Evaluation of Antibiotic Biodegradation by a Versatile and Highly Active Recombinant Laccase from the Thermoalkaliphilic Bacterium Bacillus sp. FNT
Abstract
:1. Introduction
2. Materials and Methods
2.1. Homology Modeling of Bacillus sp. FNT laccase (FNTL)
2.2. Preparation of Ligands and Receptors for Docking
2.3. Molecular Docking Experiments
2.4. Interaction Analysis and Visualization
2.5. Recombinant Overexpression and Partial Purification of FNTL
2.6. Quantitative Evaluation of Antibiotic Biodegradation by FNTL
2.7. Preliminary Evaluation of the Antimicrobial Effect of Antibiotics Treated with FNTL
2.8. Statistical Analysis
3. Results
3.1. In Silico Evaluation of Antibiotic Affinity to the Spore-Coat Laccase of Bacillus sp. FNT (FNTL)
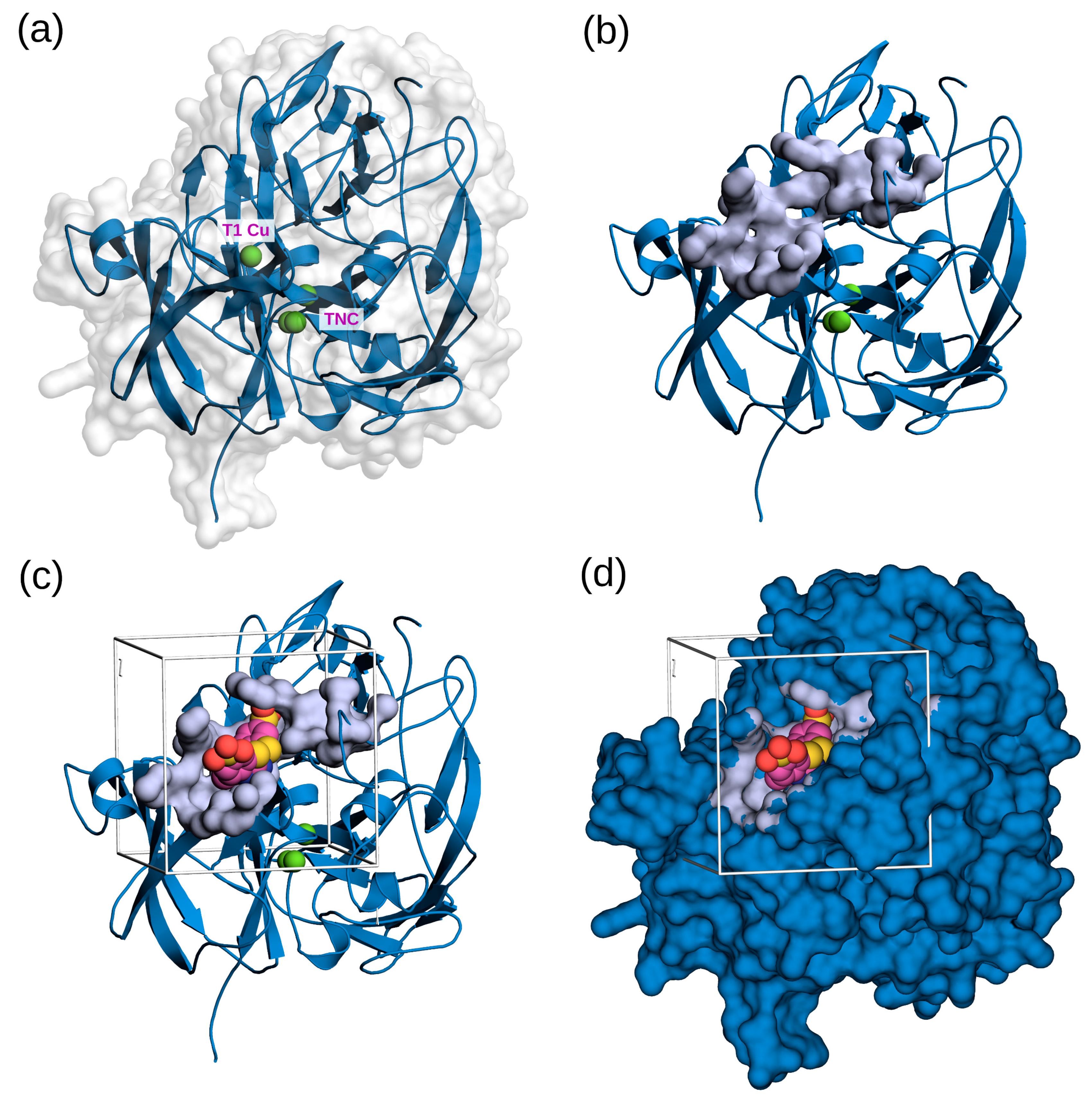
3.1.1. Homology Modeling of FNTL
3.1.2. Molecular Docking Experiments
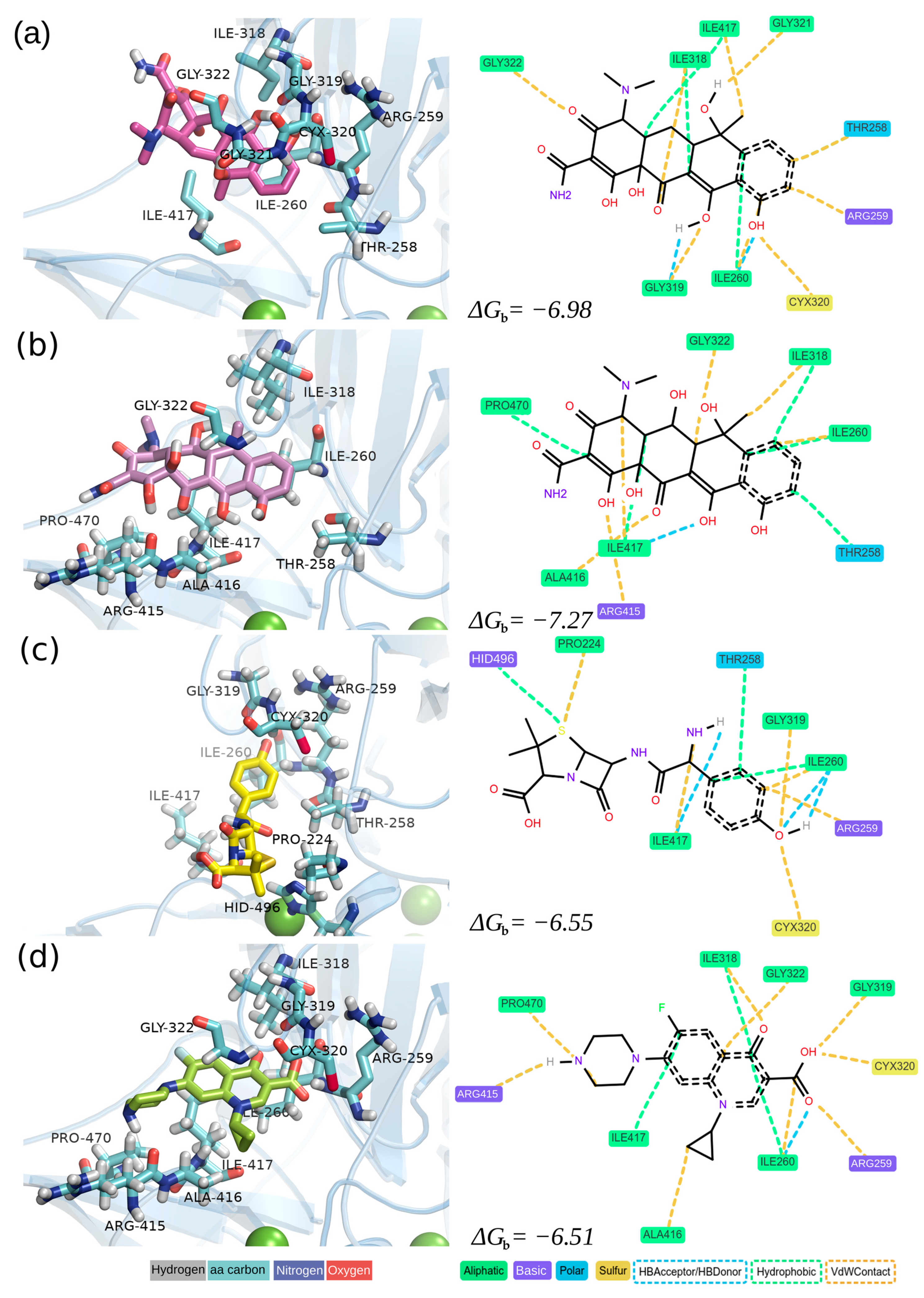
3.1.3. Analysis of Receptor–Ligand Interactions
3.2. Experimental Evaluation of Antibiotic Biodegradation by FNTL
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rout, P.R.; Zhang, T.C.; Bhunia, P.; Surampalli, R.Y. Treatment Technologies for Emerging Contaminants in Wastewater Treatment Plants: A Review. Sci. Total Environ. 2021, 753, 141990. [Google Scholar] [CrossRef]
- Hanjra, M.A.; Qureshi, M.E. Global Water Crisis and Future Food Security in an Era of Climate Change. Food Policy 2010, 35, 365–377. [Google Scholar] [CrossRef]
- Jury, W.A.; Vaux, H.J. The Emerging Global Water Crisis: Managing Scarcity and Conflict Between Water Users. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2007; Volume 95, pp. 1–76. ISBN 978-0-12-374165-3. [Google Scholar]
- Wang, Q.; Yang, Z. Industrial Water Pollution, Water Environment Treatment, and Health Risks in China. Environ. Pollut. 2016, 218, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.R.; Lee, J.T.; Cho, J.Y. Fate, Occurrence, and Toxicity of Veterinary Antibiotics in Environment. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 701–709. [Google Scholar] [CrossRef]
- Aminov, R. History of Antimicrobial Drug Discovery: Major Classes and Health Impact. Biochem. Pharmacol. 2017, 133, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; You, S.; Zhang, J.; Qi, W.; Su, R.; He, Z. An Effective In-Situ Method for Laccase Immobilization: Excellent Activity, Effective Antibiotic Removal Rate and Low Potential Ecological Risk for Degradation Products. Bioresour. Technol. 2020, 308, 123271. [Google Scholar] [CrossRef]
