Simulated Docking Predicts Putative Channels for the Transport of Long-Chain Fatty Acids in Vibrio cholerae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selecting the Vibrio cholerae Homologs
2.2. Generating the FadL Tertiary Structures
2.3. Generating the Membrane System
2.4. Equilibrating the Membrane Systems
2.5. The Docking of the FadL Proteins
3. Results
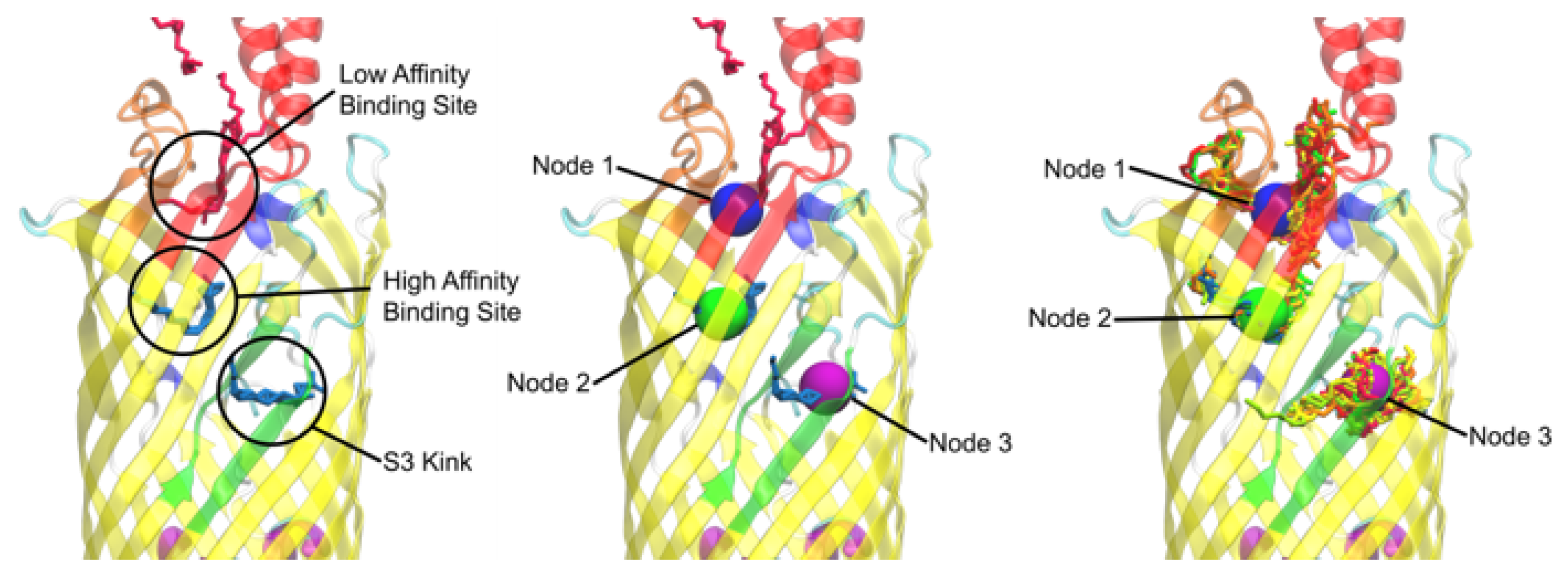
3.1. Docking Showed Viability between the Crystal Structure and Simulated results
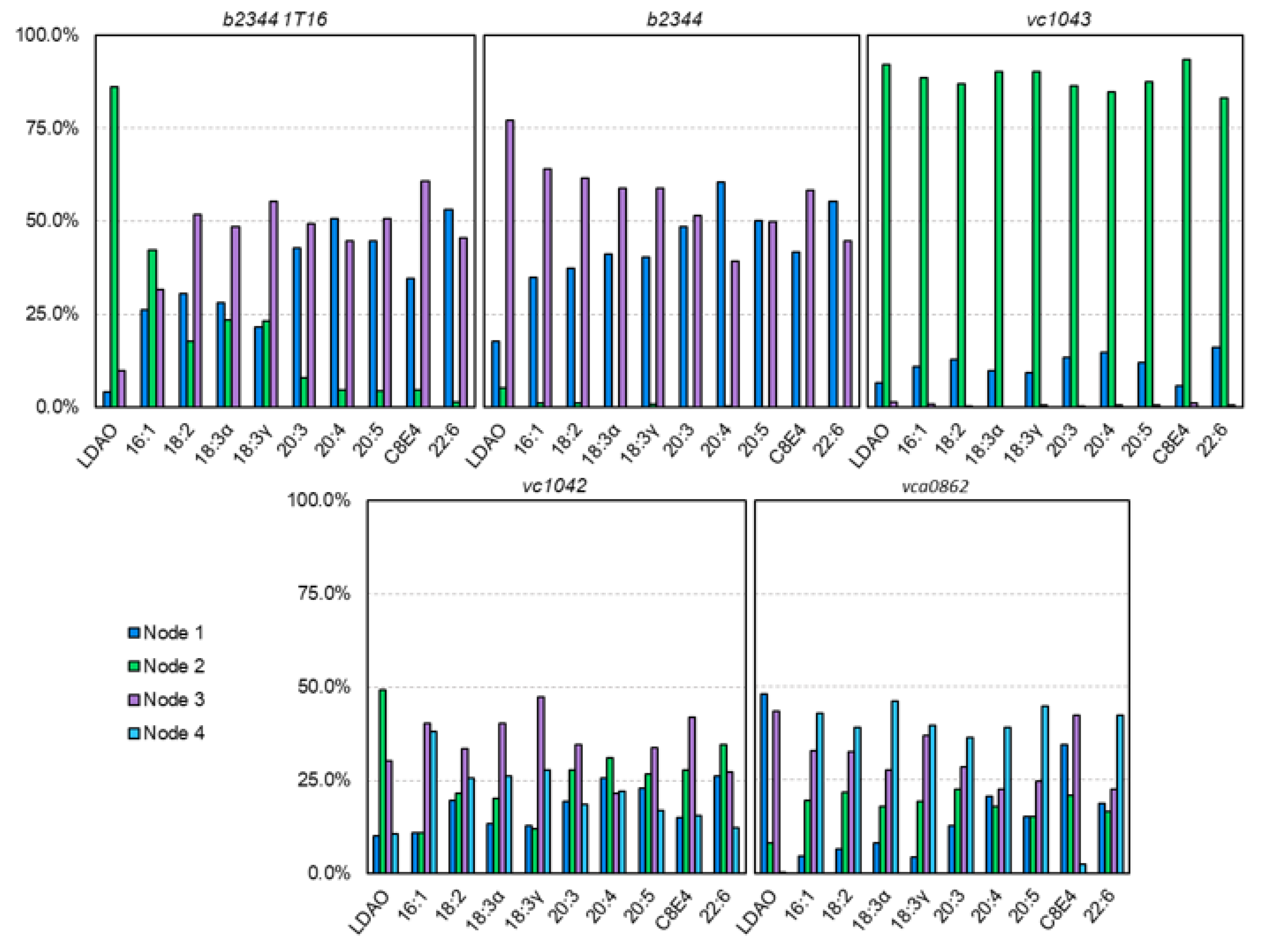
3.2. Nodal Cluster Analysis Shows Agreement with Experimental Studies
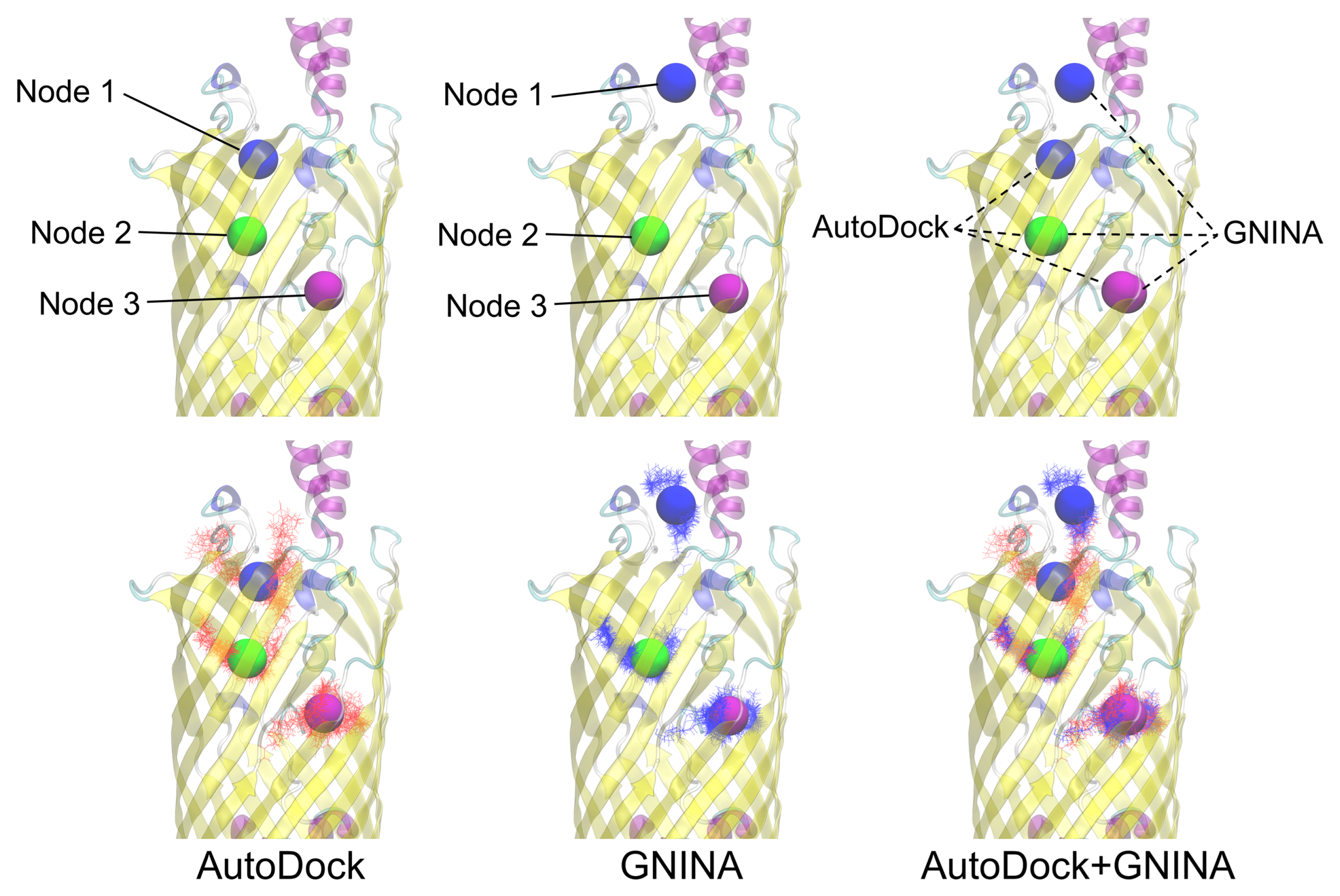
3.3. AutoDock Docking Compared to GNINA Docking
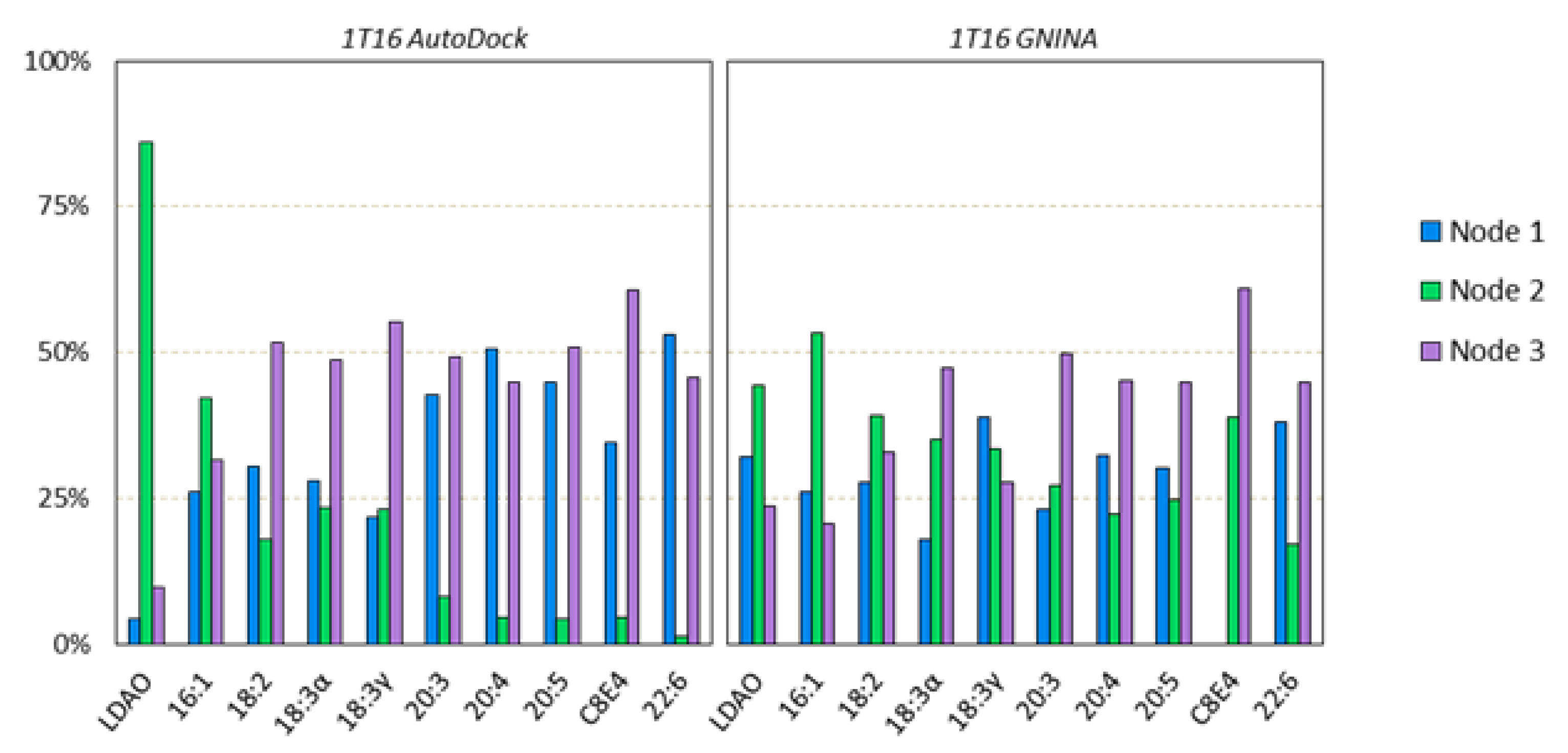
3.4. Nodal Cluster Analysis Shows Ligand Preference for Certain Binding Sites
3.5. The Nodal Analysis Shows Preference for Certain Node Loci
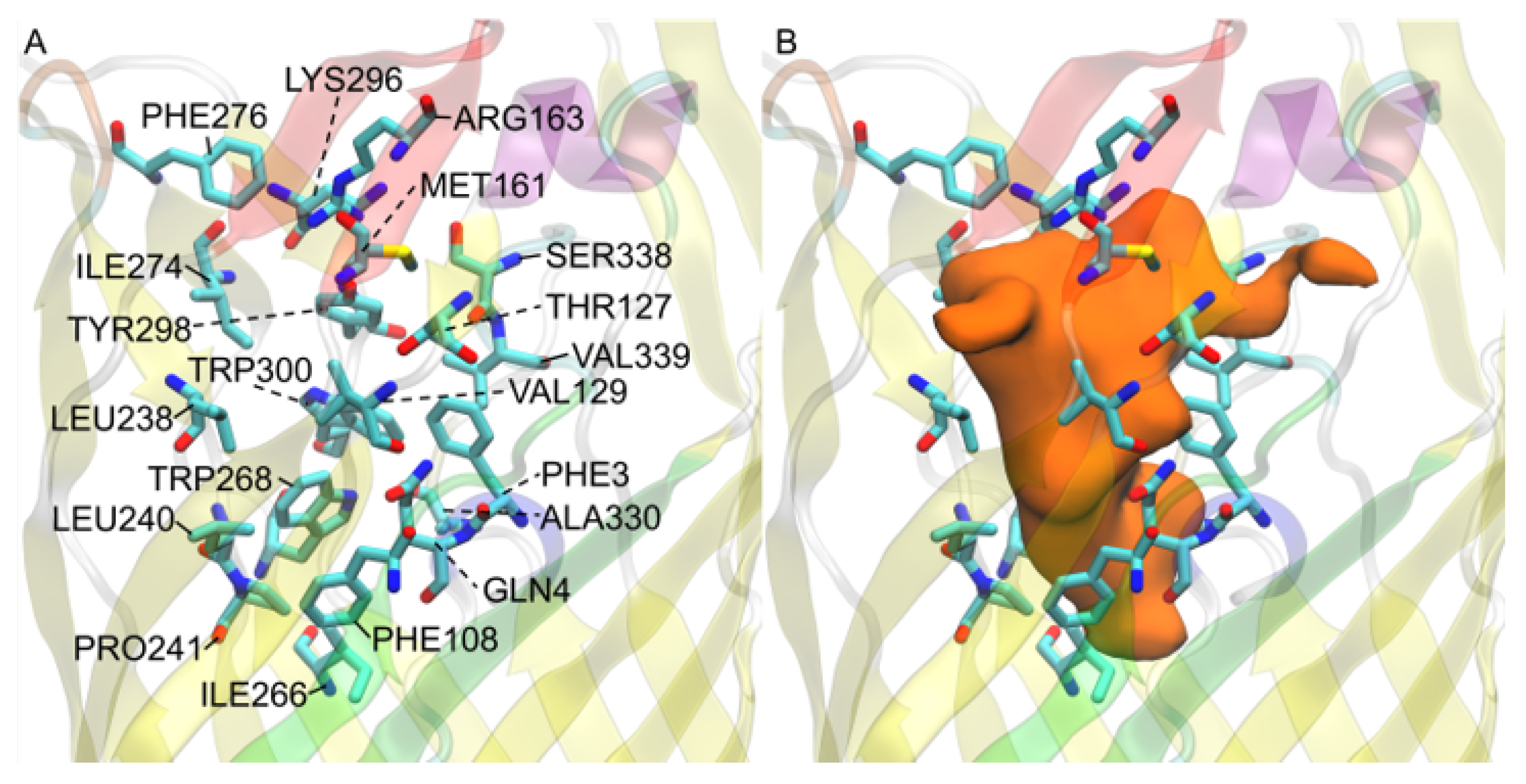
3.6. Eqiulibrated E. coli b2344 Fatty Acids show Similar Channels as the X-ray Crystal Structure 1T16
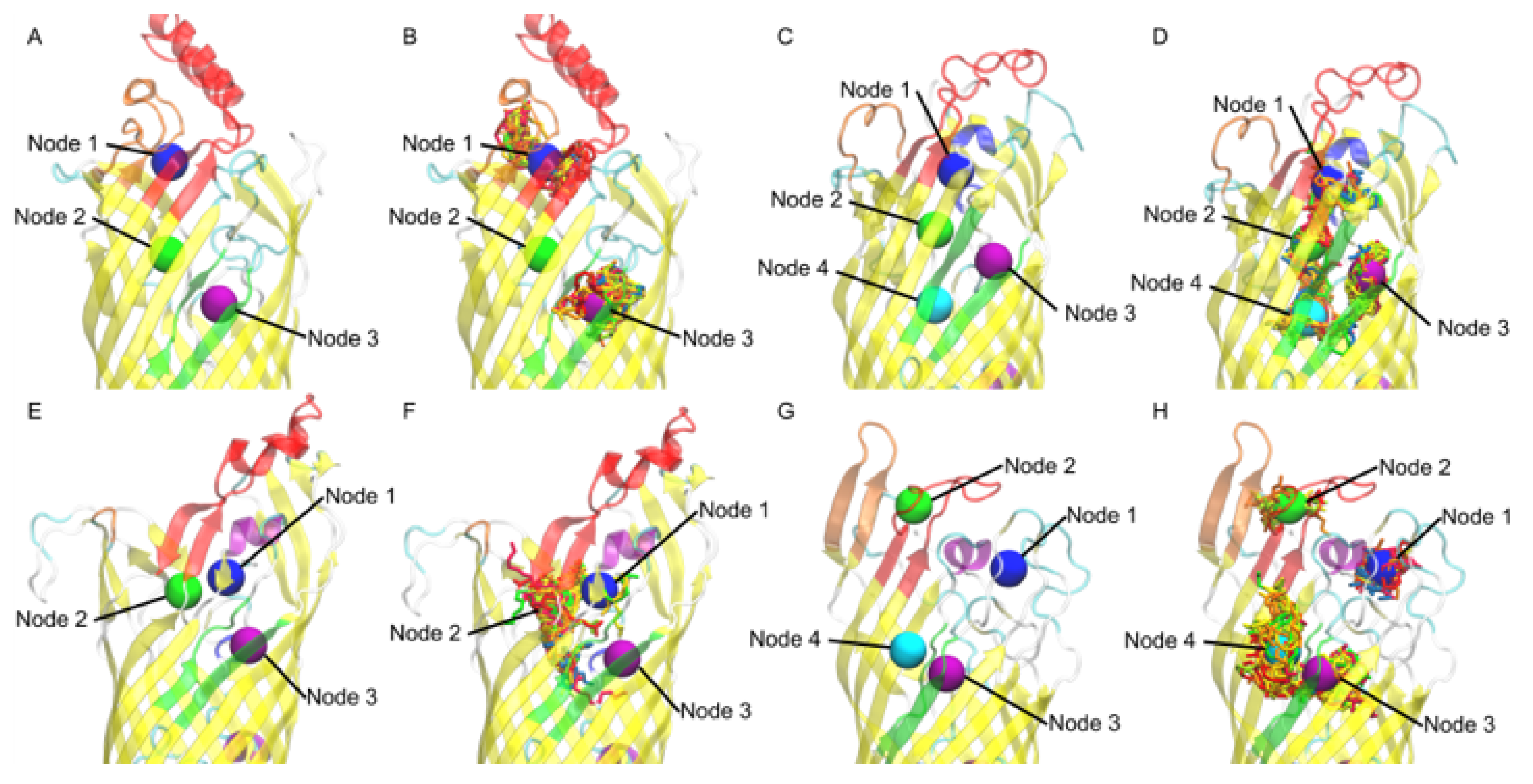
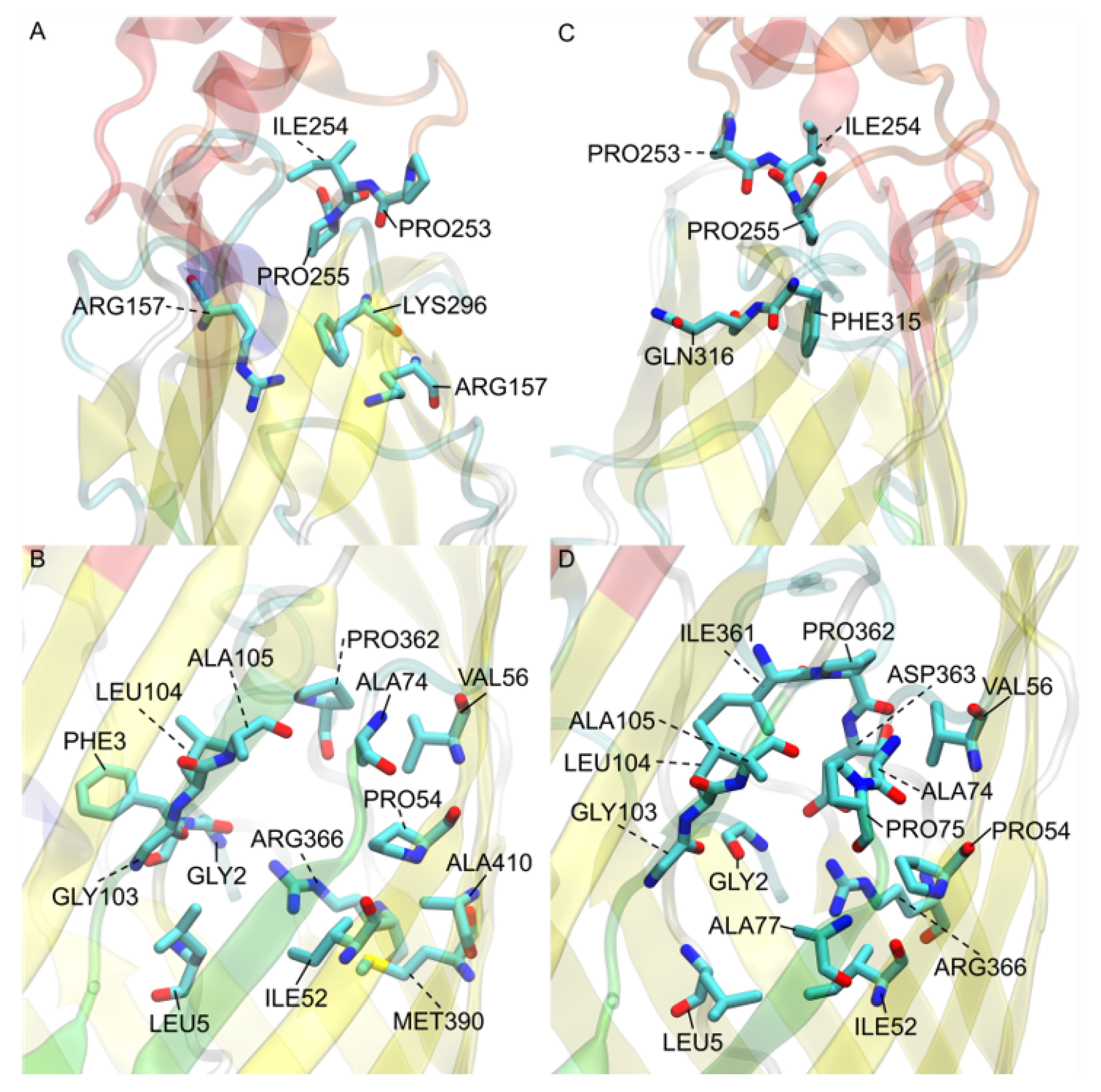
3.7. V. cholerae vc1042 Docking Generates a Well Defined Transport Channel
3.8. V. cholerae vc1043 Transport Channel Has a Discontinuity Revealing Multiple Predicted Pathways
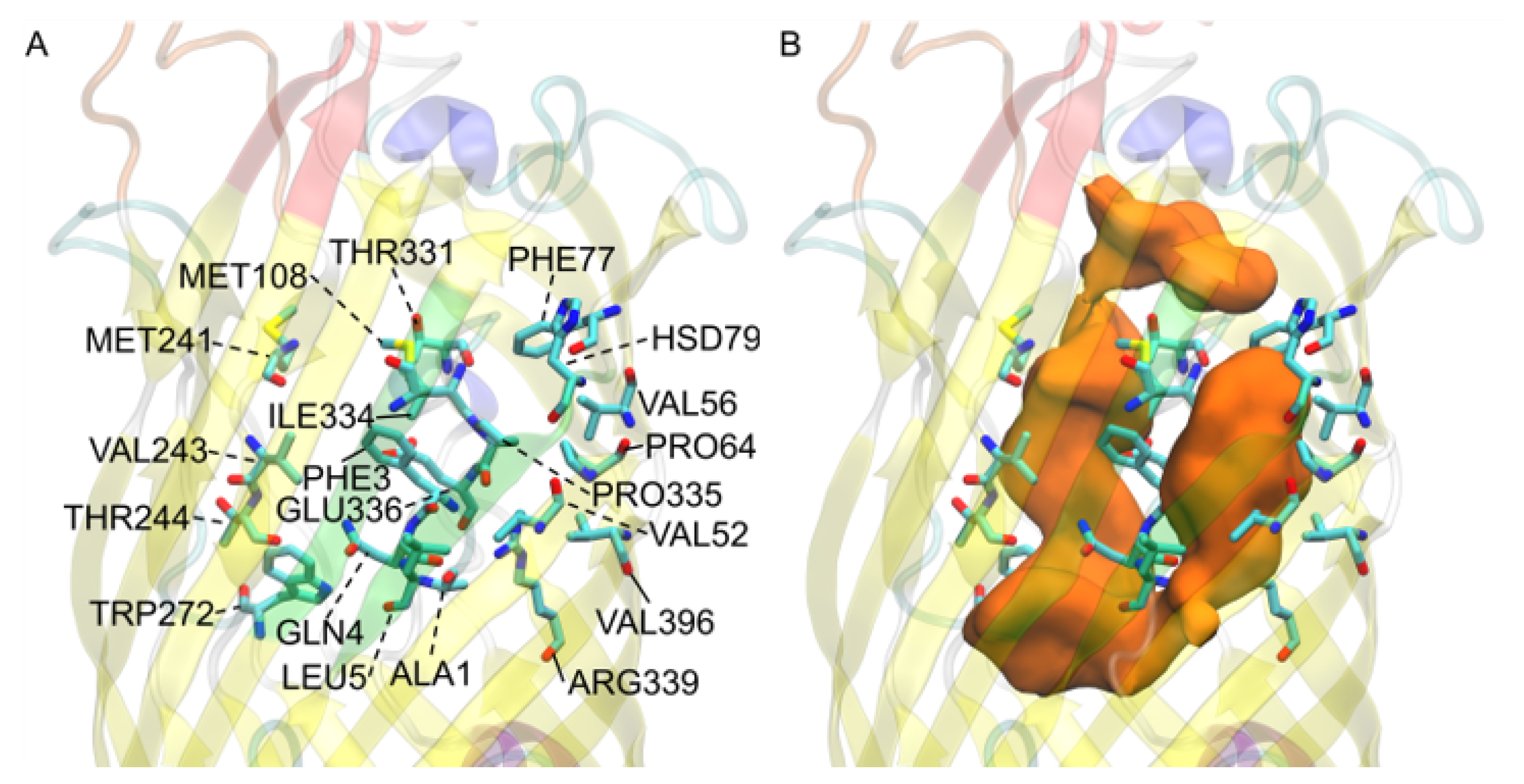
3.9. V. cholerae vca0862 Docking Reveals a Transport Channel External to the Beta Barrel
