Wheat Metabolite Interferences on Fluorescent Pseudomonas Physiology Modify Wheat Metabolome through an Ecological Feedback
Abstract
:1. Introduction
2. Results
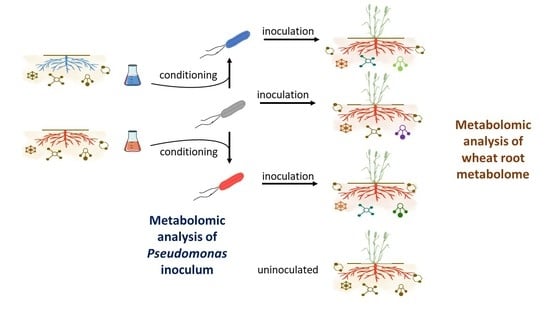
2.1. Conditioning of Pseudomonas Cells with Wheat Extracts and Evaluation of Inoculation with Conditioned and Unconditioned Bacterial Cells on Wheat Genotype Growth
2.2. Impact of the Inoculation of Conditioned and Unconditioned Bacterial Cells on Plant Growth
2.3. Impact of the Inoculation of Unconditioned Bacterial Cells on Root Metabolism of Two Wheat Genotypes
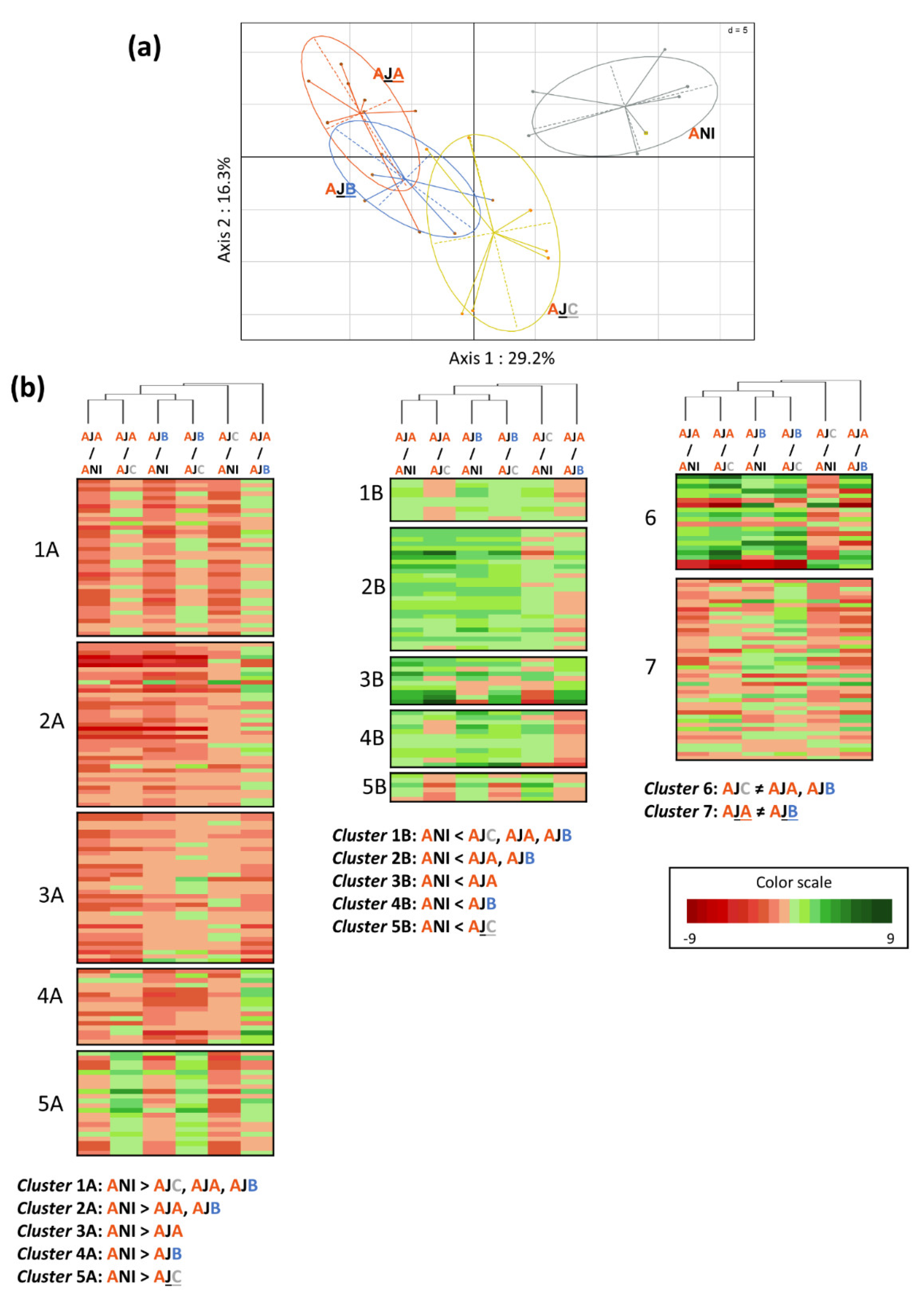
2.4. Impact of Inoculation of Conditioned Bacterial Cells on Root Metabolism of Two Wheat Genotypes
2.5. Impact of Inoculation of Conditioned Bacterial Cells on the Production of Bioactive Secondary Metabolites in the Adular Genotype
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Conditioning of Bacterial Strains before Inoculation
4.3. Bacterial Inoculation of Plants and Plant Growth
4.4. High Performance Liquid Chromatography Analysis Coupled with High Resolution Mass Spectrometry
4.5. Data Processing and Statistical Analysis
4.6. Molecular Network Analysis and Metabolite Identification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moënne-Loccoz, Y. Let the Core Microbiota Be Functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Moënne-Loccoz, Y.; Dubost, A.; Gonçalves-Martins, M.; Muller, D.; Prigent-Combaret, C. Fluorescent Pseudomonas strains with only few plant-beneficial properties are favored in the maize rhizosphere. Front. Plant Sci. 2016, 7, 1212. [Google Scholar] [CrossRef] [Green Version]
- Renoud, S.; Bouffaud, M.-L.; Dubost, A.; Prigent-Combaret, C.; Legendre, L.; Moënne-Loccoz, Y.; Muller, D. Co-Occurrence of Rhizobacteria with Nitrogen Fixation and/or 1-Aminocyclopropane-1-Carboxylate Deamination Abilities in the Maize Rhizosphere. FEMS Microbiol. Ecol. 2020, 96, fiaa062. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant Growth-Promoting Rhizobacteria and Root System Functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [Green Version]
- Besset-Manzoni, Y.; Rieusset, L.; Joly, P.; Comte, G.; Prigent-Combaret, C. Exploiting Rhizosphere Microbial Cooperation for Developing Sustainable Agriculture Strategies. Environ. Sci. Pollut. Res. 2018, 25, 29953–29970. [Google Scholar] [CrossRef]
- Venturi, V.; Keel, C. Signaling in the Rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef]
- Besset-Manzoni, Y.; Joly, P.; Brutel, A.; Gerin, F.; Soudière, O.; Langin, T.; Prigent-Combaret, C. Does in Vitro Selection of Biocontrol Agents Guarantee Success in Planta? A Study Case of Wheat Protection against Fusarium Seedling Blight by Soil Bacteria. PLoS ONE 2019, 14, e0225655. [Google Scholar] [CrossRef] [Green Version]
- Rieusset, L.; Rey, M.; Gerin, F.; Wisniewski-Dyé, F.; Prigent-Combaret, C.; Comte, G. A Cross-Metabolomic Approach Shows That Wheat Interferes with Fluorescent Pseudomonas Physiology through Its Root Metabolites. Metabolites 2021, 11, 84. [Google Scholar] [CrossRef]