- Kovalakova, P.; Cizmas, L.; McDonald, T.J.; Marsalek, B.; Feng, M.; Sharma, V.K. Occurrence and Toxicity of Antibiotics in the Aquatic Environment: A Review. Chemosphere 2020, 251, 126351. [Google Scholar] [CrossRef]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global Increase and Geographic Convergence in Antibiotic Consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Larsson, D.G.J. Antibiotics in the Environment. Upsala J. Med. Sci. 2014, 119, 108–112. [Google Scholar] [CrossRef]
- Oulton, R.L.; Kohn, T.; Cwiertny, D.M. Pharmaceuticals and Personal Care Products in Effluent Matrices: A Survey of Transformation and Removal during Wastewater Treatment and Implications for Wastewater Management. J. Environ. Monit. 2010, 12, 1956–1978. [Google Scholar] [CrossRef]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological Effects of Antibiotics on Natural Ecosystems: A Review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Berglund, B. Environmental Dissemination of Antibiotic Resistance Genes and Correlation to Anthropogenic Contamination with Antibiotics. Infect. Ecol. Epidemiol. 2015, 5, 28564. [Google Scholar] [CrossRef] [PubMed]
- Minguez, L.; Pedelucq, J.; Farcy, E.; Ballandonne, C.; Budzinski, H.; Halm-Lemeille, M.-P. Toxicities of 48 Pharmaceuticals and Their Freshwater and Marine Environmental Assessment in Northwestern France. Environ. Sci. Pollut. Res. 2016, 23, 4992–5001. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.S.; Midya, S. Growth Regulatory Pattern of Zooplankton in Herbicide and Antibiotic Contaminated Aquatic Ecosystem: An Overview. Watershed Ecol. Environ. 2023, 5, 153–160. [Google Scholar] [CrossRef]
- Kim, B.; Ji, K.; Kho, Y.; Kim, P.-G.; Park, K.; Kim, K.; Kim, Y.; Kim, K.-T.; Choi, K. Effects of Chronic Exposure to Cefadroxil and Cefradine on Daphnia Magna and Oryzias Latipes. Chemosphere 2017, 185, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Jaiswal, S.; Sodhi, K.K.; Shree, P.; Singh, D.K.; Agrawal, P.K.; Shukla, P. Antibiotics Bioremediation: Perspectives on Its Ecotoxicity and Resistance. Environ. Int. 2019, 124, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Akram, F.; Imtiaz, M.; Haq, I.U. Emergent Crisis of Antibiotic Resistance: A Silent Pandemic Threat to 21st Century. Microb. Pathog. 2023, 174, 105923. [Google Scholar] [CrossRef]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef]