3.10. Simulations and Docking Agree the S3 Kink Is the Fatty Acid Point of Egress
3.11. Docking Energies Show That Binding Is Stronger in the Presence of Fatty Acids
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nunn, W.D. A molecular view of fatty acid catabolism in Escherichia coli. Microbiol. Rev. 1986, 50, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Nunn, W.D.; Simons, R.W. Transport of long-chain fatty acids by Escherichia coli: Mapping and characterization of mutants in the fadL gene. Proc. Natl. Acad. Sci. USA 1978, 75, 3377–3381. [Google Scholar] [CrossRef] [PubMed]
- Black, P.N.; Said, B.; Ghosn, C.R.; Beach, J.V.; Nunn, W.D. Purification and characterization of an outer membrane-bound protein involved in long-chain fatty acid transport in Escherichia coli. J. Biol. Chem. 1987, 262, 1412–1419. [Google Scholar] [CrossRef]
- de Carvalho, C.; Caramujo, M.J. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef]
- Abedi, E.; Sahari, M.A. Long-chain polyunsaturated fatty acid sources and evaluation of their nutritional and functional properties. Food Sci. Nutr. 2014, 2, 443–463. [Google Scholar] [CrossRef]
- Leonard, A.E.; Pereira, S.L.; Sprecher, H.; Huang, Y.S. Elongation of long-chain fatty acids. Prog. Lipid Res. 2004, 43, 36–54. [Google Scholar] [CrossRef]
