Biomarker Research in Parkinson’s Disease Using Metabolite Profiling
Abstract
:1. Introduction
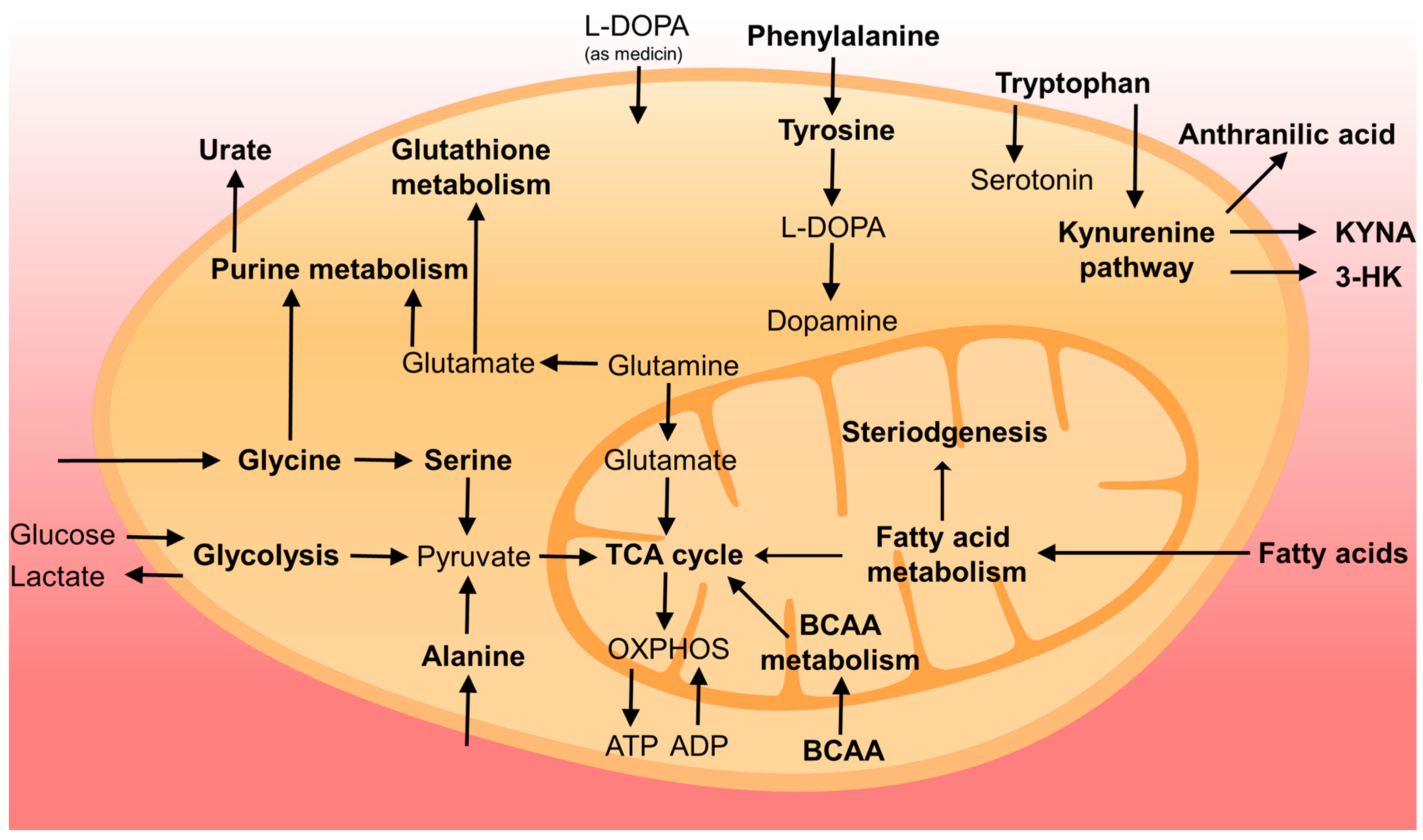
2. Metabolites as Biomarkers
3. Metabolomics in PD
3.1. Analytical Challenges to Assess the “Metabolome”
3.2. Sample Preparation, Extraction Procedures and Analytical Platforms
3.3. Identification and Quantification of Small Molecules
3.4. Untargeted versus Targeted Analysis at Acquisition or Data Analysis Levels
3.5. Statistical Analysis
4. Metabolomics Studies in PD Patients and Experimental Models
4.1. Metabolomic Studies in PD Patients (Table 1)
4.2. Metabolomic Studies in Experimental Models of PD (Table 2)
5. General Discussion
6. Conclusions and Future Perspectives
Acknowledgments
Conflicts of Interest
References
- De Lau, L.M.; Breteler, M.M. Epidemiology of parkinson’s disease. Lancet. Neurol. 2006, 5, 525–535. [Google Scholar] [CrossRef]
- Adler, C.H.; Beach, T.G.; Hentz, J.G.; Shill, H.A.; Caviness, J.N.; Driver-Dunckley, E.; Sabbagh, M.N.; Sue, L.I.; Jacobson, S.A.; Belden, C.M.; et al. Low clinical diagnostic accuracy of early vs. advanced Parkinson disease: Clinicopathologic study. Neurology 2014, 83, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Marek, K.; Innis, R.; van Dyck, C.; Fussell, B.; Early, M.; Eberly, S.; Oakes, D.; Seibyl, J. [123I] β-CIT SPECT imaging assessment of the rate of Parkinson’s disease progression. Neurology 2001, 57, 2089–2094. [Google Scholar] [CrossRef] [PubMed]
- Morrish, P.K.; Sawle, G.V.; Brooks, D.J. Clinical and [18F] dopa PET findings in early Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 1995, 59, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, J.M.; Lees, A.J. Ageing and Parkinson’s disease: Substantia nigra regional selectivity. Brain 1991, 114, 2283–2301. [Google Scholar] [CrossRef] [PubMed]
- Berg, D.; Postuma, R.B.; Adler, C.H.; Bloem, B.R.; Chan, P.; Dubois, B.; Gasser, T.; Goetz, C.G.; Halliday, G.; Joseph, L.; et al. MDS research criteria for prodromal Parkinson’s disease. Mov. Disord. 2015, 30, 1600–1611. [Google Scholar] [CrossRef] [PubMed]
- Schapira, A.H.V.; Chaudhuri, K.R.; Jenner, P. Non-motor features of Parkinson disease. Nat. Rev. Neurosci. 2017, 8. [Google Scholar] [CrossRef]
- Ory-Magne, F.; Corvol, J.C.; Azulay, J.P.; Bonnet, A.M.; Brefel-Courbon, C.; Damier, P.; Dellapina, E.; Destee, A.; Durif, F.; Galitzky, M.; et al. Withdrawing amantadine in dyskinetic patients with Parkinson disease: The amandysk trial. Neurology 2014, 82, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Bastide, M.F.; Meissner, W.G.; Picconi, B.; Fasano, S.; Fernagut, P.O.; Feyder, M.; Francardo, V.; Alcacer, C.; Ding, Y.; Brambilla, R.; et al. Pathophysiology of L-DOPA-induced motor and non-motor complications in Parkinson’s disease. Prog. Neurobiol. 2015, 132, 96–168. [Google Scholar] [CrossRef] [PubMed]
- Franco-Iborra, S.; Vila, M.; Perier, C. The Parkinson disease mitochondrial hypothesis: Where are we at? Neuroscientist 2016, 22, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Truban, D.; Hou, X.; Caulfield, T.R.; Fiesel, F.C.; Springer, W. PINK1, parkin, and mitochondrial quality control: What can we learn about Parkinson’s disease pathobiology? J. Parkinsons Dis. 2017, 7, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Schapira, A.H.; Chiasserini, D.; Beccari, T.; Parnetti, L. Glucocerebrosidase in Parkinson’s disease: Insights into pathogenesis and prospects for treatment. Mov. Disord. 2016, 31, 830–835. [Google Scholar] [CrossRef] [PubMed]
- Moors, T.; Paciotti, S.; Chiasserini, D.; Calabresi, P.; Parnetti, L.; Beccari, T.; van de Berg, W.D. Lysosomal dysfunction and alpha-synuclein aggregation in Parkinson’s disease: Diagnostic links. Mov. Disord. 2016, 31, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Shi, M.; Chung, K.A.; Quinn, J.F.; Peskind, E.R.; Galasko, D.; Jankovic, J.; Zabetian, C.P.; Leverenz, J.B.; Baird, G.; et al. DJ-1 and alpha-synuclein in human cerebrospinal fluid as biomarkers of Parkinson’s disease. Brain 2010, 133, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Furay, A.R.; Sossi, V.; Aasly, J.O.; Armaly, J.; Wang, Y.; Wszolek, Z.K.; Uitti, R.J.; Hasegawa, K.; Yokoyama, T.; et al. DJ-1 and alphaSYN in LRRK2 CSF do not correlate with striatal dopaminergic function. Neurobiol. Aging 2012, 33, e835–e837. [Google Scholar] [CrossRef] [PubMed]
