Neonatal Urine Metabolic Profiling and Development of Childhood Asthma
Abstract
:1. Introduction
2. Results
2.1. Baseline Characteristics
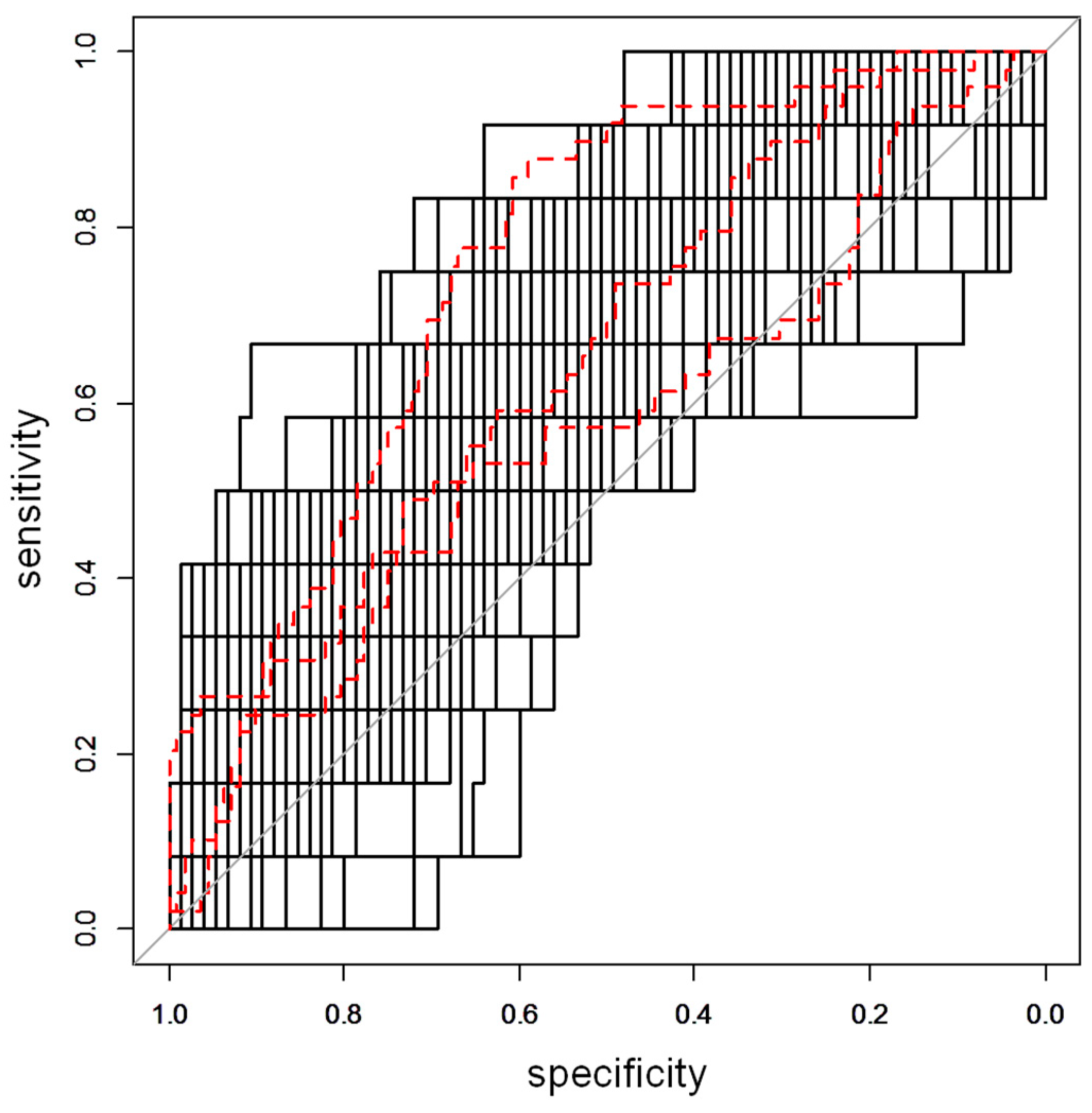
2.2. Urine Metabolomics Analysis
2.3. Metabolite Annotation
3. Discussion
4. Materials and Methods
4.1. Study Populations
4.2. Persistent Wheeze and Asthma Diagnosis
4.3. Urine Collection and Sample Preparation
4.4. UPLC-MS Analysis
4.5. Data Processing and Pre-Treatment
4.6. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
- ○
- Filter was set 0.25 on POS and NEG acquisition
- ○
- Adducts
- ○
- POS (M+H, M+Na, M+K, M+H-H2O, M+NH4, M+CH3OH+H)
- ○
- NEG (M-H, M+Na-2H, M-H2O-H, M-NH4, M+CH3CO2)
- ○
- Filter 10 ppm for mass
- ○
- Filter 0.5 min for Rt
- ○
- Filter 20 ppm for mass fragment
- ○
- Filter 10 ppm for mass
- ○
- Filter 20 ppm for mass fragment
References
- Eder, W.; Ege, M.J.; von Mutius, E. The asthma epidemic. N. Engl. J. Med. 2006, 355, 2226–2235. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.D.; Wright, A.L.; Taussig, L.M.; Holberg, C.J.; Halonen, M.; Morgan, W.J. Asthma and wheezing in the first six years of life. The group health medical associates. N. Engl. J. Med. 1995, 332, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Bisgaard, H.; Szefler, S. Prevalence of asthma-like symptoms in young children. Pediatr. Pulmonol. 2007, 42, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Kocevar, V.S.; Bisgaard, H.; Jönsson, L.; Valovirta, E.; Kristensen, F.; Yin, D.D.; Thomas, J., 3rd. Variations in pediatric asthma hospitalization rates and costs between and within Nordic countries. Chest 2004, 125, 1680–1684. [Google Scholar] [CrossRef] [PubMed]
- Prescott, S.L. Early-Life environmental determinants of allergic diseases and the wider pandemic of inflammatory noncommunicable diseases. J. Allergy Clin. Immunol. 2013, 131, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Chawes, B.L.K.; Bønnelykke, K.; Bisgaard, H. Elevated eosinophil protein X in urine from healthy neonates precedes development of atopy in the first 6 years of life. Am. J. Respir. Crit. Care Med. 2011, 184, 656–661. [Google Scholar] [CrossRef]
- Chawes, B.L.K.; Buchvald, F.; Bischoff, A.L.; Loland, L.; Hermansen, M.; Halkjaer, L.B.; Bønnelykke, K.; Bisgaard, H. Elevated exhaled nitric oxide in high-risk neonates precedes transient early but not persistent wheeze. Am. J. Respir. Crit. Care Med. 2010, 182, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Kettunen, J.; Tukiainen, T.; Sarin, A.-P.; Ortega-Alonso, A.; Tikkanen, E.; Lyytikäinen, L.