- Berge, J.P.; Barnathan, G. Fatty acids from lipids of marine organisms: Molecular biodiversity, roles as biomarkers, biologically active compounds, and economical aspects. Adv. Biochem. Eng. Biotechnol. 2005, 96, 49–125. [Google Scholar] [CrossRef]
- Harwood, J.L.; Guschina, I.A. The versatility of algae and their lipid metabolism. Biochimie 2009, 91, 679–684. [Google Scholar] [CrossRef]
- Jimenez-Diaz, L.; Caballero, A.; Segura, A. Pathways for the Degradation of Fatty Acids in Bacteria. In Aerobic Utilization of Hydrocarbons, Oils and Lipids; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–23. [Google Scholar] [CrossRef]
- Ernst, R.; Ejsing, C.S.; Antonny, B. Homeoviscous Adaptation and the Regulation of Membrane Lipids. J. Mol. Biol. 2016, 428, 4776–4791. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [Google Scholar] [CrossRef]
- Cronan, J.E.; Thomas, J. Bacterial Fatty Acid Synthesis and its Relationships with Polyketide Synthetic Pathways. Methods Enzym. 2009, 459, 395–433. [Google Scholar] [CrossRef]
- Parsons, J.B.; Rock, C.O. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr. Opin. Microbiol. 2011, 14, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Rock, C.O. Exogenous fatty acid metabolism in bacteria. Biochimie 2017, 141, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Ginsburgh, C.L.; Black, P.N.; Nunn, W.D. Transport of long chain fatty acids in Escherichia coli. Identification of a membrane protein associated with the fadL gene. J. Biol. Chem. 1984, 13, 8437–8443. [Google Scholar] [CrossRef]