- Parnetti, L.; Castrioto, A.; Chiasserini, D.; Persichetti, E.; Tambasco, N.; El-Agnaf, O.; Calabresi, P. Cerebrospinal fluid biomarkers in Parkinson disease. Nat. Rev. Neurol. 2013, 9, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Parnetti, L.; Chiasserini, D.; Persichetti, E.; Eusebi, P.; Varghese, S.; Qureshi, M.M.; Dardis, A.; Deganuto, M.; De Carlo, C.; Castrioto, A.; et al. Cerebrospinal fluid lysosomal enzymes and alpha-synuclein in Parkinson’s disease. Mov. Disord. 2014, 29, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Parnetti, L.; Cicognola, C.; Eusebi, P.; Chiasserini, D. Value of cerebrospinal fluid alpha-synuclein species as biomarker in Parkinson’s diagnosis and prognosis. Biomark. Med. 2016, 10, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Matrone, C.; Dzamko, N.; Madsen, P.; Nyegaard, M.; Pohlmann, R.; Sondergaard, R.V.; Lassen, L.B.; Andresen, T.L.; Halliday, G.M.; Jensen, P.H.; et al. Mannose 6-phosphate receptor is reduced in -synuclein overexpressing models of Parkinsons disease. PLoS ONE 2016, 11, e0160501. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, K.D.; Persichetti, E.; Chiasserini, D.; Eusebi, P.; Beccari, T.; Calabresi, P.; Berendse, H.W.; Parnetti, L.; van de Berg, W.D. Changes in endolysosomal enzyme activities in cerebrospinal fluid of patients with Parkinson’s disease. Mov. Disord. 2013, 28, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Mollenhauer, B.; Coffey, C.S.; Toledo, J.B.; Weintraub, D.; Galasko, D.R.; Irwin, D.J.; Van Deerlin, V.; Chen-Plotkin, A.S.; Caspell-Garcia, C.; et al. CSF biomarkers associated with disease heterogeneity in early Parkinson’s disease: The Parkinson’s progression markers initiative study. Acta Neuropathol. 2016, 131, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.; Surova, Y.; Ohrfelt, A.; Swedish Bio, F.S.; Blennow, K.; Zetterberg, H.; Hansson, O. Longitudinal measurements of cerebrospinal fluid biomarkers in Parkinson’s disease. Mov. Disord. 2016, 31, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Schrag, A.; Siddiqui, U.F.; Anastasiou, Z.; Weintraub, D.; Schott, J.M. Clinical variables and biomarkers in prediction of cognitive impairment in patients with newly diagnosed Parkinson’s disease: A cohort study. Lancet Neurol. 2017, 16, 66–75. [Google Scholar] [CrossRef]
- Doppler, K.; Jentschke, H.M.; Schulmeyer, L.; Vadasz, D.; Janzen, A.; Luster, M.; Hoffken, H.; Mayer, G.; Brumberg, J.; Booij, J.; et al. Dermal phospho-alpha-synuclein deposits confirm REM sleep behaviour disorder as prodromal Parkinson’s disease. Acta Neuropathol. 2017, 133, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Espay, A.J.; Schwarzschild, M.A.; Tanner, C.M.; Fernandez, H.H.; Simon, D.K.; Leverenz, J.B.; Merola, A.; Chen-Plotkin, A.; Brundin, P.; Kauffman, M.A.; et al. Biomarker-driven phenotyping in Parkinson’s disease: A translational missing link in disease-modifying clinical trials. Mov. Disord. 2017, 32, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Ascherio, A.; LeWitt, P.A.; Xu, K.; Eberly, S.; Watts, A.; Matson, W.R.; Marras, C.; Kieburtz, K.; Rudolph, A.; Bogdanov, M.B.; et al. Urate as a predictor of the rate of clinical decline in Parkinson disease. Arch Neurol. 2009, 66, 1460–1468. [Google Scholar] [CrossRef] [PubMed]
- LeWitt, P.; Schultz, L.; Auinger, P.; Lu, M.; Parkinson Study Group, Datatop Investigators. CSF xanthine, homovanillic acid, and their ratio as biomarkers of Parkinson’s disease. Brain Res. 2011, 1408, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Mischley, L.K.; Standish, L.J.; Weiss, N.S.; Padowski, J.M.; Kavanagh, T.J.; White, C.C.; Rosenfeld, M.E. Glutathione as a biomarker in Parkinson’s disease: Associations with aging and disease severity. Oxid. Med. Cell Longev. 2016, 9409363. [Google Scholar] [CrossRef] [PubMed]
- LeWitt, P.A.; Galloway, M.P.; Matson, W.; Milbury, P.; McDermott, M.; Srivastava, D.K.; Oakes, D. Markers of dopamine metabolism in Parkinson’s disease. Neurology 1992, 42, 2111–2117. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.S.; Holmes, C.; Sharabi, Y. Cerebrospinal fluid biomarkers of central catecholamine deficiency in Parkinson’s disease and other synucleinopathies. Brain 2012, 135, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.D.; Blaabjerg, M.; Binzer, M.; Kamal, A.; Thagesen, H.; Kjaer, T.W.; Stenager, E.; Gramsbergen, J.B.P. Cerebrospinal fluid levels of catecholamines and its metabolites in Parkinson’s disease: Effect of L-DOPA treatment and changes in levodopa-induced dyskinesia. J. Neurochem. 2017, 141, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.S. Biomarkers, mechanisms, and potential prevention of catecholamine neuron loss in Parkinson disease. Adv. Pharmacol. 2013, 68, 235–272. [Google Scholar] [PubMed]