-P.; Kangas, A.J.; Soininen, P.; Würtz, P.; Silander, K.; et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012, 44, 269–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carraro, S.; Giordano, G.; Reniero, F.; Perilongo, G.; Baraldi, E. Metabolomics: A new frontier for research in pediatrics. J. Pediatr. 2009, 154, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.S.; Dahlin, A.; McGeachie, M.J.; Qiu, W.; Sordillo, J.; Wan, E.S.; Wu, A.C.; Lasky-Su, J. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest 2017, 151, 262–277. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.S.; Chawes, B.L.; Blighe, K.; Virkud, Y.V.; Croteau-Chonka, D.C.; McGeachie, M.J.; Clish, C.B.; Bullock, K.; Celedón, J.C.; Weiss, S.T.; et al. An integrative transcriptomic and metabolomic study of lung function in children with asthma. Chest 2018, 154, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Saude, E.J.; Skappak, C.D.; Regush, S.; Cook, K.; Ben-Zvi, A.; Becker, A.; Moqbel, R.; Sykes, B.D.; Rowe, B.H.; Adamko, D.J. Metabolomic profiling of asthma: Diagnostic utility of urine nuclear magnetic resonance spectroscopy. J. Allergy Clin. Immunol. 2011, 127, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Mattarucchi, E.; Baraldi, E.; Guillou, C. Metabolomics applied to urine samples in childhood asthma; differentiation between asthma phenotypes and identification of relevant metabolites. Biomed. Chromatogr. BMC 2012, 26, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Carraro, S.; Bozzetto, S.; Giordano, G.; El Mazloum, D.; Stocchero, M.; Pirillo, P.; Zanconato, S.; Baraldi, E. Wheezing preschool children with early-onset asthma reveal a specific metabolomic profile. Pediatr. Allergy Immunol. 2018, 29, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Bisgaard, H. The Copenhagen Prospective Study on Asthma in Childhood (COPSAC): Design, rationale, and baseline data from a longitudinal birth cohort study. Ann. Allergy Asthma Immunol. 2004, 93, 381–389. [Google Scholar] [CrossRef]
- Loland, L.; Bisgaard, H. Feasibility of repetitive lung function measurements by raised volume rapid thoracoabdominal compression during methacholine challenge in young infants. Chest 2008, 133, 115–122. [Google Scholar] [CrossRef]
- Bisgaard, H.; Vissing, N.H.; Carson, C.G.; Bischoff, A.L.; Følsgaard, N.V.; Kreiner-Møller, E.; Chawes, B.L.K.; Stokholm, J.; Pedersen, L.; Bjarnadóttir, E.; et al. Deep phenotyping of the unselected COPSAC2010 birth cohort study. Clin. Exp. Allergy 2013, 43, 1384–1394. [Google Scholar] [CrossRef]