- Weimar, J.D.; DiRusso, C.C.; Delio, R.; Black, P.N. Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. Amino acid residues within the ATP/AMP signature motif of Escherichia coli FadD are required for enzyme activity and fatty acid transport. J. Biol. Chem. 2002, 277, 29369–29376. [Google Scholar] [CrossRef]
- Giles, D.K.; Hankins, J.V.; Guan, Z.; Trent, M.S. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol. Microbiol. 2011, 79, 716–728. [Google Scholar] [CrossRef]
- Hobby, C.R.; Herndon, J.L.; Morrow, C.A.; Peters, R.E.; Symes, S.J.K.; Giles, D.K. Exogenous fatty acids alter phospholipid composition, membrane permeability, capacity for biofilm formation, and antimicrobial peptide susceptibility in Klebsiella pneumoniae. Microbiologyopen 2019, 8, e00635. [Google Scholar] [CrossRef]
- Eder, A.E.; Munir, S.A.; Hobby, C.R.; Anderson, D.M.; Herndon, J.L.; Siv, A.W.; Symes, S.J.K.; Giles, D.K. Exogenous polyunsaturated fatty acids (PUFAs) alter phospholipid composition, membrane permeability, biofilm formation and motility in Acinetobacter baumannii. Microbiology 2017, 163, 1626–1636. [Google Scholar] [CrossRef]
