Conservation of Glutathione Transferase mRNA and Protein Sequences Similar to Human and Horse Alpha Class GST A3-3 across Dog, Goat, and Opossum Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Testis Tissue Samples and RNA Preparation
2.2.2. Reverse Transcription
2.2.3. Cloning GSTA3 cDNAs into the Vector pCR2.1
2.2.4. Sequencing and Structure Analyses
2.2.5. Cloning GSTA3 cDNA in the Bacterial Expression Vector pET-21a(+)
2.2.6. Quantitative Reverse Transcription-PCR
3. Results
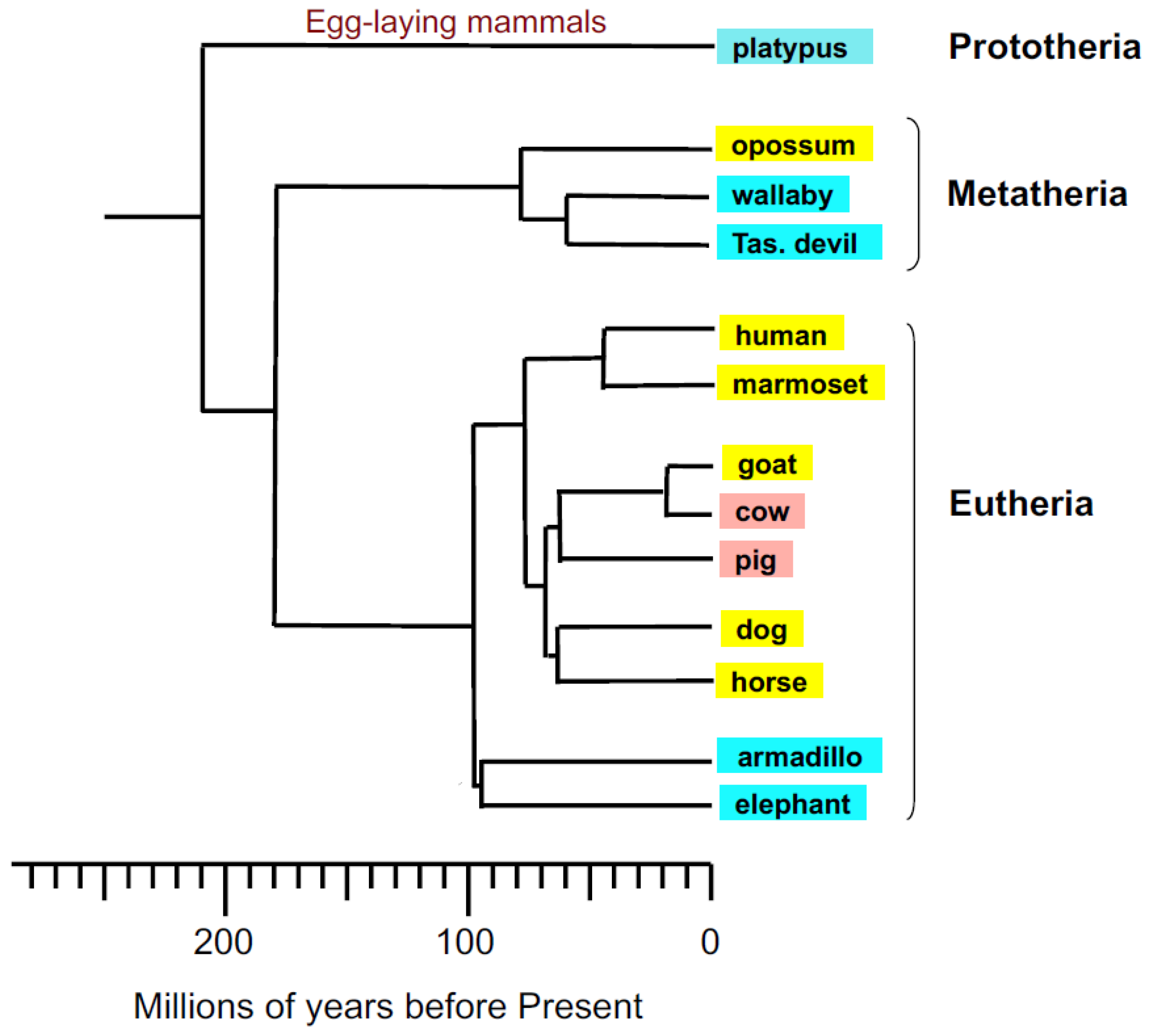
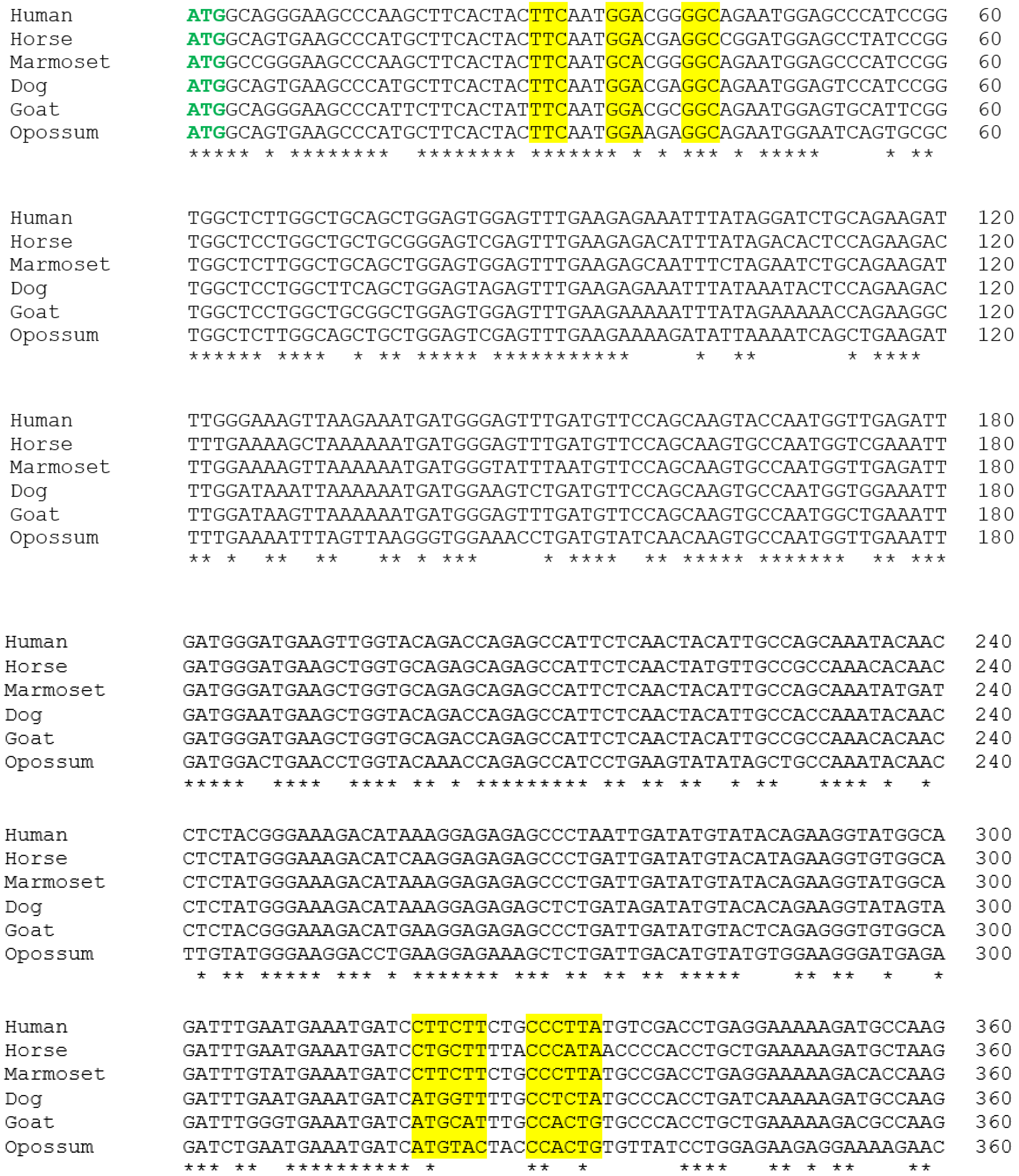
3.1. Alignment of the Cloned GSTA3 mRNA Sequences to Those of Human and Horse GSTA3 mRNAs
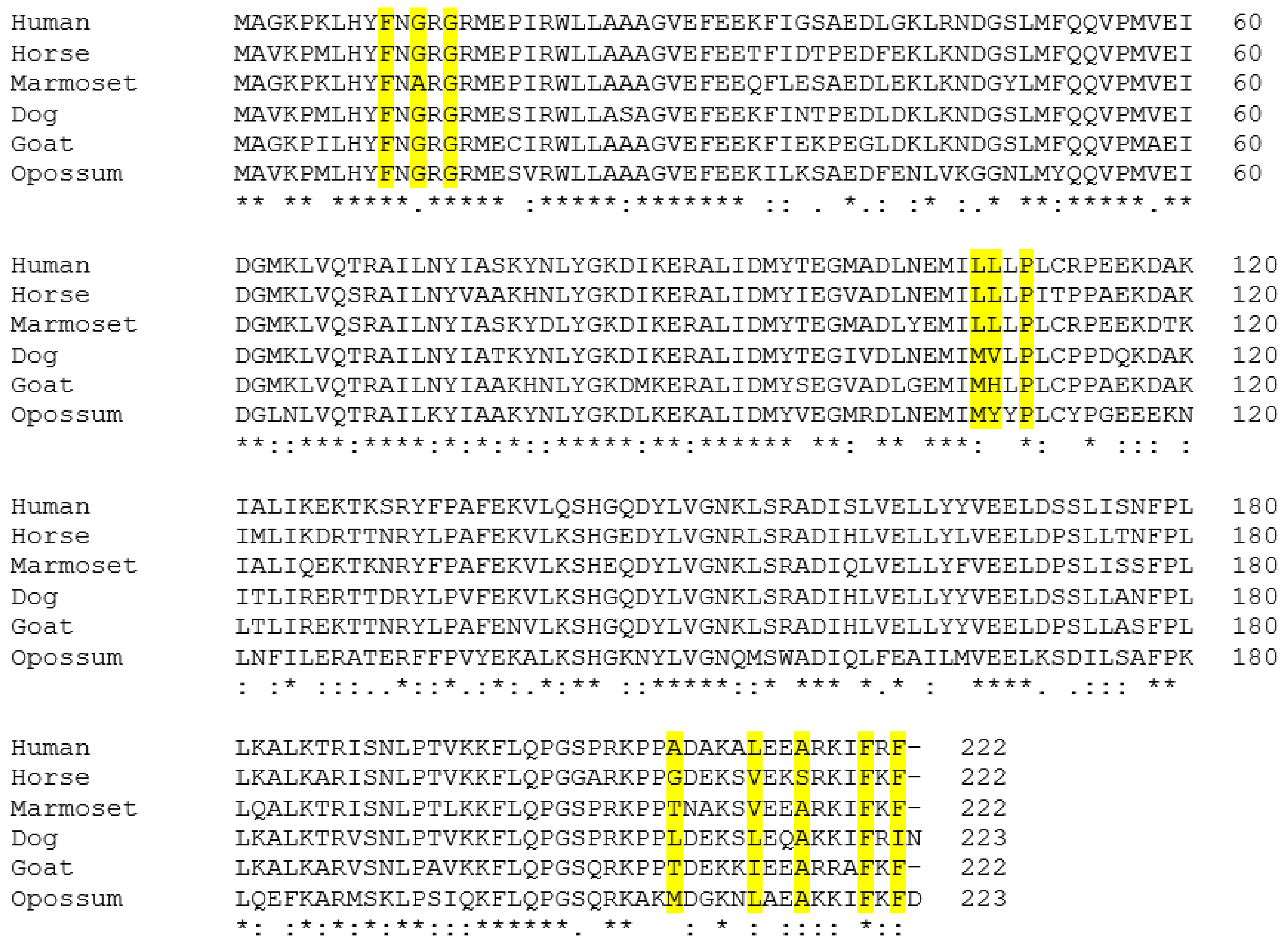
3.2. Alignment of the Predicted GSTA3 Protein Sequences from Six Mammals
3.3. Conservation of Amino Acids Critical for Steroid Isomerase Activity and Binding of the Cofactor Glutathione
3.4. Modeling the Structures of the Newly Described GST A3 Sequences
3.5. Expression of Dog GSTA3 mRNA in Dog Tissues
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diemern, T.; Hales, D.B.; Weidner, W. Immune-endocrine interactions and Leydig cell function: The role of cytokines. Andrologia 2003, 35, 55–63. [Google Scholar] [CrossRef]
- Payne, A.H.; Hales, D.B. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 2004, 25, 947–970. [Google Scholar] [CrossRef]
- Herrera-Luna, C.V.; Budik, S.; Aurich, C. Gene expression of ACTH, glucocorticoid receptors, 11BHSD enzymes, LH-, FSH-, GH receptors and aromatase in equine epididymal and testicular tissue. Reprod. Dom. Anim. 2012, 47, 928–935. [Google Scholar] [CrossRef]
- Ing, N.H. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of messenger RNAs. Biol. Reprod. 2005, 72, 1290–1296. [Google Scholar] [CrossRef]
- Lonard, D.M.; O’Malley, B.W. Nuclear receptor coregulators: Modulators of pathology and therapeutic targets. Nat. Rev. Endocrinol. 2012, 8, 598–604. [Google Scholar] [CrossRef]