- Storey, J.D. A direct approach to false discovery rates. J. R. Stat. Soc. 2002, 64, 479–498. [Google Scholar] [CrossRef] [Green Version]
- Arredouani, A.; Stocchero, M.; Culeddu, N.; Moustafa, J.E.-S.; Tichet, J.; Balkau, B.; Brousseau, T.; Manca, M.; Falchi, M. Metabolomic profile of low-copy number carriers at the salivary α-amylase gene suggests a metabolic shift toward lipid-based energy production. Diabetes 2016, 65, 3362–3368. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metab. Off. J. Metab. Soc. 2007, 3, 211–221. [Google Scholar]
- Chiu, C.-Y.; Lin, G.; Cheng, M.-L.; Chiang, M.-H.; Tsai, M.-H.; Su, K.-W.; Hua, M.-C.; Liao, S.-L.; Lai, S.-H.; Yao, T.-C.; et al. Longitudinal urinary metabolomic profiling reveals metabolites for asthma development in early childhood. Pediatr. Allergy Immunol. 2018, 29, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Turi, K.N.; Romick-Rosendale, L.; Gebretsadik, T.; Watanabe, M.; Brunwasser, S.; Anderson, L.J.; Moore, M.L.; Larkin, E.K.; Peebles, R.S.; Hartert, T.V. Using urine metabolomics to understand the pathogenesis of infant respiratory syncytial virus (RSV) infection and its role in childhood wheezing. Metab. Off. J. Metab. Soc. 2018, 14, 135. [Google Scholar] [CrossRef] [PubMed]
- Bathena, S.P.R.; Mukherjee, S.; Olivera, M.; Alnouti, Y. The profile of bile acids and their sulfate metabolites in human urine and serum. J. Chromatogr. B 2013, 942, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Xun, P.; Pike, K.; Wills, A.K.; Chawes, B.L.; Bisgaard, H.; Cai, W.; Wan, Y.; He, K. In utero exposure to 25-hydroxyvitamin D and risk of childhood asthma, wheeze, and respiratory tract infections: A meta-analysis of birth cohort studies. J. Allergy Clin. Immunol. 2017, 139, 1508–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddesha, J.M.; Nakada, E.M.; Mihavics, B.R.; Hoffman, S.M.; Rattu, G.K.; Chamberlain, N.; Cahoon, J.M.; Lahue, K.G.; Daphtary, N.; Aliyeva, M.; et al. Effect of a chemical chaperone, tauroursodeoxycholic acid, on HDM-induced allergic airway disease. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 310, L1243–L1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakada, E.M.; Bhakta, N.R.; Korwin-Mihavics, B.R.; Kumar, A.; Chamberlain, N.; Bruno, S.R.; Chapman, D.G.; Hoffman, S.M.; Daphtary, N.; Aliyeva, M.; et al. Conjugated bile acids attenuate allergen-induced airway inflammation and hyperresposiveness by inhibiting UPR transducers. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Shaik, F.B.; Panati, K.; Narasimha, V.R.; Narala, V.R. Chenodeoxycholic acid attenuates ovalbumin-induced airway inflammation in murine model of asthma by inhibiting the T(H)2 cytokines. Biochem. Biophys. Res. Commun. 2015, 463, 600–605. [Google Scholar] [CrossRef]