- Mathesius, U.; Mulders, S.; Gao, M.; Teplitski, M.; Caetano-Anolles, G.; Rolfe, B.G.; Bauer, W.D. Extensive and Specific Responses of a Eukaryote to Bacterial Quorum-Sensing Signals. Proc. Natl. Acad. Sci. USA 2003, 100, 1444–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, D.W.; Gong, F.; Daykin, M.M.; Williams, P.; Pierson, L.S. N-Acyl-Homoserine Lactone-Mediated Regulation of Phenazine Gene Expression by Pseudomonas Aureofaciens 30–84 in the Wheat Rhizosphere. J. Bacteriol. 1997, 179, 7663–7670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, A. Quorum sensing N-acyl-homoserine lactone signal molecules of plant beneficial Gram-negative rhizobacteria support plant growth and resistance to pathogens. Rhizosphere 2020, 16, 100258. [Google Scholar] [CrossRef]
- Moshynets, O.V.; Babenko, L.M.; Rogalsky, S.P.; Iungin, O.S.; Foster, J.; Kosakivska, I.V.; Potters, G.; Spiers, A.J. Priming winter wheat seeds with the bacterial quorum sensing signal N-hexanoyl-L-homoserine lactone (C6-HSL) shows potential to improve plant growth and seed yield. PLoS ONE 2019, 14, e0209460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morohoshi, T.; Yamaguchi, T.; Xie, X.; Wang, W.; Takeuchi, K.; Someya, N. Complete Genome Sequence of Pseudomonas Chlororaphis Subsp. Aurantiaca Reveals a Triplicate Quorum-Sensing Mechanism for Regulation of Phenazine Production. Microbes Environ. 2017, 32, 47–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price-Whelan, A.; Dietrich, L.E.P.; Newman, D.K. Rethinking “secondary” Metabolism: Physiological Roles for Phenazine Antibiotics. Nat. Chem. Biol. 2006, 2, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Pierson, L.S.; Pierson, E.A. Metabolism and Function of Phenazines in Bacteria: Impacts on the Behavior of Bacteria in the Environment and Biotechnological Processes. Appl. Microbiol. Biotechnol. 2010, 86, 1659–1670. [Google Scholar] [CrossRef] [Green Version]
- Brazelton, J.N.; Pfeufer, E.E.; Sweat, T.A.; Gardener, B.B.M.; Coenen, C. 2,4-Diacetylphloroglucinol Alters Plant Root Development. Mol. Plant-Microbe Interact. 2008, 21, 1349–1358. [Google Scholar] [CrossRef] [Green Version]
- Vacheron, J.; Desbrosses, G.; Renoud, S.; Padilla, R.; Walker, V.; Muller, D.; Prigent-Combaret, C. Differential Contribution of Plant-Beneficial Functions from Pseudomonas Kilonensis F113 to Root System Architecture Alterations in Arabidopsis Thaliana and Zea Mays. Mol. Plant-Microbe Interact. 2018, 31, 212–223. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, T.R.; Yu, J.M.; Liu, S.; Pierson, L.S.; Pierson, E.A. Drought-Stress Tolerance in Wheat Seedlings Conferred by Phenazine-Producing Rhizobacteria. Front. Microbiol. 2019, 10, 1590. [Google Scholar] [CrossRef] [Green Version]