- Guo, X.; Yan, Z.; Zhang, Y.; Xu, W.; Kong, D.; Shan, Z.; Wang, N. Behavior of Antibiotic Resistance Genes under Extremely High-Level Antibiotic Selection Pressures in Pharmaceutical Wastewater Treatment Plants. Sci. Total Environ. 2018, 612, 119–128. [Google Scholar] [CrossRef]
- Bueno, I.; Verdugo, C.; Jimenez-Lopez, O.; Alvarez, P.P.; Gonzalez-Rocha, G.; Lima, C.A.; Travis, D.A.; Wass, B.; Zhang, Q.; Ishii, S.; et al. Role of Wastewater Treatment Plants on Environmental Abundance of Antimicrobial Resistance Genes in Chilean Rivers. Int. J. Hyg. Environ. Health 2020, 223, 56–64. [Google Scholar] [CrossRef]
- Fresia, P.; Antelo, V.; Salazar, C.; Giménez, M.; D’Alessandro, B.; Afshinnekoo, E.; Mason, C.; Gonnet, G.H.; Iraola, G. Urban Metagenomics Uncover Antibiotic Resistance Reservoirs in Coastal Beach and Sewage Waters. Microbiome 2019, 7, 35. [Google Scholar] [CrossRef]
- Bird, K.; Boopathy, R.; Nathaniel, R.; LaFleur, G. Water Pollution and Observation of Acquired Antibiotic Resistance in Bayou Lafourche, a Major Drinking Water Source in Southeast Louisiana, USA. Environ. Sci. Pollut. Res. 2019, 26, 34220–34232. [Google Scholar] [CrossRef]
- Lyu, J.; Yang, L.; Zhang, L.; Ye, B.; Wang, L. Antibiotics in Soil and Water in China—A Systematic Review and Source Analysis. Environ. Pollut. 2020, 266, 115147. [Google Scholar] [CrossRef]
- Fick, J.; Söderström, H.; Lindberg, R.H.; Phan, C.; Tysklind, M.; Larsson, D.G.J. Contamination of Surface, Ground, and Drinking Water from Pharmaceutical Production. Environ. Toxicol. Chem. 2009, 28, 2522–2527. [Google Scholar] [CrossRef]
- Son, D.; Aleta, P.; Park, M.; Yoon, H.; Cho, K.H.; Kim, Y.M.; Kim, S. Seasonal Changes in Antibiotic Resistance Genes in Rivers and Reservoirs in South Korea. J. Environ. Qual. 2018, 47, 1079–1085. [Google Scholar] [CrossRef]
- Burke, V.; Richter, D.; Greskowiak, J.; Mehrtens, A.; Schulz, L.; Massmann, G. Occurrence of Antibiotics in Surface and Groundwater of a Drinking Water Catchment Area in Germany. Water Environ. Res. 2016, 88, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, M.J.; Paíga, P.; Silva, A.; Llaguno, C.P.; Carvalho, M.; Vázquez, F.M.; Delerue-Matos, C. Antibiotics and Antidepressants Occurrence in Surface Waters and Sediments Collected in the North of Portugal. Chemosphere 2020, 239, 124729. [Google Scholar] [CrossRef] [PubMed]