- Baker, L.Y.; Hobby, C.R.; Siv, A.W.; Bible, W.C.; Glennon, M.S.; Anderson, D.M.; Symes, S.J.; Giles, D.K. Pseudomonas aeruginosa responds to exogenous polyunsaturated fatty acids (PUFAs) by modifying phospholipid composition, membrane permeability, and phenotypes associated with virulence. BMC Microbiol. 2018, 18, 117. [Google Scholar] [CrossRef] [Green Version]
- Herndon, J.L.; Peters, R.E.; Hofer, R.N.; Simmons, T.B.; Symes, S.J.; Giles, D.K. Exogenous polyunsaturated fatty acids (PUFAs) promote changes in growth, phospholipid composition, membrane permeability and virulence phenotypes in Escherichia coli. BMC Microbiol. 2020, 20, 305. [Google Scholar] [CrossRef]
- Moravec, A.R.; Siv, A.W.; Hobby, C.R.; Lindsay, E.N.; Norbash, L.V.; Shults, D.J.; Symes, S.J.K.; Giles, D.K. Exogenous Polyunsaturated Fatty Acids Impact Membrane Remodeling and Affect Virulence Phenotypes among Pathogenic Vibrio Species. Appl. Environ. Microbiol. 2017, 83, e01415-17. [Google Scholar] [CrossRef] [PubMed]
- van Berge Henegouwen, G.P.; van der Werf, S.D.J.; Ruben, A.T. Fatty acid composition of phospholipids in bile in man: Promoting effect of deoxycholate on arachidonate. Clin. Chim. Acta 1987, 165, 27–37. [Google Scholar] [CrossRef]