- Schenk, P.M.; Carvalhais, L.C.; Kazan, K. Unraveling Plant–Microbe Interactions: Can Multi-Species Transcriptomics Help? Trends Biotechnol. 2012, 30, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Valente, J.; Gerin, F.; Le Gouis, J.; Moënne-Loccoz, Y.; Prigent–Combaret, C. Ancient Wheat Varieties Have a Higher Ability to Interact with Plant Growth-promoting Rhizobacteria. Plant Cell Environ. 2020, 43, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Souard, F.; Delporte, C.; Stoffelen, P.; Thévenot, E.A.; Noret, N.; Dauvergne, B.; Kauffmann, J.-M.; Van Antwerpen, P.; Stévigny, C. Metabolomics Fingerprint of Coffee Species Determined by Untargeted-Profiling Study Using LC-HRMS. Food Chem. 2018, 245, 603–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinelli, G.; Segura-Carretero, A.; Di Silvestro, R.; Marotti, I.; Arráez-Román, D.; Benedettelli, S.; Ghiselli, L.; Fernadez-Gutierrez, A. Profiles of Phenolic Compounds in Modern and Old Common Wheat Varieties Determined by Liquid Chromatography Coupled with Time-of-Flight Mass Spectrometry. J. Chromatogr. A 2011, 1218, 7670–7681. [Google Scholar] [CrossRef]
- Rieusset, L.; Rey, M.; Muller, D.; Vacheron, J.; Gerin, F.; Dubost, A.; Comte, G.; Prigent-Combaret, C. Secondary Metabolites from Plant-associated Pseudomonas Are Overproduced in Biofilm. Microb. Biotechnol. 2020, 13, 1562–1580. [Google Scholar] [CrossRef]
- Almario, J.; Bruto, M.; Vacheron, J.; Prigent-Combaret, C.; Moënne-Loccoz, Y.; Muller, D. Distribution of 2,4-Diacetylphloroglucinol Biosynthetic Genes among the Pseudomonas spp. Reveals Unexpected Polyphyletism. Front. Microbiol. 2017, 8, 1218. [Google Scholar] [CrossRef] [Green Version]
- Walker, V.; Bertrand, C.; Bellvert, F.; Moënne-Loccoz, Y.; Bally, R.; Comte, G. Host Plant Secondary Metabolite Profiling Shows a Complex, Strain-dependent Response of Maize to Plant Growth-promoting Rhizobacteria of the Genus Azospirillum. New Phytol. 2011, 189, 494–506. [Google Scholar] [CrossRef]
- Chamam, A.; Sanguin, H.; Bellvert, F.; Meiffren, G.; Comte, G.; Wisniewski-Dyé, F.; Bertrand, C.; Prigent-Combaret, C. Plant Secondary Metabolite Profiling Evidences Strain-Dependent Effect in the Azospirillum–Oryza Sativa Association. Phytochemistry 2013, 87, 65–77. [Google Scholar] [CrossRef]
- Drogue, B.; Sanguin, H.; Borland, S.; Prigent-Combaret, C.; Wisniewski-Dyé, F. Genome Wide Profiling of Azospirillum Lipoferum 4B Gene Expression during Interaction with Rice Roots. FEMS Microbiol. Ecol. 2014, 87, 543–555. [Google Scholar] [CrossRef] [Green Version]
- Valette, M.; Rey, M.; Gerin, F.; Comte, G.; Wisniewski-Dyé, F. A Common Metabolomic Signature Is Observed upon Inoculation of Rice Roots with Various Rhizobacteria. J. Integr. Plant Biol. 2020, 62, 228–246. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Daniels, R.; Vanderleyden, J.; Michiels, J. Quorum Sensing and Swarming Migration in Bacteria. FEMS Microbiol. Rev. 2004, 28, 261–289. [Google Scholar] [CrossRef] [PubMed]