- Andersen, A.D.; Binzer, M.; Stenager, E.; Gramsbergen, J.B. Cerebrospinal fluid biomarkers for Parkinson’s disease—A systematic review. Acta Neurol. Scand. 2017, 135, 34–56. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Jimenez, F.J.; Alonso-Navarro, H.; Garcia-Martin, E.; Agundez, J.A. Cerebrospinal fluid biochemical studies in patients with Parkinson’s disease: Toward a potential search for biomarkers for this disease. Front. Cell Neurosci. 2014, 8, 369. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.A.; Jimenez-Jimenez, F.J.; Gomez, P.; Vargas, C.; Navarro, J.A.; Orti-Pareja, M.; Gasalla, T.; Benito-Leon, J.; Bermejo, F.; Arenas, J. Decreased cerebrospinal fluid levels of neutral and basic amino acids in patients with Parkinson’s disease. J. Neurol. Sci. 1997, 150, 123–127. [Google Scholar] [CrossRef]
- Bowen, B.C.; Block, R.E.; Sanchez-Ramos, J.; Pattany, P.M.; Lampman, D.A.; Murdoch, J.B.; Quencer, R.M. Proton MR spectroscopy of the brain in 14 patients with Parkinson disease. AJNR Am. J. Neuroradiol. 1995, 16, 61–68. [Google Scholar] [PubMed]
- Henchcliffe, C.; Shungu, D.C.; Mao, X.; Huang, C.; Nirenberg, M.J.; Jenkins, B.G.; Beal, M.F. Multinuclear magnetic resonance spectroscopy for in vivo assessment of mitochondrial dysfunction in Parkinson’s disease. Ann. N. Y. Acad. Sci. 2008, 1147, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Ciurleo, R.; Di Lorenzo, G.; Bramanti, P.; Marino, S. Magnetic resonance spectroscopy: An in vivo molecular imaging biomarker for Parkinson’s disease? Biomed. Res. Int. 2014, 519816. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.E.; Katzen, H.L.; Maudsley, A.; Post, J.; Myerson, C.; Govind, V.; Nahab, F.; Scanlon, B.; Mittel, A. Whole-brain proton MR spectroscopic imaging in Parkinson’s disease. J. Neuroimaging 2014, 24, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Seraji-Bozorgzad, N.; Bao, F.; George, E.; Krstevska, S.; Gorden, V.; Chorostecki, J.; Santiago, C.; Zak, I.; Caon, C.; Khan, O. Longitudinal study of the substantia nigra in Parkinson disease: A high-field 1H-MR spectroscopy imaging study. Mov. Disord. 2015, 30, 1400–1404. [Google Scholar] [CrossRef] [PubMed]
- Weiduschat, N.; Mao, X.; Beal, M.F.; Nirenberg, M.J.; Shungu, D.C.; Henchcliffe, C. Usefulness of proton and phosphorus MR spectroscopic imaging for early diagnosis of Parkinson’s disease. J. Neuroimaging 2015, 25, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, S.M.; Arnold, J.; Sreekumar, A. Metabolomic profiling of hormone-dependent cancers: A bird’s eye view. Trends Endocrinol. Metab. 2015, 26, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Powers, R. NMR metabolomics analysis of Parkinson’s disease. Curr. Metabolomics 2013, 1, 191–209. [Google Scholar] [CrossRef] [PubMed]
- Jove, M.; Portero-Otin, M.; Naudi, A.; Ferrer, I.; Pamplona, R. Metabolomics of human brain aging and age-related neurodegenerative diseases. J Neuropathol. Exp. Neurol. 2014, 73, 640–657. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, C.; Cifuentes, A.; Simo, C. Recent advances and applications of metabolomics to investigate neurodegenerative diseases. Int. Rev. Neurobiol. 2015, 122, 95–132. [Google Scholar] [PubMed]
- Kori, M.; Aydin, B.; Unal, S.; Arga, K.Y.; Kazan, D. Metabolic biomarkers and neurodegeneration: A pathway enrichment analysis of Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis. Omics 2016, 20, 645–661. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics-the link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Villas-Boas, S.G.; Mas, S.; Akesson, M.; Smedsgaard, J.; Nielsen, J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Michell, A.W.; Mosedale, D.; Grainger, D.J.; Barker, R.A. Metabolomic analysis of urine and serum in parkinson’s disease. Metabolomics 2008, 4, 191–201. [Google Scholar] [CrossRef]