- Agular, B.M.; Vinggaard, A.M.; Vind, C. Regulation by dexamethasone of the 3β-hydroxysteroid dehydrogenase activity in adult rat Leydig cells. J. Steroid Biochem. Mol. Biol. 1992, 43, 565–571. [Google Scholar] [CrossRef]
- Conley, A.J.; Bird, I.M. The role of cytochrome P450 17α-hydroxylase and 3ß-hydroxysteroid dehydrogenase in the integration of gonadal and adrenal steroidogenesis via the Δ5 and Δ4 pathways of steroidogenesis in mammals. Biol. Reprod. 1997, 56, 789–799. [Google Scholar] [CrossRef]
- Johansson, A.-S.; Mannervik, B. Human glutathione transferase A3-3, a highly efficient catalyst of double-bond isomerization in the biosynthetic pathway of steroid hormones. J. Biol. Chem. 2001, 276, 33061–33065. [Google Scholar] [CrossRef]
- Johansson, A.-S.; Mannervik, B. Active-site residues governing high steroid isomerase activity in human glutathione transferase A3-3. J. Biol. Chem. 2002, 277, 16648–16654. [Google Scholar] [CrossRef]
- Fedulova, N.; Raffalli-Mathieu, F.; Mannervik, B. Porcine glutathione transferase Alpha 2-2 is a human GST A3-3 analogue catalyzing steroid double-bond isomerization. J. Biochem. 2010, 431, 159–167. [Google Scholar] [CrossRef]
- Larsson, E.; Mannervik, B.; Raffalli-Mathieu, F. Quantitative and selective polymerase chain reaction analysis of highly similar human alpha-class glutathione transferases. Anal. Biochem. 2011, 412, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Calvaresi, M.; Stenta, M.; Garavelli, M.; Altoe, P.; Bottoni, A. Computational evidence for the catalytic mechanism of human glutathione transferase A3-3: A QM/MM investigation. ACS Catal. 2012, 2, 280–286. [Google Scholar] [CrossRef]
- Matsumura, T.; Imamichi, Y.; Mizutani, T.; Ju, Y.; Yazawa, T.; Kawabe, S.; Kanno, M.; Ayabe, T.; Katsumata, N.; Fukami, M.; et al. Human glutathione transferase A (GSTA) family genes are regulated by steroidogenic factor 1 (SF-1) and are involved in steroidogenesis. FASEB J. 2013, 27, 3198–3208. [Google Scholar] [CrossRef] [PubMed]