- Pride, A.C.; Guan, Z.; Trent, M.S. Characterization of the Vibrio cholerae VolA surface-exposed lipoprotein lysophospholipase. J. Bacteriol. 2014, 196, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- van den Berg, B.; Black, P.N.; Clemons, W.M.J.; Rapoport, T.A. Crystal structure of the long-chain fatty acid transporter FadL. Science 2004, 304, 1506–1509. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Z’idek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Davila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.J.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: AnN log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- McNutt, A.T.; Francoeur, P.; Aggarwal, R.; Masuda, T.; Meli, R.; Ragoza, M.; Sunseri, J.; Koes, D.R. GNINA 1.0: Molecular docking with deep learning. J. Cheminform. 2021, 13, 43. [Google Scholar] [CrossRef]
- Hearn, E.M.; Patel, D.R.; Lepore, B.W.; Indic, M.; van den Berg, B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature 2009, 458, 367–370. [Google Scholar] [CrossRef]
- Black, P.N. Primary sequence of the Escherichia coli fadL gene encoding an outer membrane protein required for long-chain fatty acid transport. J. Bacteriol. 1991, 173, 435–442. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Xu, Q.; Dziejman, M.; Mekalanos, J.J. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 1286–1291. [Google Scholar] [CrossRef] [Green Version]
- Hottes, A.K.; Freddolino, P.L.; Khare, A.; Donnell, Z.N.; Liu, J.C.; Tavazoie, S. Bacterial adaptation through loss of function. PLoS Genet. 2013, 9, e1003617. [Google Scholar] [CrossRef]