- Kelly, R.S.; Sordillo, J.E.; Lasky-Su, J.; Dahlin, A.; Perng, W.; Rifas-Shiman, S.L.; Weiss, S.T.; Gold, D.R.; Litonjua, A.A.; Hivert, M.-F.; et al. Plasma metabolite profiles in children with current asthma. Clin. Exp. Allergy 2018, 48, 1297–1304. [Google Scholar] [CrossRef]
- Comhair, S.A.A.; McDunn, J.; Bennett, C.; Fettig, J.; Erzurum, S.C.; Kalhan, S.C. Metabolomic endotype of asthma. J. Immunol. 2015, 195, 643–650. [Google Scholar] [CrossRef]
- Klaassen, C.D.; Cui, J.Y. Review: Mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab. Dispos. 2015, 43, 1505–1521. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsland, B.J.; Trompette, A.; Gollwitzer, E.S. The gut-lung axis in respiratory disease. Ann. Am. Thorac. Soc. 2015, 12 (Suppl. 2), S150–S156. [Google Scholar] [PubMed]
- Duran, M.; Wanders, R.J.; de Jager, J.P.; Dorland, L.; Bruinvis, L.; Ketting, D.; Ijlst, L.; van Sprang, F.J. 3-Hydroxydicarboxylic aciduria due to long-chain 3-hydroxyacyl-coenzyme A dehydrogenase deficiency associated with sudden neonatal death: Protective effect of medium-chain triglyceride treatment. Eur. J. Pediatr. 1991, 150, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Bergoffen, J.; Kaplan, P.; Hale, D.E.; Bennett, M.J.; Berry, G.T. Marked elevation of urinary 3-hydroxydecanedioic acid in a malnourished infant with glycogen storage disease, mimicking long-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency. J. Inherit. Metab. Dis. 1993, 16, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Turi, K.N.; Romick-Rosendale, L.; Ryckman, K.K.; Hartert, T.V. A review of metabolomics approaches and their application in identifying causal pathways of childhood asthma. J. Allergy Clin. Immunol. 2018, 141, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- McGeachie, M.J.; Dahlin, A.; Qiu, W.; Croteau-Chonka, D.C.; Savage, J.; Wu, A.C.; Wan, E.S.; Sordillo, J.E.; Al-Garawi, A.; Martinez, F.D.; et al. The metabolomics of asthma control: A promising link between genetics and disease. Immun. Inflamm. Dis. 2015, 3, 224–238. [Google Scholar] [CrossRef]
- Dennis, E.A.; Norris, P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015, 15, 511–523. [Google Scholar] [CrossRef] [Green Version]