- Johansen, K.K.; Wang, L.; Aasly, J.O.; White, L.R.; Matson, W.R.; Henchcliffe, C.; Beal, M.F.; Bogdanov, M. Metabolomic profiling in LRRK2-related parkinson’s disease. PLoS ONE 2009, 4, e7551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havelund, J.F.; Andersen, A.D.; Binzer, M.; Blaabjerg, M.; Heegaard, N.H.H.; Stenager, E.; Faergeman, N.J.; Gramsbergen, J.B. Changes in Kynurenine Pathway Metabolism in Parkinson Patients with L-DOPA-Induced Dyskinesia. Available online: http://onlinelibrary.wiley.com/doi/10.1111/jnc.14104/full (accessed on 10 August 2017).
- Lewitt, P.A.; Li, J.; Lu, M.; Beach, T.G.; Adler, C.H.; Guo, L.; The Arizona Parkinson’s Disease Consortium. 3-hydroxykynurenine and other Parkinson’s disease biomarkers discovered by metabolomic analysis. Mov. Disord. 2013, 28, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Trupp, M.; Jonsson, P.; Ohrfelt, A.; Zetterberg, H.; Obudulu, O.; Malm, L.; Wuolikainen, A.; Linder, J.; Moritz, T.; Blennow, K.; et al. Metabolite and peptide levels in plasma and CSF differentiating healthy controls from patients with newly diagnosed Parkinson’s disease. J. Parkinsons Dis. 2014, 4, 549–560. [Google Scholar] [PubMed]
- Ahmed, S.S.; Santosh, W.; Kumar, S.; Christlet, H.T. Metabolic profiling of Parkinson’s disease: Evidence of biomarker from gene expression analysis and rapid neural network detection. J. Biomed. Sci. 2009, 16, 63. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, M.; Matson, W.R.; Wang, L.; Matson, T.; Saunders-Pullman, R.; Bressman, S.S.; Flint Beal, M. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain 2008, 131, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Trushina, E.; Mielke, M.M. Recent advances in the application of metabolomics to Alzheimer’s disease. Biochim. Biophys. Acta 2014, 1842, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Kaddurah-Daouk, R.; Kristal, B.S.; Weinshilboum, R.M. Metabolomics: A global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 653–683. [Google Scholar] [CrossRef] [PubMed]
- Kristal, B.S.; Vigneau-Callahan, K.; Matson, W.R. Simultaneous analysis of multiple redox-active metabolites from biological matrices. Methods Mol. Biol. 2002, 186, 185–194. [Google Scholar] [PubMed]
- Shi, H.; Vigneau-Callahan, K.E.; Matson, W.R.; Kristal, B.S. Attention to relative response across sequential electrodes improves quantitation of coulometric array. Anal. Biochem. 2002, 302, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Marion, D. An introduction to biological NMR spectroscopy. Mol. Cell Proteom. 2013, 12, 3006–3025. [Google Scholar] [CrossRef] [PubMed]
- De Hoffmann, E.; Stroobant, V. Mass Spectrometry: Principles and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Dias, D.A.; Jones, O.A.; Beale, D.J.; Boughton, B.A.; Benheim, D.; Kouremenos, K.A.; Wolfender, J.L.; Wishart, D.S. Current and future perspectives on the structural identification of small molecules in biological systems. Metabolites 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Dunand, M.; Gubian, D.; Stauffer, M.; Abid, K.; Grouzmann, E. High-throughput and sensitive quantitation of plasma catecholamines by ultraperformance liquid chromatography-tandem mass spectrometry using a solid phase microwell extraction plate. Anal. Chem. 2013, 85, 3539–3544. [Google Scholar] [CrossRef] [PubMed]
- Barganska, Z.; Namiesnik, J. Pesticide analysis of bee and bee product samples. Crit. Rev. Anal. Chem. 2010, 40, 159–171. [Google Scholar] [CrossRef]
- Hatano, T.; Saiki, S.; Okuzumi, A.; Mohney, R.P.; Hattori, N. Identification of novel biomarkers for Parkinson’s disease by metabolomic technologies. J. Neurol. Neurosurg. Psychiatry 2016, 87, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Liu, L.F.; Meng, N.; Tang, Z.; Chua, K.K.; Chen, L.L.; Song, J.X.; Mok, V.C.; Xie, L.X.; Li, M.; et al. LC-MS-based urinary metabolite signatures in idiopathic Parkinson’s disease. J. Proteome Res. 2015, 14, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Salek, R.M.; Neumann, S.; Schober, D.; Hummel, J.; Billiau, K.; Kopka, J.; Correa, E.; Reijmers, T.; Rosato, A.; Tenori, L.; et al. Coordination of standards in metabolomics (COSMOS): Facilitating integrated metabolomics data access. Metabolomics 2015, 11, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Roede, J.R.; Uppal, K.; Park, Y.; Lee, K.; Tran, V.; Walker, D.; Strobel, F.H.; Rhodes, S.L.; Ritz, B.; Jones, D.P. Serum metabolomics of slow vs. Rapid motor progression Parkinson’s disease: A pilot study. PLoS ONE 2013, 8, e77629. [Google Scholar] [CrossRef] [PubMed]