| b2344 1T16 | b2344 | vc1042 | vc1043 | vca0862 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Proximity | Residue | Proximity | Residue | Proximity | Residue | Proximity | Residue | Proximity | Residue |
| 2958 | LEU104 | 2852 | LEU104 | 2654 | PHE3 | 4253 | PHE108 | 2627 | ALA102 |
| 2156 | ARG366 ** | 2705 | ALA105 | 2488 | GLN4 * | 3751 | PHE3 | 2522 | GLN74 * |
| 2083 | PHE3 | 2683 | ARG366 ** | 2075 | LEU5 | 3586 | TRP300 | 1761 | PRO72 |
| 2044 | PRO54 | 2651 | ALA74 | 1981 | MET108 | 3415 | TYR298 * | 1732 | SER119 * |
| 1977 | ALA74 | 2624 | PRO54 | 1797 | ARG339 ** | 3342 | GLN4* | 1716 | LYS151 ** |
| 1943 | ALA105 | 2331 | PRO362 | 1709 | PHE77 | 3249 | LEU238 | 1501 | VAL5 |
| 1910 | PRO362 | 2274 | ASP363 * | 1646 | PRO54 | 3182 | TRP268 | 1498 | VAL101 |
| 1849 | LEU5 | 2178 | GLY2 * | 1626 | VAL52 | 3074 | ARG163 ** | 1456 | VAL52 |
| 1789 | VAL56 | 2154 | LEU5 | 1568 | VAL396 | 2858 | MET161 | 1417 | VAL76 |
| 1680 | PHE315 | 2087 | ILE361 | 1538 | PRO335 | 2504 | ILE274 | 1401 | PRO54 |
| 1673 | PRO255 | 2078 | ILE52 | 1523 | VAL56 | 2277 | PRO241 | 1396 | ILE121 |
| 1639 | PRO253 | 2058 | GLY103 * | 1503 | THR331 * | 2187 | VAL129 | 1237 | ALA71 |
| 1626 | GLY2 * | 2049 | PRO255 | 1491 | TRP272 | 1697 | VAL339 | 1168 | ASP327 * |