- Hernández, F.; Calısto-Ulloa, N.; Gómez-Fuentes, C.; Gómez, M.; Ferrer, J.; González-Rocha, G.; Bello-Toledo, H.; Botero-Coy, A.M.; Boıx, C.; Ibáñez, M.; et al. Occurrence of Antibiotics and Bacterial Resistance in Wastewater and Sea Water from the Antarctic. J. Hazard. Mater. 2019, 363, 447–456. [Google Scholar] [CrossRef]
- Rodriguez-Narvaez, O.M.; Peralta-Hernandez, J.M.; Goonetilleke, A.; Bandala, E.R. Treatment Technologies for Emerging Contaminants in Water: A Review. Chem. Eng. J. 2017, 323, 361–380. [Google Scholar] [CrossRef]
- Ajala, O.J.; Tijani, J.O.; Bankole, M.T.; Abdulkareem, A.S. Wastewater Treatment Technologies. In Inorganic-Organic Composites for Water and Wastewater Treatment; Lichtfouse, E., Muthu, S.S., Khadir, A., Eds.; Springer: Singapore, 2022; pp. 1–28. ISBN 9789811659157. [Google Scholar]
- Bamforth, S.M.; Singleton, I. Bioremediation of Polycyclic Aromatic Hydrocarbons: Current Knowledge and Future Directions. J. Chem. Technol. Biotechnol. 2005, 80, 723–736. [Google Scholar] [CrossRef]
- Cabrera, M.Á.; Márquez, S.L.; Quezada, C.P.; Osorio, M.I.; Castro-Nallar, E.; González-Nilo, F.D.; Pérez-Donoso, J.M. Biotransformation of 2,4,6-Trinitrotoluene by Pseudomonas sp. TNT3 Isolated from Deception Island, Antarctica. Environ. Pollut. 2020, 262, 113922. [Google Scholar] [CrossRef]
- Cabrera, M.Á.; Márquez, S.L.; Pérez-Donoso, J.M. Comparative Genomic Analysis of Antarctic Pseudomonas Isolates with 2,4,6-Trinitrotoluene Transformation Capabilities Reveals Their Unique Features for Xenobiotics Degradation. Genes 2022, 13, 1354. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, M.Á.; Márquez, S.L.; Pérez-Donoso, J.M. New Insights into Xenobiotic Tolerance of Antarctic Bacteria: Transcriptomic Analysis of Pseudomonas Sp. TNT3 during 2,4,6-Trinitrotoluene Biotransformation. Environ. Sci. Pollut. Res. 2024, 31, 17256–17274. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Dangi, A.K.; Shukla, P. Contemporary Enzyme Based Technologies for Bioremediation: A Review. J. Environ. Manag. 2018, 210, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Karigar, C.S.; Rao, S.S. Role of Microbial Enzymes in the Bioremediation of Pollutants: A Review. Enzym. Res. 2011, 2011, 805187. [Google Scholar] [CrossRef]