- Dourado, D.F.A.R.; Fernandes, P.A.; Mannervik, B.; Ramos, M.J. Isomerization of ∆5-androstene-3,17-dione into ∆4-androstene-3, 17-dione catalyzed by human glutathione transferase A3-3: A computational study identifies a dual role for glutathione. J. Phys. Chem. A 2014, 31, 5790–5800. [Google Scholar] [CrossRef]
- Lindström, H.; Peer, S.M.; Ing, N.H.; Mannervik, B. Characterization of equine GST A3-3 as a steroid isomerase. J. Steroid Biochem. Mol. Biol. 2018, 178, 117–128. [Google Scholar] [CrossRef]
- Mannervik, B.; Ismail, A.; Lindström, H.; Sjödin, B.; Ing, N.H. Glutathione transferases as efficient ketosteroid isomerases. Front. Mol. Biosci. 2021, 8, 765970. [Google Scholar] [CrossRef]
- Islam, M.Q.; Platz, A.; Szpirer, J.; Szpirer, C.; Levan, G.; Mannervik, B. Chromosomal localization of human glutathione transferase genes of classes Alpha, Mu and Pi. Hum. Genet. 1989, 82, 338–342. [Google Scholar] [CrossRef]
- Raffalli-Mathieu, F.; Orre, C.; Stridsberg, M.; Edalat, M.; Mannervik, B. Targeting human glutathione transferase A3-3 attenuates progesterone production in human steroidogenic cells. Biochem. J. 2008, 414, 103–109. [Google Scholar] [CrossRef]
- Ing, N.H.; Forrest, D.W.; Riggs, P.K.; Loux, S.; Love, C.C.; Brinsko, S.P.; Varner, D.D.; Welsh, T.H., Jr. Dexamethasone acutely down-regulates genes involved in steroidogenesis in stallion testes. J. Steroid Biochem. Mol. Biol. 2014, 143, 451–459. [Google Scholar] [CrossRef]
- Ing, N.; Brinsko, S.; Curley, K.; Forrest, D.; Love, C.; Hinrichs, K.; Vogelsang, M.; Varner, D.; Welsh, T. Dexamethasone acutely regulates endocrine parameters in stallions and subsequently affects gene expression in testicular germ cells. Anim. Reprod. Sci. 2015, 152, 47–54. [Google Scholar] [CrossRef]