- Elias, S.; Banin, E. Multi-Species Biofilms: Living with Friendly Neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Ferreira, N.; Hayashi Sant’ Anna, F.; Massena Reis, V.; Ambrosini, A.; Gazolla Volpiano, C.; Rothballer, M.; Schwab, S.; Baura, V.A.; Balsanelli, E.; Pedrosa, F.d.O.; et al. Genome-Based Reclassification of Azospirillum Brasilense Sp245 as the Type Strain of Azospirillum Baldaniorum sp. Nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 6203–6212. [Google Scholar] [CrossRef] [PubMed]
- Couillerot, O.; Combes-Meynet, E.; Pothier, J.F.; Bellvert, F.; Challita, E.; Poirier, M.-A.; Rohr, R.; Comte, G.; Moënne-Loccoz, Y.; Prigent-Combaret, C. The Role of the Antimicrobial Compound 2,4-Diacetylphloroglucinol in the Impact of Biocontrol Pseudomonas Fluorescens F113 on Azospirillum Brasilense Phytostimulators. Microbiology 2011, 157, 1694–1705. [Google Scholar] [CrossRef] [Green Version]
- Combes-Meynet, E.; Pothier, J.F.; Moënne-Loccoz, Y.; Prigent-Combaret, C. The Pseudomonas Secondary Metabolite 2,4-Diacetylphloroglucinol Is a Signal Inducing Rhizoplane Expression of Azospirillum Genes Involved in Plant-Growth Promotion. Mol. Plant-Microbe Interact. 2011, 24, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Rozier, C. Xylem Sap Metabolite Profile Changes During Phytostimulation of Maize by the Plant Growth-Promoting Rhizobacterium, Azospirillum Lipoferum CRT1. Mol. Biol. 2016, 6, 182. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Haig, T.; Pratley, J.; Lemerle, D.; An, M. Allelochemicals in Wheat (Triticum Aestivum L.): Cultivar Difference in the Exudation of Phenolic Acids. J. Agric. Food Chem. 2001, 49, 3742–3745. [Google Scholar] [CrossRef]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin Biosynthesis and Its Integration into Metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Macoy, D.M.; Kim, W.-Y.; Lee, S.Y.; Kim, M.G. Biosynthesis, Physiology, and Functions of Hydroxycinnamic Acid Amides in Plants. Plant Biotechnol. Rep. 2015, 9, 269–278. [Google Scholar] [CrossRef]
- Häusler, R.E.; Ludewig, F.; Krueger, S. Amino Acids—A Life between Metabolism and Signaling. Plant Sci. 2014, 229, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Yoshida, M.; Nakajima, T.; Murai, A. Accumulation of Hydroxycinnamic Acid Amides in Winter Wheat under Snow. Biosci. Biotechnol. Biochem. 2003, 67, 1245–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zacarés, L.; López-Gresa, M.P.; Fayos, J.; Primo, J.; Bellés, J.M.; Conejero, V. Induction of p-Coumaroyldopamine and Feruloyldopamine, Two Novel Metabolites, in Tomato by the Bacterial Pathogen Pseudomonas Syringae. Mol. Plant-Microbe Interact. 2007, 20, 1439–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ube, N.; Harada, D.; Katsuyama, Y.; Osaki-Oka, K.; Tonooka, T.; Ueno, K.; Taketa, S.; Ishihara, A. Identification of Phenylamide Phytoalexins and Characterization of Inducible Phenylamide Metabolism in Wheat. Phytochemistry 2019, 167, 112098. [Google Scholar] [CrossRef]