- Naghdi, M.; Taheran, M.; Brar, S.K.; Kermanshahi-pour, A.; Verma, M.; Surampalli, R.Y. Removal of Pharmaceutical Compounds in Water and Wastewater Using Fungal Oxidoreductase Enzymes. Environ. Pollut. 2018, 234, 190–213. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, W.; Ng, T.B.; Deng, X.; Lin, J.; Ye, X. Laccases: Production, Expression Regulation, and Applications in Pharmaceutical Biodegradation. Front. Microbiol. 2017, 8, 832. [Google Scholar] [CrossRef]
- Singh, D.; Gupta, N. Microbial Laccase: A Robust Enzyme and Its Industrial Applications. Biologia 2020, 75, 1183–1193. [Google Scholar] [CrossRef]
- Riva, S. Laccases: Blue Enzymes for Green Chemistry. Trends Biotechnol. 2006, 24, 219–226. [Google Scholar] [CrossRef]
- Mate, D.M.; Alcalde, M. Laccase: A Multi-purpose Biocatalyst at the Forefront of Biotechnology. Microb. Biotechnol. 2017, 10, 1457–1467. [Google Scholar] [CrossRef] [PubMed]
- Morozova, O.V.; Shumakovich, G.P.; Shleev, S.V.; Yaropolov, Y.I. Laccase-Mediator Systems and Their Applications: A Review. Appl. Biochem. Microbiol. 2007, 43, 523–535. [Google Scholar] [CrossRef]
- ur Rahman, M.; Ullah, M.W.; Shah, J.A.; Sethupathy, S.; Bilal, H.; Abdikakharovich, S.A.; Khan, A.U.; Khan, K.A.; Elboughdiri, N.; Zhu, D. Harnessing the Power of Bacterial Laccases for Xenobiotic Degradation in Water: A 10-Year Overview. Sci. Total Environ. 2024, 918, 170498. [Google Scholar] [CrossRef]
- Unuofin, J.O.; Falade, A.O.; Aladekoyi, O.J. Applications of Microbial Laccases in Bioremediation of Environmental Pollutants: Potential Issues, Challenges, and Prospects. In Bioremediation for Environmental Sustainability: Toxicity, Mechanisms of Contaminants Degradation, Detoxification and Challenges; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; Santos, M.H.D.L.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.N.; et al. Laccases: Structure, Function, and Potential Application in Water Bioremediation. Microb. Cell Factories 2019, 18, 200. [Google Scholar] [CrossRef]
- Wang, X.; Meng, F.; Zhang, B.; Xia, Y. Elimination of Tetracyclines in Seawater by Laccase-Mediator System. Chemosphere 2023, 333, 138916. [Google Scholar] [CrossRef]
- Bhatt, S.; Choudhary, P.; Chatterjee, S.; Akhter, Y. Comparative Analysis of SilA-Laccase Mediated Degradation of Ciprofloxacin, Norfloxacin and Ofloxacin and Interpretation of the Possible Catalytic Mechanism. J. Biomol. Struct. Dyn. 2024, 42, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Espina, G.; Cáceres-Moreno, P.; Mejías-Navarrete, G.; Ji, M.; Sun, J.; Blamey, J.M. A Novel and Highly Active Recombinant Spore-Coat Bacterial Laccase, Able to Rapidly Biodecolorize Azo, Triarylmethane and Anthraquinonic Dyestuffs. Int. J. Biol. Macromol. 2021, 170, 29–306. [Google Scholar] [CrossRef]