- Burte, F.; Houghton, D.; Lowes, H.; Pyle, A.; Nesbitt, S.; Yarnall, A.; Yu-Wai-Man, P.; Burn, D.J.; Santibanez-Koref, M.; Hudson, G. Metabolic Profiling of Parkinson’s Disease and Mild Cognitive Impairment. Available online: http://onlinelibrary.wiley.com/doi/10.1002/mds.26992/full (accessed on 10 August 2017).
- Wuolikainen, A.; Jonsson, P.; Ahnlund, M.; Antti, H.; Marklund, S.L.; Moritz, T.; Forsgren, L.; Andersen, P.M.; Trupp, M. Multi-platform mass spectrometry analysis of the CSF and plasma metabolomes of rigorously matched amyotrophic lateral sclerosis, Parkinson’s disease and control subjects. Mol. Biosyst. 2016, 12, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Bruschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Huhman, D.V.; Sumner, L.W. Mass spectrometry strategies in metabolomics. J. Biol. Chem. 2011, 286, 25435–25442. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Liu, L.F.; Tang, Z.; Zhang, M.; Chua, K.K.; Song, J.X.; Mok, V.C.; Li, M.; Cai, Z. Comprehensive urinary metabolomic profiling and identification of potential noninvasive marker for idiopathic Parkinson’s disease. Sci. Rep. 2015, 5, 13888. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS models. J. Chemometr. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Xia, J.G.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Zweig, M.H.; Campbell, G. Receiver operating characteristic (ROC) plots—A fundamental evaluation tool in clinical medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; Potter, J.D.; Thompson, M.L.; Thornquist, M.; Winget, M.; Yasui, Y. Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst. 2001, 93, 1054–1061. [Google Scholar] [CrossRef] [PubMed]
- Obuchowski, N.A.; Lieber, M.L.; Wians, F.H., Jr. ROC curves in clinical chemistry: Uses, misuses, and possible solutions. Clin. Chem. 2004, 50, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Søreide, K. Receiver-operating characteristic curve analysis in diagnostic, prognostic and predictive biomarker research. J. Clin. Pathol. 2009, 62, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Nyamundanda, G.; Gormley, I.C.; Fan, Y.; Gallagher, W.M.; Brennan, L. Metsizer: Selecting the optimal sample size for metabolomic studies using an analysis based approach. Bioinformatics 2013, 14, 338. [Google Scholar] [CrossRef] [PubMed]
- LeWitt, P.A.; Li, J.; Lu, M.; Guo, L.; Auinger, P.; Parkinson Study Group, Datatop Investigators. Metabolomic biomarkers as strong correlates of Parkinson disease progression. Neurology 2017, 88, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Ohman, A.; Forsgren, L. NMR metabonomics of cerebrospinal fluid distinguishes between Parkinson’s disease and controls. Neurosci. Lett. 2015, 594, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Davidson, D.F.; Grosset, K.; Grosset, D. Parkinson’s disease: The effect of l-dopa therapy on urinary free catecholamines and metabolites. Ann. Clin. Biochem. 2007, 44, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, G.; Brown, S.; Peitzsch, M.; Pelzel, D.; Lattke, P.; Glockner, S.; Stell, A.; Prejbisz, A.; Fassnacht, M.; Beuschlein, F.; et al. Levodopa therapy in Parkinson’s disease: Influence on liquid chromatographic tandem mass spectrometric-based measurements of plasma and urinary normetanephrine, metanephrine and methoxytyramine. Ann. Clin. Biochem. 2014, 51, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Zhang, S.N.; Lu, F.; Liu, C.F.; Wang, Y.; Bai, Y.; Wang, N.; Liu, S.M. Cerebral metabonomics study on Parkinson’s disease mice treated with extract of acanthopanax senticosus harms. Phytomedicine 2013, 20, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Poliquin, P.O.; Chen, J.; Cloutier, M.; Trudeau, L.E.; Jolicoeur, M. Metabolomics and in-silico analysis reveal critical energy deregulations in animal models of Parkinson’s disease. PLoS ONE 2013, 8, e69146. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Zavala-Flores, L.; Garcia-Garcia, A.; Nandakumar, R.; Huang, Y.; Madayiputhiya, N.; Stanton, R.C.; Dodds, E.D.; Powers, R.; Franco, R. Alterations in energy/redox metabolism induced by mitochondrial and environmental toxins: A specific role for glucose-6-phosphate-dehydrogenase and the pentose phosphate pathway in paraquat toxicity. ACS Chem. Biol. 2014, 9, 2032–2048. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Wang, J.; Li, M.; Liu, Q.; Wei, D.; Yang, M.; Kong, L. 1H-NMR based metabolomics study on a goldfish model of Parkinson’s disease induced by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP). Chem. Biol. Interact. 2014, 223, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xie, C.; Sun, L.; Ding, J.; Cai, H. Longitudinal metabolomics profiling of Parkinson’s disease-related alpha-synuclein a53t transgenic mice. PLoS ONE 2015, 10, e0136612. [Google Scholar] [CrossRef]
- Farmer, K.; Smith, C.A.; Hayley, S.; Smith, J. Major alterations of phosphatidylcholine and lysophosphotidylcholine lipids in the substantia nigra using an early stage model of Parkinson’s disease. Int. J. Mol. Sci. 2015, 16, 18865–18877. [Google Scholar] [CrossRef] [PubMed]
- Tyurina, Y.Y.; Polimova, A.M.; Maciel, E.; Tyurin, V.A.; Kapralova, V.I.; Winnica, D.E.; Vikulina, A.S.; Domingues, M.R.; McCoy, J.; Sanders, L.H.; et al. LC/MS analysis of cardiolipins in substantia nigra and plasma of rotenone-treated rats: Implication for mitochondrial dysfunction in Parkinson’s disease. Free Radic. Res. 2015, 49, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Ratnasekhar, C.; Pragya, P.; Chaouhan, H.S.; Patel, D.K.; Chowdhuri, D.K.; Mudiam, M.K. Metabolomic analysis provides insights on paraquat-induced Parkinson-like symptoms in drosophila melanogaster. Mol. Neurobiol. 2016, 53, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Jenner, P. Functional models of Parkinson’s disease: A valuable tool in the development of novel therapies. Ann. Neurol. 2008, 64, S16–S29. [Google Scholar] [CrossRef] [PubMed]
- Chesselet, M.F.; Fleming, S.; Mortazavi, F.; Meurers, B. Strengths and limitations of genetic mouse models of Parkinson’s disease. Parkinsonism Relat. Disord. 2008, 14, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Dehay, B.; Vila, M.; Bezard, E.; Brundin, P.; Kordower, J.H. Alpha-synuclein propagation: New insights from animal models. Mov. Disord. 2016, 31, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Blesa, J.; Przedborski, S. Parkinson’s disease: Animal models and dopaminergic cell vulnerability. Front. Neuroanat. 2014, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Bannon, D.; Landau, A.M.; Doudet, D.J. How Relevant Are Imaging Findings in Animal Models of Movement Disorders to Human Disease? Available online: https://link.springer.com/article/10.1007/s11910-015-0571-z (accessed on 10 August 2017).
- Larsen, T.R.; Soderling, A.S.; Caidahl, K.; Roepstorff, P.; Gramsbergen, J.B. Nitration of soluble proteins in organotypic culture models of Parkinson’s disease. Neurochem. Int. 2008, 52, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Playne, R.; Connor, B. Understanding Parkinson’s disease through the use of cell reprogramming. Stem cell. Rev. 2017, 13, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Torrent, R.; De Angelis Rigotti, F.; Dell’Era, P.; Memo, M.; Raya, A.; Consiglio, A. Using ips cells toward the understanding of Parkinson’s disease. J. Clin. Med. 2015, 4, 548–566. [Google Scholar] [CrossRef] [PubMed]
- Fernstrom, J.D. Large neutral amino acids: Dietary effects on brain neurochemistry and function. Amino Acids 2013, 45, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.; Xiao, R.; Place, E.; Zhang, Z.; Sondheimer, N.; Bennett, M.; Yudkoff, M.; Falk, M.J. Mitochondrial respiratory chain disease discrimination by retrospective cohort analysis of blood metabolites. Mol. Genet. Metab. 2013, 110, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Connelly, M.A.; Wolak-Dinsmore, J.; Dullaart, R.P.F. Branched chain amino acids are associated with insulin resistance independent of leptin and adiponectin in subjects with varying degrees of glucose tolerance. Metab. Syndr. Relat. Disord. 2017, 15, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Toledo, J.B.; Arnold, M.; Kastenmuller, G.; Chang, R.; Baillie, R.A.; Han, X.; Thambisetty, M.; Tenenbaum, J.D.; Suhre, K.; Thompson, J.W.; et al. Metabolic Network Failures in Alzheimer’s Disease—A Biochemical Road Map. Available online: http://www.sciencedirect.com/science/article/pii/S1552526017300468 (accessed on 10 August 2017).