- Calder, P.C. Omega-3 polyunsaturated fatty acids and inflammatory processes: Nutrition or pharmacology? Br. J. Clin. Pharmacol. 2013, 75, 645–662. [Google Scholar] [CrossRef]
- Reinke, S.N.; Gallart-Ayala, H.; Gómez, C.; Checa, A.; Fauland, A.; Naz, S.; Kamleh, M.A.; Djukanović, R.; Hinks, T.S.C.; Wheelock, C.E. Metabolomics analysis identifies different metabotypes of asthma severity. Eur. Respir. J. 2017, 49, 1601740. [Google Scholar] [CrossRef] [Green Version]
- Chawes, B.L.K.; Stokholm, J.; Bønnelykke, K.; Brix, S.; Bisgaard, H. Neonates with reduced neonatal lung function have systemic low-grade inflammation. J. Allergy Clin. Immunol. 2015, 135, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Bisgaard, H.; Pipper, C.B.; Bønnelykke, K. Endotyping early childhood asthma by quantitative symptom assessment. J. Allergy Clin. Immunol. 2011, 127, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Fiehn, O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2010, 2, 23–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holcapek, M.; Jirásko, R.; Lísa, M. Basic rules for the interpretation of atmospheric pressure ionization mass spectra of small molecules. J. Chromatogr. A 2010, 1217, 3908–3921. [Google Scholar] [CrossRef] [PubMed]
- Chawes, B.L.; Bønnelykke, K.; Stokholm, J.; Vissing, N.H.; Bjarnadóttir, E.; Schoos, A.-M.M.; Wolsk, H.M.; Pedersen, T.M.; Vinding, R.K.; Thorsteinsdóttir, S.; et al. Effect of vitamin D3 supplementation during pregnancy on risk of persistent wheeze in the offspring: A randomized clinical trial. JAMA 2016, 315, 353–361. [Google Scholar] [CrossRef]
- Bisgaard, H.; Stokholm, J.; Chawes, B.L.; Vissing, N.H.; Bjarnadóttir, E.; Schoos, A.-M.M.; Wolsk, H.M.; Pedersen, T.M.; Vinding, R.K.; Thorsteinsdóttir, S.; et al. Fish oil-derived fatty acids in pregnancy and wheeze and asthma in offspring. N. Engl. J. Med. 2016, 375, 2530–2539. [Google Scholar] [CrossRef]
- Bisgaard, H.; Hermansen, M.N.; Loland, L.; Halkjaer, L.B.; Buchvald, F. Intermittent inhaled corticosteroids in infants with episodic wheezing. N. Engl. J. Med. 2006, 354, 1998–2005. [Google Scholar] [CrossRef]
| Metabolite ID | Higher in asthma/wheeze 3 | t-Test p-Value | Mann–Whitney Test p-Value | AUC 4 | |
|---|---|---|---|---|---|
| Rt 2 | Mass | ||||
| 2.38 | 425.0667 m/z | N > Y | 8.1 × 10-11 | 3.9 × 10-7 | 0.849 |
| 1.77 | 215.0312 m/z | N > Y | 1.7 × 10-9 | 1.2 × 10-7 | 0.864 |
| 1.77 | 102.0554 m/z | N > Y | 7.9 × 10-7 | 5.3 × 10-4 | 0.739 |
| 4.98 | 394.1348 m/z | Y > N | 1.2 × 10-5 | 1.0 × 10-3 | 0.726 |
| 2.77 | 278.1021 m/z | Y > N | 1.3 × 10-5 | 9.2 × 10-6 | 0.805 |