- Bueno-Nieto, C.; Cortés-Antiquera, R.; Espina, G.; Atalah, J.; Villanueva, J.; Aliaga, C.; Zuñiga, G.E.; Blamey, J.M. Biochemical and Spectroscopic Characterization of a Recombinant Laccase from Thermoalkaliphilic Bacillus sp. FNT with Potential for Degradation of Polycyclic Aromatic Hydrocarbons (PAHs). Catalysts 2023, 13, 763. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and Upgrading Automated Preparation of Biomolecular Structures for Molecular Simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.B.; Barton, G.J. Multiple Protein Sequence Alignment from Tertiary Structure Comparison: Assignment of Global and Residue Confidence Levels. Proteins 1992, 14, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Bouysset, C.; Fiorucci, S. ProLIF: A Library to Encode Molecular Interactions as Fingerprints. J. Cheminform. 2021, 13, 72. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, S.; Sasse, J. Protein Analysis by SDS-PAGE and Detection by Coomassie Blue or Silver Staining. Curr. Protoc. Pharmacol. 1998, 2, A-3B. [Google Scholar] [CrossRef]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Brooke, D.; Movahed, N.; Bothner, B. Universal Buffers for Use in Biochemistry and Biophysical Experiments. AIMS Biophys. 2015, 2, 336–342. [Google Scholar] [CrossRef]
- Yang, X.-Q.; Yang, C.-X.; Yan, X.-P. Zeolite Imidazolate Framework-8 as Sorbent for on-Line Solid-Phase Extraction Coupled with High-Performance Liquid Chromatography for the Determination of Tetracyclines in Water and Milk Samples. J. Chromatogr. A 2013, 1304, 28–33. [Google Scholar] [CrossRef]
- Oka, H.; Ito, Y.; Matsumoto, H. Chromatographic Analysis of Tetracycline Antibiotics in Foods. J. Chromatogr. A 2000, 882, 109–133. [Google Scholar] [CrossRef]
- Ding, H.; Wu, Y.; Zou, B.; Lou, Q.; Zhang, W.; Zhong, J.; Lu, L.; Dai, G. Simultaneous Removal and Degradation Characteristics of Sulfonamide, Tetracycline, and Quinolone Antibiotics by Laccase-Mediated Oxidation Coupled with Soil Adsorption. J. Hazard. Mater. 2016, 307, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Available online: https://asm.org:443/Protocols/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Pro (accessed on 17 February 2024).