| 1623 | ILE52 | 2006 | GLN316 * | 1462 | VAL243 | 1588 | LYS296 ** | 1137 | ILE75 |
| 1594 | GLY103 * | 1965 | PHE315 | 1418 | THR244 * | 1567 | PHE276 | 1035 | ARG330 ** |
| 1588 | ARG157 ** | 1923 | VAL56 | 1385 | GLU336 * | 1413 | ILE266 | 1019 | GLY100 * |
| 1580 | ILE254 | 1870 | ILE254 | 1384 | ALA1 | 1319 | LEU240 | 1013 | GLY2 * |
| 1526 | LYS317 ** | 1756 | PRO75 | 1268 | HSD79 ** | 1293 | THR127 * | 1004 | THR103 * |
| 1440 | MET390 | 1673 | PRO253 | 1231 | ILE334 | 1272 | SER338 * | 977 | ASP117 * |
| 1426 | ALA410 | 1643 | ALA77 | 1187 | MET241 | 1256 | ALA330 | 943 | ALA379 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turgeson, A.; Morley, L.; Giles, D.; Harris, B. Simulated Docking Predicts Putative Channels for the Transport of Long-Chain Fatty Acids in Vibrio cholerae. Biomolecules 2022, 12, 1269. https://doi.org/10.3390/biom12091269
Turgeson A, Morley L, Giles D, Harris B. Simulated Docking Predicts Putative Channels for the Transport of Long-Chain Fatty Acids in Vibrio cholerae. Biomolecules. 2022; 12(9):1269. https://doi.org/10.3390/biom12091269
Chicago/Turabian StyleTurgeson, Andrew, Lucas Morley, David Giles, and Bradley Harris. 2022. "Simulated Docking Predicts Putative Channels for the Transport of Long-Chain Fatty Acids in Vibrio cholerae" Biomolecules 12, no. 9: 1269. https://doi.org/10.3390/biom12091269