- Valdez, R.; Cavinder, C.A.; Varner, D.D.; Welsh, T.W.; Vogelsang, M.M.; Ing, N.H. Dexamethasone downregulates expression of several genes encoding orphan nuclear receptors that are important to steroidogenesis in stallion testes. J. Biochem. Mol. Toxicol. 2019, 33, e22309. [Google Scholar] [CrossRef] [PubMed]
- Shiota, M.; Endo, S.; Blas, L.; Fujimoto, N.; Eto, M. Steroidogenesis in castration-resistant prostate cancer. Urologic Oncol. 2023, 41, 240–251. [Google Scholar] [CrossRef]
- Ismail, A.; Sawmi, J.; Mannervik, B. Marmoset glutathione transferases with ketosteroid isomerase activity. Biochem. Biophys. Rep. 2021, 27, 101078. [Google Scholar] [CrossRef]
- Raffalli-Mathieu, F.; Persson, D.; Mannervik, B. Differences between bovine and human steroid double-bond isomerase activities of Alpha-class glutathione transferases selectively expressed in steroidogenic tissues. Biochim. Biophys. Acta 2007, 1770, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Foley, N.M.; Bredmeyer, K.R.; Gatesy, J.; Springer, M.S. Phylogenetics and the genetic architecture of the placental mammal radiation. Annu. Rev. Anim. Biosci. 2021, 9, 29–53. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Suleski, M.; Craig, J.M.; Kasprowicz, A.E.; Sanderford, M.; Li, M.; Stecher, G.; Hedges, S.B. TimeTree 5: An expanded resource for species divergence times. Mol. Biol. Evol. 2022, 39, msac174. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W267–W279. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Tars, K.; Olin, B.; Mannervik, B. Structural basis for featuring of steroid isomerase activity in Alpha class glutathione transferases. J. Mol. Biol. 2010, 397, 332–340. [Google Scholar] [CrossRef]
- Benbrahim-Tallaa, L.; Tabone, E.; Tosser-Klopp, G.; Hatey, F.; Benamed, M. Glutathione transferase alpha expressed in porcine Sertoli cells is under the control of follicle-stimulating hormone and testosterone. Biol. Reprod. 2002, 66, 1734–1742. [Google Scholar] [CrossRef] [PubMed]