- Weston, L.A.; Mathesius, U. Flavonoids: Their Structure, Biosynthesis and Role in the Rhizosphere, Including Allelopathy. J. Chem. Ecol. 2013, 39, 283–297. [Google Scholar] [CrossRef]
- Wojakowska, A.; Perkowski, J.; Góral, T.; Stobiecki, M. Structural Characterization of Flavonoid Glycosides from Leaves of Wheat (Triticum Aestivum L.) Using LC/MS/MS Profiling of the Target Compounds: Secondary Metabolite Profiles in Wheat Leaves. J. Mass Spectrom. 2013, 48, 329–339. [Google Scholar] [CrossRef]
- Shaw, L.J.; Morris, P.; Hooker, J.E. Perception and Modification of Plant Flavonoid Signals by Rhizosphere Microorganisms. Environ. Microbiol. 2006, 8, 1867–1880. [Google Scholar] [CrossRef]
- Webster, G.; Jain, V.; Davey, M.R.; Gough, C.; Vasse, J.; Denarie, J.; Cocking, E.C. The Flavonoid Naringenin Stimulates the Intercellular Colonization of Wheat Roots by Azorhizobium Caulinodans. Plant Cell Environ. 1998, 21, 373–383. [Google Scholar] [CrossRef]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stévigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The Flavanone Naringenin Reduces the Production of Quorum Sensing-Controlled Virulence Factors in Pseudomonas aeruginosa PAO1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef] [Green Version]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum Quenching: Role in Nature and Applied Developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef]
- Vanholme, B.; El Houari, I.; Boerjan, W. Bioactivity: Phenylpropanoids’ Best Kept Secret. Curr. Opin. Biotechnol. 2019, 56, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Cotton, T.E.A.; Pétriacq, P.; Cameron, D.D.; Meselmani, M.A.; Schwarzenbacher, R.; Rolfe, S.A.; Ton, J. Metabolic Regulation of the Maize Rhizobiome by Benzoxazinoids. ISME J. 2019, 13, 1647–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Richter, A.; Jander, G. Beyond Defense: Multiple Functions of Benzoxazinoids in Maize Metabolism. Plant Cell Physiol. 2018, 59, 1528–1537. [Google Scholar] [CrossRef] [PubMed]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in Root Exudates of Maize Attract Pseudomonas Putida to the Rhizosphere. PLoS ONE 2012, 7, e35498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glauser, G.; Marti, G.; Villard, N.; Doyen, G.A.; Wolfender, J.-L.; Turlings, T.C.J.; Erb, M. Induction and Detoxification of Maize 1,4-Benzoxazin-3-Ones by Insect Herbivores: Defense Induction and Detoxification in Maize. Plant J. 2011, 68, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Schlaeppi, K.; Gross, J.J.; Hapfelmeier, S.; Erb, M. Plant Chemistry and Food Web Health. New Phytol. 2021, 231, 957–962. [Google Scholar] [CrossRef]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root Exudate Metabolites Drive Plant-Soil Feedbacks on Growth and Defense by Shaping the Rhizosphere Microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef] [Green Version]