| 0.85 | 187.0604 m/z | Y > N | 3.3 × 10-5 | 8.1 × 10-4 | 0.731 |
| 3.87 | 506.2357 n 5 | Y > N | 3.8 × 10-5 | 2.6 × 10-3 | 0.707 |
| 1.92 | 232.0570 m/z | Y > N | 2.0 × 10-4 | 3.6 × 10-3 | 0.700 |
| 0.77 | 266.6361 n | N > Y | 3.1 × 10-4 | 1.9 × 10-4 | 0.757 |
| 0.77 | 615.2553 m/z | N > Y | 6.0 × 10-4 | 6.0 × 10-4 | 0.736 |
| 1.93 | 91.0519 n | Y > N | 6.1 × 10-4 | 4.3 × 10-3 | 0.696 |
| 2.63 | 240.0830 n | N > Y | 6.6 × 10-4 | 1.1 × 10-3 | 0.725 |
| 1.81 | 322.2205 n | Y > N | 8.0 × 10-4 | 3.4 × 10-3 | 0.701 |
| 4.95 | 520.1083 n | N > Y | 8.6 × 10-4 | 1.9 × 10-3 | 0.714 |
| 1.80 | 394.1729 n | Y > N | 9.4 × 10-4 | 1.0 × 10-3 | 0.726 |
| 0.77 | 614.2596 m/z | N > Y | 1.4 × 10-3 | 2.5 × 10-3 | 0.708 |
| 1.92 | 308.1753 n | Y > N | 1.4 × 10-3 | 8.2 × 10-3 | 0.682 |
| 4.95 | 472.0821 n | N > Y | 2.4 × 10-3 | 1.4 × 10-3 | 0.720 |
| 4.95 | 425.0693 m/z | N > Y | 2.9 × 10-3 | 5.5 × 10-3 | 0.691 |
| 1.80 | 417.1577 m/z | Y > N | 4.1 × 10-3 | 6.7 × 10-3 | 0.687 |
| 3.10 | 200.0651 m/z | Y > N | 4.4 × 10-3 | 5.0 × 10-3 | 0.693 |
| 1.90 | 277.1282 m/z | Y > N | 4.7 × 10-3 | 3.1 × 10-3 | 0.703 |
| 1.86 | 376.1704 n | Y > N | 5.8 × 10-3 | 1.1 × 10-3 | 0.725 |
| 1.94 | 249.1164 n | Y > N | 5.9 × 10-3 | 7.4 × 10-3 | 0.684 |
| 1.84 | 384.1464 m/z | Y > N | 1.5 × 10-2 | 6.5 × 10-3 | 0.687 |
| 1.93 | 238.1074 m/z | Y > N | 2.2 × 10-2 | 7.4 × 10-3 | 0.684 |
| 2.34 | 320.0695 n | N > Y | 2.6 × 10-2 | 5.6 × 10-3 | 0.691 |
| Metabolite ID | Higher in asthma/wheeze 3 | t-Test p-Value | Mann–Whitney Test p-Value | AUC 4 | |
|---|---|---|---|---|---|
| Rt 2 | Mass | ||||
| 0.83 | 197.0309 m/z | N > Y | 5.7 × 10-9 | 7.9 × 10-7 | 0.745 |
| 3.34 | 579.2516 n 5 | Y > N | 5.2 × 10-6 | 2.8 × 10-4 | 0.680 |
| 3.18 | 287.6108 m/z | Y > N | 8.9 × 10-6 | 3.0 × 10-5 | 0.707 |
| 0.75 | 526.1535 n | Y > N | 3.0 × 10-5 | 8.2 × 10-4 | 0.666 |
| 0.77 | 266.6361 n | N > Y | 4.8 × 10-5 | 1.0 × 10-3 | 0.663 |
| 0.97 | 113.0466 n | Y > N | 6.6 × 10-5 | 7.7 × 10-5 | 0.696 |
| 0.97 | 273.1696 m/z | Y > N | 2.9 × 10-4 | 1.3 × 10-3 | 0.659 |
| 0.75 | 524.1340 n | Y > N | 3.5 × 10-4 | 7.7 × 10-4 | 0.666 |
| 2.51 | 296.0547 m/z | Y > N | 3.7 × 10-4 | 7.2 × 10-4 | 0.668 |
| 2.42 | 507.2270 m/z | N > Y | 4.7 × 10-4 | 5.5 × 10-3 | 0.637 |
| 1.86 | 376.1704 n | Y > N | 5.2 × 10-4 | 7.6 × 10-3 | 0.632 |
| 1.82 | 310.1155 m/z | Y > N | 6.8 × 10-4 | 1.1 × 10-3 | 0.661 |
| 4.88 | 1022.5875 m/z | N > Y | 6.9 × 10-4 | 2.8 × 10-3 | 0.648 |
| 4.22 | 568.1558 m/z | N > Y | 7.1 × 10-4 | 3.0 × 10-3 | 0.647 |
| 6.82 | 298.0351 m/z | N > Y | 7.7 × 10-4 | 1.7 × 10-3 | 0.656 |