- Škerlová, J.; Ismail, A.; Lindström, H.; Sjödin, B.; Mannervik, B.; Stenmark, P. Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants. Environ. Pollut. 2021, 268, 115960. [Google Scholar] [CrossRef] [PubMed]



| Species | Primer Set | Orientation | Sequence |
|---|---|---|---|
| Dog | Outside | Sense | GGAGACCTGCATCATGGCAGTGAAGCCCATG |
| Antisense | AGGAGATTGGCCCTGCATGTGCT | ||
| Inside | Sense | ATGGCGGGGAAGCCCAAGCTTCACTACTTCAATGG | |
| Antisense | CTGGGCATCCATTCCTGTTCAGTTAATCCT | ||
| Goat | Outside | Sense | GGAGACTGCATCATGGCAGTGAAGCCCATG |
| Antisense | TCAAATTTGTCCCAAACAGCCCC | ||
| Inside | Sense | ATGGCAGGGAAGCCCATTCTTCACTATTTCAATGG | |
| Antisense | CCCCGCCAGCCGCCAGCTTTATTAAAACTT | ||
| Opossum | Outside | Sense | GGAGACTGCCATCATGGCAGTGAAGCCCATG |
| Antisense | TGTGTTTAAGAAACACAGAGTCA | ||
| Inside | Sense | GGTAAAGAAGATATTCAAGGTTGATGA | |
| Antisense | TCATCAACCTTGAATATCTTCTTTACC |
| Species | Primer Set | Orientation | Sequence |
|---|---|---|---|
| Dog | Outside | Sense | CCAGAGACTACCATGGCGGGGAAGCCCAAG |
| Antisense | TCTCAGGAGATTGGCCCTGCATG | ||
| Inside | Sense | gcgaattcATGGCGGGGAAGCCCAAGCTTCACTACTTCAATGG | |
| Antisense | cgctcgagCTGGGCATCCATTCCTGTTCAGTTAATCCT | ||
| Goat | Outside | Sense | AGAACTGCTATTATGGCAGGGAAGCCCAT |
| Antisense | TCAAATTTGTCCCAGACAGCCC | ||
| Inside | Sense | gcgaattcATGGCAGGGAAGCCCATTCTTCACTATTTCAATGG | |
| Antisense | cgctcgagCCCCGCCAGCCGCCAGCTTTATTAAAACTT | ||
| Opossum | Outside | Sense | GAATGGAAGATCATGTCTGGGAAGCCCAT |
| Antisense | TTGCATTACTTAGAACTCTTCCTGAATATTCAGCT | ||
| Inside | Sense | gcgaattcGGTAAAGAAGATATTCAAGGTTGATGA | |
| Antisense | cgctcgagTCATCAACCTTGAATATCTTCTTTACC |
| Human | Marmoset | Dog | Horse | Goat | Opossum | |
|---|---|---|---|---|---|---|
| Human | 100 | 94 | 88 | 85 | 84 | 70 |
| Marmoset | 94 | 100 | 86 | 86 | 85 | 70 |
| Dog | 88 | 86 | 100 | 87 | 87 | 71 |
| Horse | 85 | 86 | 87 | 100 | 85 | 70 |
| Goat | 84 | 85 | 87 | 85 | 100 | 68 |