- Enguita, F.J.; Martins, L.O.; Henriques, A.O.; Carrondo, M.A. Crystal Structure of a Bacterial Endospore Coat Component. J. Biol. Chem. 2003, 278, 19416–19425. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.A.; Weichsel, A.; Grass, G.; Thakali, K.; Hazzard, J.T.; Tollin, G.; Rensing, C.; Montfort, W.R. Crystal Structure and Electron Transfer Kinetics of CueO, a Multicopper Oxidase Required for Copper Homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 2766–2771. [Google Scholar] [CrossRef]
- Jones, S.M.; Solomon, E.I. Electron Transfer and Reaction Mechanism of Laccases. Cell. Mol. Life Sci. 2015, 72, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Liu, Z.; Wang, G. Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase. ChemBioChem 2018, 19, 1502–1506. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Bhardwaj, P.; Ishqi, H.M.; Shahid, M.; Islam, A. Laccase Engineering: Redox Potential Is Not the Only Activity-Determining Feature in the Metalloproteins. Molecules 2023, 28, 6209. [Google Scholar] [CrossRef]
- Langbehn, R.K.; Michels, C.; Soares, H.M. Antibiotics in Wastewater: From Its Occurrence to the Biological Removal by Environmentally Conscious Technologies. Environ. Pollut. 2021, 275, 116603. [Google Scholar] [CrossRef] [PubMed]
- Scaria, J.; Anupama, K.V.; Nidheesh, P.V. Tetracyclines in the Environment: An Overview on the Occurrence, Fate, Toxicity, Detection, Removal Methods, and Sludge Management. Sci. Total Environ. 2021, 771, 145291. [Google Scholar] [CrossRef]
- Cáceres-Moreno, P.; Muñoz-Ibacache, S.A.; Monsalves, M.T.; Amenabar, M.J.; Blamey, J.M. Functional Approach for the Development and Production of Novel Extreme Biocatalysts. In ACS Symposium Series; Rathinam, N.K., Sani, R.K., Eds.; American Chemical Society: Washington, DC, USA, 2019; Volume 1329, pp. 1–22. ISBN 978-0-8412-3500-7. [Google Scholar]
- Espina, G.; Cáceres-Moreno, P.; Correa-Llantén, D.; Sarmiento, F.; Blamey, J.M. 4 Extremozymes: From Discovery to Novel Bio-Products. In 4 Extremozymes: From Discovery to Novel Bio-Products; De Gruyter: Berlin, Germany, 2020; pp. 97–120. ISBN 978-3-11-042433-1. [Google Scholar]
- Camarero, S.; Ibarra, D.; Martínez, Á.T.; Romero, J.; Gutiérrez, A.; Del Río, J.C. Paper Pulp Delignification Using Laccase and Natural Mediators. Enzym. Microb. Technol. 2007, 40, 1264–1271. [Google Scholar] [CrossRef]
- Mani, P.; Kumar, V.T.F.; Keshavarz, T.; Chandra, T.S.; Kyazze, G. The Role of Natural Laccase Redox Mediators in Simultaneous Dye Decolorization and Power Production in Microbial Fuel Cells. Energies 2018, 11, 3455. [Google Scholar] [CrossRef]
- Becker, D.; Varela Della Giustina, S.; Rodriguez-Mozaz, S.; Schoevaart, R.; Barceló, D.; de Cazes, M.; Belleville, M.-P.; Sanchez-Marcano, J.; de Gunzburg, J.; Couillerot, O.; et al. Removal of Antibiotics in Wastewater by Enzymatic Treatment with Fungal Laccase—Degradation of Compounds Does Not Always Eliminate Toxicity. Bioresour. Technol. 2016, 219, 500–509. [Google Scholar] [CrossRef]