| 1.19 | 365.1172 m/z | N > Y | 1.1 × 10-3 | 4.5 × 10-3 | 0.641 |
| 4.88 | 938.6252 m/z | N > Y | 1.2 × 10-3 | 3.7 × 10-3 | 0.644 |
| 2.55 | 185.0551 m/z | Y > N | 1.2 × 10-3 | 5.9 × 10-3 | 0.636 |
| 1.78 | 159.0757 n | N > Y | 1.2 × 10-3 | 2.2 × 10-3 | 0.652 |
| 0.76 | 509.1502 m/z | Y > N | 1.4 × 10-3 | 7.5 × 10-3 | 0.633 |
| 6.84 | 268.1436 n | N > Y | 1.6 × 10-3 | 3.0 × 10-3 | 0.647 |
| 0.75 | 254.0712 m/z | Y > N | 2.4 × 10-3 | 9.5 × 10-3 | 0.628 |
| 4.10 | 399.0603 m/z | Y > N | 4.2 × 10-3 | 8.7 × 10-3 | 0.630 |
| 0.99 | 626.2139 m/z | Y > N | 4.8 × 10-3 | 7.6 × 10-3 | 0.632 |
| 0.76 | 561.2416 n | Y > N | 4.9 × 10-3 | 2.1 × 10-3 | 0.652 |
| 0.76 | 269.0584 m/z | Y > N | 5.4 × 10-3 | 8.7 × 10-3 | 0.630 |
| 2.25 | 90.0214 n | N > Y | 6.3 × 10-3 | 4.3 × 10-3 | 0.641 |
| 0.75 | 253.5653 m/z | Y > N | 1.1 × 10-2 | 7.2 × 10-3 | 0.633 |
| 4.87 | 633.7972 n | N > Y | 1.3 × 10-2 | 8.5 × 10-3 | 0.630 |
| 5.07 | 307.1007 m/z | Y > N | 1.4 × 10-2 | 5.7 × 10-3 | 0.637 |
| Metabolite ID | Higher in asthma/wheeze 3 | COPSAC2000 4 | COPSAC2010 4 | |
|---|---|---|---|---|
| Rt 2 | Mass | |||
| 0.83 | 197.0309 m/z | N > Y | 2.5 × 10-4 | 7.9 × 10-7 |
| 4.95 | 520.1083 n 5 | N > Y | 1.9 × 10-3 | 1.8 × 10-4 |
| 2.63 | 240.0830 n | N > Y | 1.1 × 10-3 | 5.9 × 10-4 |
| 2.62 | 323.1058 m/z | N > Y | 1.3 × 10-6 | 6.2 × 10-4 |
| 0.77 | 266.6361 n | N > Y | 1.9 × 10-4 | 1.0 × 10-3 |
| 4.95 | 425.0693 m/z | N > Y | 5.5 × 10-3 | 1.7 × 10-3 |
| 4.95 | 472.0821 n | N > Y | 1.4 × 10-3 | 3.3 × 10-3 |
| 2.34 | 320.0695 n | N > Y | 5.6 × 10-3 | 4.0 × 10-3 |
| 2.84 | 671.1053 m/z | N > Y | 1.3 × 10-4 | 8.1 × 10-3 |
| 2.84 | 503.0769 m/z | N > Y | 1.3 × 10-4 | 8.9 × 10-3 |
| 2.02 | 314.0735 m/z | Y > N | 9.4 × 10-4 | 6.9 × 10-4 |
| 5.07 | 480.2963 m/z | Y > N | 7.8 × 10-4 | 4.2 × 10-3 |
| 0.95 | 611.2039 m/z | Y > N | 8.2 × 10-4 | 4.1 × 10-3 |
| 1.86 | 376.1704 n | Y > N | 1.1 × 10-3 | 7.6 × 10-3 |
| Dataset | Metabolite ID | N2 | |
|---|---|---|---|
| Rt 1 | Mass | ||
| COPSAC2010 | 6.52 | 130.9831 m/z | 77 |
| COPSAC2010 | 6.46 | 164.9354 m/z | 91 |
| COPSAC2010 | 5.77 | 171.0055 m/z | 52 |
| COPSAC2010 | 0.83 | 197.0309 m/z | 81 |
| COPSAC2010 | 6.82 | 208.9162 m/z | 60 |
| COPSAC2010 | 6.75 | 239.0211 n 3 | 52 |
| COPSAC2010 | 6.51 | 272.9581 m/z | 91 |
| COPSAC2010 | 6.47 | 278.9172 m/z | 85 |
| COPSAC2010 | 6.68 | 288.9358 m/z | 81 |
| COPSAC2010 | 3.80 | 303.6059 m/z | 66 |
| COPSAC2010 | 6.69 | 304.9131 m/z | 75 |
| COPSAC2010 | 6.69 | 273.9368 n | 88 |
| COPSAC2010 | 6.40 | 279.8984 n | 74 |