| Opossum | 70 | 70 | 71 | 70 | 68 | 100 |
| Human | Marmoset | Dog | Horse | Goat | Opossum | |
|---|---|---|---|---|---|---|
| Human | 100 | 88 | 85 | 81 | 81 | 64 |
| Marmoset | 88 | 100 | 79 | 82 | 79 | 63 |
| Dog | 85 | 79 | 100 | 81 | 83 | 62 |
| Horse | 81 | 82 | 81 | 100 | 82 | 61 |
| Goat | 81 | 79 | 83 | 82 | 100 | 62 |
| Opossum | 64 | 63 | 62 | 61 | 62 | 100 |
| Residue | Human | Marmoset | Horse | Dog | Goat | Opossum | Pig | Cattle |
|---|---|---|---|---|---|---|---|---|
| 10 * | F | F | F | F | F | F | F | F |
| 12 * | G | A | G | G | G | G | G | G |
| 14 | G | G | G | G | G | G | G | G |
| 107 | L | L | L | M | M | M | L | M |
| 108 | L | L | L | V | H | Y | L | H |
| 110 | P | P | P | P | P | P | P | P |
| 111 * | L | L | L | L | L | L | L | L |
| 208 * | A | T | G | L | T | M | T | T |
| 213 | L | V | V | L | I | L | L | I |
| 216 * | A | A | S | A | A | V | A | A |
| 220 | F | F | F | F | F | F | F | F |
| 222 | F | F | F | I | F | V | F | F |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hubert, S.M.; Samollow, P.B.; Lindström, H.; Mannervik, B.; Ing, N.H. Conservation of Glutathione Transferase mRNA and Protein Sequences Similar to Human and Horse Alpha Class GST A3-3 across Dog, Goat, and Opossum Species. Biomolecules 2023, 13, 1420. https://doi.org/10.3390/biom13091420
Hubert SM, Samollow PB, Lindström H, Mannervik B, Ing NH. Conservation of Glutathione Transferase mRNA and Protein Sequences Similar to Human and Horse Alpha Class GST A3-3 across Dog, Goat, and Opossum Species. Biomolecules. 2023; 13(9):1420. https://doi.org/10.3390/biom13091420
Chicago/Turabian StyleHubert, Shawna M., Paul B. Samollow, Helena Lindström, Bengt Mannervik, and Nancy H. Ing. 2023. "Conservation of Glutathione Transferase mRNA and Protein Sequences Similar to Human and Horse Alpha Class GST A3-3 across Dog, Goat, and Opossum Species" Biomolecules 13, no. 9: 1420. https://doi.org/10.3390/biom13091420