- Ruiz, H.H.; Chi, T.; Shin, A.C.; Lindtner, C.; Hsieh, W.; Ehrlich, M.; Gandy, S.; Buettner, C. Increased susceptibility to metabolic dysregulation in a mouse model of Alzheimer’s disease is associated with impaired hypothalamic insulin signaling and elevated bcaa levels. Alzheimer’s Dement. 2016, 12, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Valerio, A.; D’Antona, G.; Nisoli, E. Branched-chain amino acids, mitochondrial biogenesis, and healthspan: An evolutionary perspective. Aging 2011, 3, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Bove, J.; Martinez-Vicente, M.; Vila, M. Fighting neurodegeneration with rapamycin: Mechanistic insights. Nat. Rev. Neurosci. 2011, 12, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Malagelada, C.; Jin, Z.H.; Jackson-Lewis, V.; Przedborski, S.; Greene, L.A. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J. Neurosci. 2010, 30, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Santini, E.; Feyder, M.; Gangarossa, G.; Bateup, H.S.; Greengard, P.; Fisone, G. Dopamine- and cAMP-regulated phosphoprotein of 32-kDa (DARPP-32)-dependent activation of extracellular signal-regulated kinase (ERK) and mammalian target of rapamycin complex 1 (mtorc1) signaling in experimental Parkinsonism. J. Biol. Chem. 2012, 287, 27806–27812. [Google Scholar] [CrossRef] [PubMed]
- Decressac, M.; Bjorklund, A. mTOR inhibition alleviates L-DOPA-induced dyskinesia in Parkinsonian rats. J. Parkinsons Dis. 2013, 3, 13–17. [Google Scholar] [PubMed]
- Hafizi Abu Bakar, M.; Kian Kai, C.; Wan Hassan, W.N.; Sarmidi, M.R.; Yaakob, H.; Zaman Huri, H. Mitochondrial dysfunction as a central event for mechanisms underlying insulin resistance: The roles of long chain fatty acids. Diabetes Metab. Res. Rev. 2015, 31, 453–475. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, I.; Schlesinger, N. Uric acid in Parkinson’s disease. Mov. Disord. 2008, 23, 1653–1657. [Google Scholar] [CrossRef] [PubMed]
- Wills, A.M.; Eberly, S.; Tennis, M.; Lang, A.E.; Messing, S.; Togasaki, D.; Tanner, C.M.; Kamp, C.; Chen, J.F.; Oakes, D.; et al. Caffeine consumption and risk of dyskinesia in CALM-PD. Mov. Disord. 2013, 28, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Palacios, N.; Gao, X.; McCullough, M.L.; Schwarzschild, M.A.; Shah, R.; Gapstur, S.; Ascherio, A. Caffeine and risk of Parkinson’s disease in a large cohort of men and women. Mov. Disord. 2012, 27, 1276–1282. [Google Scholar] [CrossRef] [PubMed]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.Q. Kynurenines in the mammalian brain: When physiology meets pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Balu, D.T.; Coyle, J.T. The NMDA receptor ‘glycine modulatory site’ in schizophrenia: D-serine, glycine, and beyond. Curr. Opin. Pharmacol. 2015, 20, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Connor, M.; Vaughan, C.W.; Vandenberg, R.J. N-acyl amino acids and N-acyl neurotransmitter conjugates: Neuromodulators and probes for new drug targets. Br. J. Pharmacol. 2010, 160, 1857–1871. [Google Scholar] [CrossRef] [PubMed]
- Scheperjans, F. Gut microbiota, 1013 new pieces in the Parkinson’s disease puzzle. Curr. Opin. Neurol. 2016, 29, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Goto, S.; Tsuji, H.; Okuno, T.; Asahara, T.; Nomoto, K.; Shibata, A.; Fujisawa, Y.; Minato, T.; Okamoto, A.; et al. Intestinal dysbiosis and lowered serum lipopolysaccharide-binding protein in Parkinson’s disease. PLoS ONE 2015, 10, e0142164. [Google Scholar] [CrossRef] [PubMed]
- Unger, M.M.; Spiegel, J.; Dillmann, K.U.; Grundmann, D.; Philippeit, H.; Burmann, J.; Fassbender, K.; Schwiertz, A.; Schafer, K.H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.D.; Nicholson, J.K. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 2017, 179, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 2016, 167, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
| Article | Analytical Platform | Statistics | Patients (#) | Pathway/Compound | |
|---|---|---|---|---|---|
| Increased in PD | Decreased in PD | ||||
| Bogdanov et al., 2008 [56] | LCECA | PLS-DA t-test | PD (60), controls (25) | B: Glutathione metabolism (GSH) | B: Purine metabolism (uric acid ) |
| Johansen et al., 2009 [51] | LCECA | PLS-DA | PD (41), PD-LRRK2 (12), healthy-LRRK2 (21), controls (25) | B: Purine metabolism (uric acid, hypoxanthine) | |
| Ahmed et al., 2009 [55] | H-NMR | PLS-DA, ANOVA | PD (43), controls (37) | B: Energy metabolism (pyruvate) | B: Energy metabolism (TCA metabolites, creatinine) |
| Roede et al., 2013 [70] | LC-MS2 + | (O)PLS-DA t-test | PD rapid progress (39), PD slow progress (41), Controls (20) | B: Polyamine metabolism (N8-acetylspermidine) | |
| LeWitt et al., 2013 [53] | LC-MS2 +/− GC-MS | t-test ROC curve | PD (48), controls (57) | C: Kynurenine metabolism (3-HK/KYNA) | C: Acetylated amino acids, C: glutathione metabolism (GSSG) |
| Trupp et al., 2014 [54] | GC-MS | (O)PLS-DA | PD (20), controls (20) | B: Amino acids (methionine, threonine, alanine, serine), glutathione metabolism (pyroglutamate), ketoleucine | C: Energy metabolism (creatinine), tryptophan metabolism (tryptophan) B: FA metabolism (C16 and C18) |