| COPSAC2010 | 6.52 | 327.0678 m/z | 77 |
| COPSAC2010 | 4.95 | 329.1580 n | 54 |
| COPSAC2010 | 6.65 | 357.0611 m/z | 86 |
| COPSAC2010 | 6.83 | 386.1231 n | 56 |
| COPSAC2010 | 6.65 | 372.0835 n | 84 |
| COPSAC2010 | 6.75 | 368.9785 n | 70 |
| COPSAC2010 | 6.69 | 434.8710 m/z | 89 |
| COPSAC2010 | 3.31 | 452.3010 m/z | 72 |
| COPSAC2010 | 6.62 | 472.2043 m/z | 56 |
| COPSAC2010 | 5.07 | 480.2963 m/z | 84 |
| COPSAC2010 | 2.51 | 534.1231 m/z | 55 |
| COPSAC2010 | 3.94 | 544.2579 m/z | 55 |
| COPSAC2010 | 4.95 | 520.1083 n | 52 |
| COPSAC2010 | 3.34 | 579.2516 n | 57 |
| COPSAC2010 | 0.94 | 595.2081 m/z | 70 |
| COPSAC2010 | 0.95 | 611.2039 m/z | 70 |
| COPSAC2010 | 6.65 | 305.0493 n | 79 |
| COPSAC2010 | 6.59 | 888.2750 m/z | 84 |
| COPSAC2010 | 6.40 | 891.1497 m/z | 52 |
| COPSAC2010 | 6.58 | 915.2940 m/z | 70 |
| COPSAC2010 | 2.84 | 1007.1621 m/z | 56 |
| COPSAC2010 | 2.84 | 1091.1767 m/z | 55 |
| COPSAC2000 | 1.77 | 215.0312 m/z | 80 |
| COPSAC2000 | 5.07 | 480.2963 m/z | 50 |
| COPSAC2000 | 2.84 | 503.0769 m/z | 53 |
| COPSAC2000 | 0.94 | 595.2081 m/z | 63 |
| COPSAC2000 | 0.51 | 655.0471 m/z | 51 |
| COPSAC2000 | 2.84 | 671.1053 m/z | 59 |
| COPSAC2000 | 2.83 | 839.1337 m/z | 70 |
| COPSAC2000 | 2.84 | 923.1478 m/z | 69 |
| COPSAC2000 | 2.84 | 1007.1621 m/z | 83 |
| COPSAC2000 | 2.84 | 1091.1767 m/z | 74 |
| COPSAC2000 | 2.83 | 1175.1912 m/z | 79 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chawes, B.L.; Giordano, G.; Pirillo, P.; Rago, D.; Rasmussen, M.A.; Stokholm, J.; Bønnelykke, K.; Bisgaard, H.; Baraldi, E. Neonatal Urine Metabolic Profiling and Development of Childhood Asthma. Metabolites 2019, 9, 185. https://doi.org/10.3390/metabo9090185
Chawes BL, Giordano G, Pirillo P, Rago D, Rasmussen MA, Stokholm J, Bønnelykke K, Bisgaard H, Baraldi E. Neonatal Urine Metabolic Profiling and Development of Childhood Asthma. Metabolites. 2019; 9(9):185. https://doi.org/10.3390/metabo9090185
Chicago/Turabian StyleChawes, Bo L., Giuseppe Giordano, Paola Pirillo, Daniela Rago, Morten A. Rasmussen, Jakob Stokholm, Klaus Bønnelykke, Hans Bisgaard, and Eugenio Baraldi. 2019. "Neonatal Urine Metabolic Profiling and Development of Childhood Asthma" Metabolites 9, no. 9: 185. https://doi.org/10.3390/metabo9090185
APA StyleChawes, B. L., Giordano, G., Pirillo, P., Rago, D., Rasmussen, M. A., Stokholm, J., Bønnelykke, K., Bisgaard, H., & Baraldi, E. (2019). Neonatal Urine Metabolic Profiling and Development of Childhood Asthma. Metabolites, 9(9), 185. https://doi.org/10.3390/metabo9090185






