 2016, 9, 4645–4652. [Google Scholar]
- Vennin, C.; Spruyt, N.; Robin, Y.M.; Chassat, T.; Le Bourhis, X.; Adriaenssens, E. The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/Igf2 expression through epigenetic modifications. Cancer Lett. 2017, 385, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.G.; Court, F.; Duputie, A.; Antoine, E.; Aptel, N.; Milligan, L.; Carbonell, F.; Lelay-Taha, M.N.; Piette, J.; Weber, M.; et al. H19 antisense RNA can up-regulate Igf2 transcription by activation of a novel promoter in mouse myoblasts. PLoS ONE 2012, 7, e37923. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pope, C.; Mishra, S.; Russell, J.; Zhou, Q.; Zhong, X.-B. Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases. Diseases 2017, 5, 11. https://doi.org/10.3390/diseases5010011
Pope C, Mishra S, Russell J, Zhou Q, Zhong X-B. Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases. Diseases. 2017; 5(1):11. https://doi.org/10.3390/diseases5010011
Chicago/Turabian StylePope, Chad, Shashank Mishra, Joshua Russell, Qingqing Zhou, and Xiao-Bo Zhong. 2017. "Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases" Diseases 5, no. 1: 11. https://doi.org/10.3390/diseases5010011
APA StylePope, C., Mishra, S., Russell, J., Zhou, Q., & Zhong, X.-B. (2017). Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases. Diseases, 5(1), 11. https://doi.org/10.3390/diseases5010011