| Öhman and Forsgren 2015 [87] | H-NMR | Multivariate, univariate | PD (10), controls (10) | C: Amino acids (alanine), energy metabolism (creatinine), sugars (mannose) | |
| Luan et al., 2015 [67] | LC-MS +/− | (O)PLS-DA ROC curve | PD (106), controls (104) | U: BCAA, Glycine derivatives, histidine metabolism, tryptophan/kynurenine metabolism, phenylalanine metabolism , purine metabolism, steroidogenesis | |
| Luan et al., 2015 [76] | LC-MS +/− GC-MS | (O)PLS-DA | PD (92), controls (65) | ||
| Hatano et al., 2016 [66] | LC-MS +/− GC-MS | t-test | PD (35), controls (15) | U: Phenylalanine metabolism (phenylacetate) | B: Bilirubin/biliverdin, tryptophan metabolism |
| Wuolikainen et al., 2016 [72] | LC-MS +/− GC-MS | (O)PLS-DA | PD (22), ALS (22), controls (28) | B and C: Amino acids (alanine) C: BCAA (leucine, isoleucine) | |
| LeWitt et al., 2017 [86] | LC-MS +/− GC-MS | Multivariate t-test | PD (49); collected twice with an interval of up to 2 years | B: FA metabolism (medium-long chain FA), phenylalanine metabolism (aspartylphenylalanine, benzoate), serine metabolism (serine) | B: Purine metabolism (inosine) |
| Burté et al., 2017 [71] | LC-MS2 +/− GC-MS | PCA (O)PLS-DA | PD early stage (41), Controls (40) | B: FA metabolism (acylcarnitine), histidine metabolism (1-methylhistamine) | |
| Havelund et al., 2017 [52] | LC-MS+ | ANOVA | PD (26), PD-LID (10), controls (14) | B: Kynurenine metabolism (3-HK/KYNA) | B and C: Kynurenine metabolism (anthranilic acid ) |
| Article | Analytical Platform | Statistics | Model (Treatment) | Tissue or Cells | Pathway/Compound | |
|---|---|---|---|---|---|---|
| Increased in PD | Decreased in PD | |||||
| Li et al., 2013 [91] | LC-MS + | PCA, PLS-DA, t-test | Mice (MPTP-treated mice, Acanthopanax senticosus harms) | Midbrain | Ceramide (d18:0/18:0), FA metabolism (lysoPC 20:4), methionine metabolism (5-methylthioadenosine), morphiceptin, sphingolipid metabolism (phytosphingosine-1-P), tetracosanoylglycine, tyrosine metabolism (L-DOPA) | |
| Poliquin et al., 2013 [92] | LC-MS +/− | Not reported | Parkin KO mice (complex I inhibitor) | Brain slices | Energy metabolism (ATP) | |
| Lei et al., 2014 [93] | H-NMR MS | PLS-DA, ANOVA | Neuroblastoma cells (6-OHDA, MPP+, rotenone, or paraquat) | Cells | Energy metabolism (pentose phosphate pathway), sugars (heptose, hexose) | Energy metabolism (TCA cycle), amino acid (glutamate), tyrosine metabolism (dopamine) |
| Lu et al., 2014 [94] | H-NMR LC-MS +/− | PCA, OPLS-DA, t-test, Mann−Whitney U-test | Goldfish (MPTP-treated) | Whole brain | Alanine metabolism (alanine, alanylalanine), amino acids (taurine), BCAA (leucine, isoleucine, valine), energy metabolism (creatinine), FA metabolism (18:2, total FA), sugars (myo-inositol, a glial marker) | Energy metabolism (TCA metabolites), FA metabolism (n-3 FA, unsaturated FA), glutamine metabolism, neuronal injury markers, tyrosine metabolism (dopamine, DOPAC) |
| Chen et al., 2015 [95] | LC-MS2 +/− GC-MS | PCA, ANOVA, Random forest | Mice (alpha-syn A53T transgenic) | Forebrain and midbrain | Alanine metabolism, acetyl-CoA biosynthesis pathways | Purine metabolism (guanosine) |
| Farmer et al., 2015 [96] | LC-MS2 + | t-test | Rats (6-OHDA) | Substantia nigra | Lysophosphatidylcholine (C16:0, 18:1) | Lysophosphatidylcholine, phosphatidylcholine species (12 species) |
| Tyurina et al., 2015 [97] | LC-MS − | Rats (rotenone) | Substantia nigra (SN) Blood (B) | SN: Mono-oxygenated cardiolipin metabolism, B: Polyunsaturated FA cardiolipin | SN : unsaturated FA cardiolipin species | |
| Luan et al., 2015 [67] | LC-MS + | Mann−Whitney U-test | Drosophila (alpha-syn overexpressing) | Whole flies | Kynurenine metabolism (kynurenine/KYNA) | |
| Shukla et al., 2016 [98] | LCECA GC-MS | t-test | Drosophila (paraquat) | Fly heads | Alanine metabolism (alanine), energy metabolism (lactate acid), FA metabolism (hexadecanoic acid, oleic acid), glycerolipid metabolism, glycine metabolism (glycine), sugars (inositol, myo-inositol) | Amino acids (γ-aminobutyric acid, proline), BCAA (isoleucine, leucine, valine), purine metabolism (uric acid), sugars (glucose, galactose, trehalose), tyrosine metabolism (dopamine) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Havelund, J.F.; Heegaard, N.H.H.; Færgeman, N.J.K.; Gramsbergen, J.B. Biomarker Research in Parkinson’s Disease Using Metabolite Profiling. Metabolites 2017, 7, 42. https://doi.org/10.3390/metabo7030042
Havelund JF, Heegaard NHH, Færgeman NJK, Gramsbergen JB. Biomarker Research in Parkinson’s Disease Using Metabolite Profiling. Metabolites. 2017; 7(3):42. https://doi.org/10.3390/metabo7030042
Chicago/Turabian StyleHavelund, Jesper F., Niels H. H. Heegaard, Nils J. K. Færgeman, and Jan Bert Gramsbergen. 2017. "Biomarker Research in Parkinson’s Disease Using Metabolite Profiling" Metabolites 7, no. 3: 42. https://doi.org/10.3390/metabo7030042