- Marti, G.; Erb, M.; Boccard, J.; Glauser, G.; Doyen, G.R.; Villard, N.; Robert, C.A.M.; Turlings, T.C.J.; Rudaz, S.; Wolfender, J.-L. Metabolomics Reveals Herbivore-Induced Metabolites of Resistance and Susceptibility in Maize Leaves and Roots: Plant-Insect Metabolomics. Plant Cell Environ. 2013, 36, 621–639. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Koprivova, A.; Kopriva, S. Pinpointing Secondary Metabolites That Shape the Composition and Function of the Plant Microbiome. J. Exp. Bot. 2021, 72, 57–69. [Google Scholar] [CrossRef]
- Neal, A.; Ton, J. Systemic Defense Priming by Pseudomonas Putida KT2440 in Maize Depends on Benzoxazinoid Exudation from the Roots. Plant Signal. Behav. 2013, 8, e22655. [Google Scholar] [CrossRef] [Green Version]
- Wouters, F.C.; Gershenzon, J.; Vassão, D.G. Benzoxazinoids: Reactivity and Modes of Action of a Versatile Class of Plant Chemical Defenses. J. Braz. Chem. Soc. 2016, 27, 1379–1397. [Google Scholar] [CrossRef]
- Shanahan, P.; O’Sullivan, D.J.; Simpson, P.; Glennon, J.D.; O’Gara, F. Isolation of 2,4-Diacetylphloroglucinol from a Fluorescent Pseudomonad and Investigation of Physiological Parameters Influencing Its Production. Appl. Environ. Microbiol. 1992, 58, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- De Mendiburu, F.; Simon, R. Agricolae—Ten Years of an Open Source Statistical Tool for Experiments in Breeding, Agriculture and Biology. PeerJ PrePrints 2015, 3, e1404v1. [Google Scholar] [CrossRef]
- Thioulouse, J.; Chessel, D.; Dec, S.D.; Olivier, J.-M. ADE-4: A Multivariate Analysis and Graphical Display Software. Stat. Comput. 1997, 7, 75–83. [Google Scholar] [CrossRef]
- Caraux, G.; Pinloche, S. PermutMatrix: A Graphical Environment to Arrange Gene Expression Profiles in Optimal Linear Order. Bioinformatics 2005, 21, 1280–1281. [Google Scholar] [CrossRef] [Green Version]
- Olivon, F.; Elie, N.; Grelier, G.; Roussi, F.; Litaudon, M.; Touboul, D. MetGem Software for the Generation of Molecular Networks Based on the T-SNE Algorithm. Anal. Chem. 2018, 90, 13900–13908. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rieusset, L.; Rey, M.; Wisniewski-Dyé, F.; Prigent-Combaret, C.; Comte, G. Wheat Metabolite Interferences on Fluorescent Pseudomonas Physiology Modify Wheat Metabolome through an Ecological Feedback. Metabolites 2022, 12, 236. https://doi.org/10.3390/metabo12030236
Rieusset L, Rey M, Wisniewski-Dyé F, Prigent-Combaret C, Comte G. Wheat Metabolite Interferences on Fluorescent Pseudomonas Physiology Modify Wheat Metabolome through an Ecological Feedback. Metabolites. 2022; 12(3):236. https://doi.org/10.3390/metabo12030236
Chicago/Turabian StyleRieusset, Laura, Marjolaine Rey, Florence Wisniewski-Dyé, Claire Prigent-Combaret, and Gilles Comte. 2022. "Wheat Metabolite Interferences on Fluorescent Pseudomonas Physiology Modify Wheat Metabolome through an Ecological Feedback" Metabolites 12, no. 3: 236. https://doi.org/10.3390/metabo12030236