- Chmelová, D.; Ondrejovič, M.; Miertuš, S. Laccases as Effective Tools in the Removal of Pharmaceutical Products from Aquatic Systems. Life 2024, 14, 230. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.; Hao, X.; Zhang, R.; Wang, J.; Liu, R.; Liu, C. Tetracycline Removal and Effect on the Formation and Degradation of Extracellular Polymeric Substances and Volatile Fatty Acids in the Process of Hydrogen Fermentation. Bioresour. Technol. 2016, 212, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, K.G.; Meyer, M.T. Occurrence of Antibiotics in Wastewater Treatment Facilities in Wisconsin, USA. Sci. Total Environ. 2006, 361, 196–207. [Google Scholar] [CrossRef]
- Suda, T.; Hata, T.; Kawai, S.; Okamura, H.; Nishida, T. Treatment of Tetracycline Antibiotics by Laccase in the Presence of 1-Hydroxybenzotriazole. Bioresour. Technol. 2012, 103, 498–501. [Google Scholar] [CrossRef]
- Llorca, M.; Rodríguez-Mozaz, S.; Couillerot, O.; Panigoni, K.; De Gunzburg, J.; Bayer, S.; Czaja, R.; Barceló, D. Identification of New Transformation Products during Enzymatic Treatment of Tetracycline and Erythromycin Antibiotics at Laboratory Scale by an On-Line Turbulent Flow Liquid-Chromatography Coupled to a High Resolution Mass Spectrometer LTQ-Orbitrap. Chemosphere 2015, 119, 90–98. [Google Scholar] [CrossRef]
- Tian, Q.; Dou, X.; Huang, L.; Wang, L.; Meng, D.; Zhai, L.; Shen, Y.; You, C.; Guan, Z.; Liao, X. Characterization of a Robust Cold-Adapted and Thermostable Laccase from Pycnoporus Sp. SYBC-L10 with a Strong Ability for the Degradation of Tetracycline and Oxytetracycline by Laccase-Mediated Oxidation. J. Hazard. Mater. 2020, 382, 121084. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wang, H.; Zheng, J.; Wang, S.; Yu, S.; Lu, L. Ultrafast Synthesis of Laccase-Copper Phosphate Hybrid Nanoflowers for Efficient Degradation of Tetracycline Antibiotics. Environ. Res. 2023, 216, 114690. [Google Scholar] [CrossRef]
- De Cazes, M.; Belleville, M.-P.; Petit, E.; Llorca, M.; Rodríguez-Mozaz, S.; De Gunzburg, J.; Barceló, D.; Sanchez-Marcano, J. Design and Optimization of an Enzymatic Membrane Reactor for Tetracycline Degradation. Catal. Today 2014, 236, 146–152. [Google Scholar] [CrossRef]
- Sun, K.; Huang, Q.; Li, S. Transformation and Toxicity Evaluation of Tetracycline in Humic Acid Solution by Laccase Coupled with 1-Hydroxybenzotriazole. J. Hazard. Mater. 2017, 331, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.-F.; Yang, B.; Xiong, Q.; Cai, W.-W.; Lan, Z.-G.; Ying, G.-G. Hydrolytic Transformation Mechanism of Tetracycline Antibiotics: Reaction Kinetics, Products Identification and Determination in WWTPs. Ecotoxicol. Environ. Saf. 2022, 229, 113063. [Google Scholar] [CrossRef]
- Shao, B.; Liu, Z.; Zeng, G.; Liu, Y.; Yang, X.; Zhou, C.; Chen, M.; Liu, Y.; Jiang, Y.; Yan, M. Immobilization of Laccase on Hollow Mesoporous Carbon Nanospheres: Noteworthy Immobilization, Excellent Stability and Efficacious for Antibiotic Contaminants Removal. J. Hazard. Mater. 2019, 362, 318–326. [Google Scholar] [CrossRef]
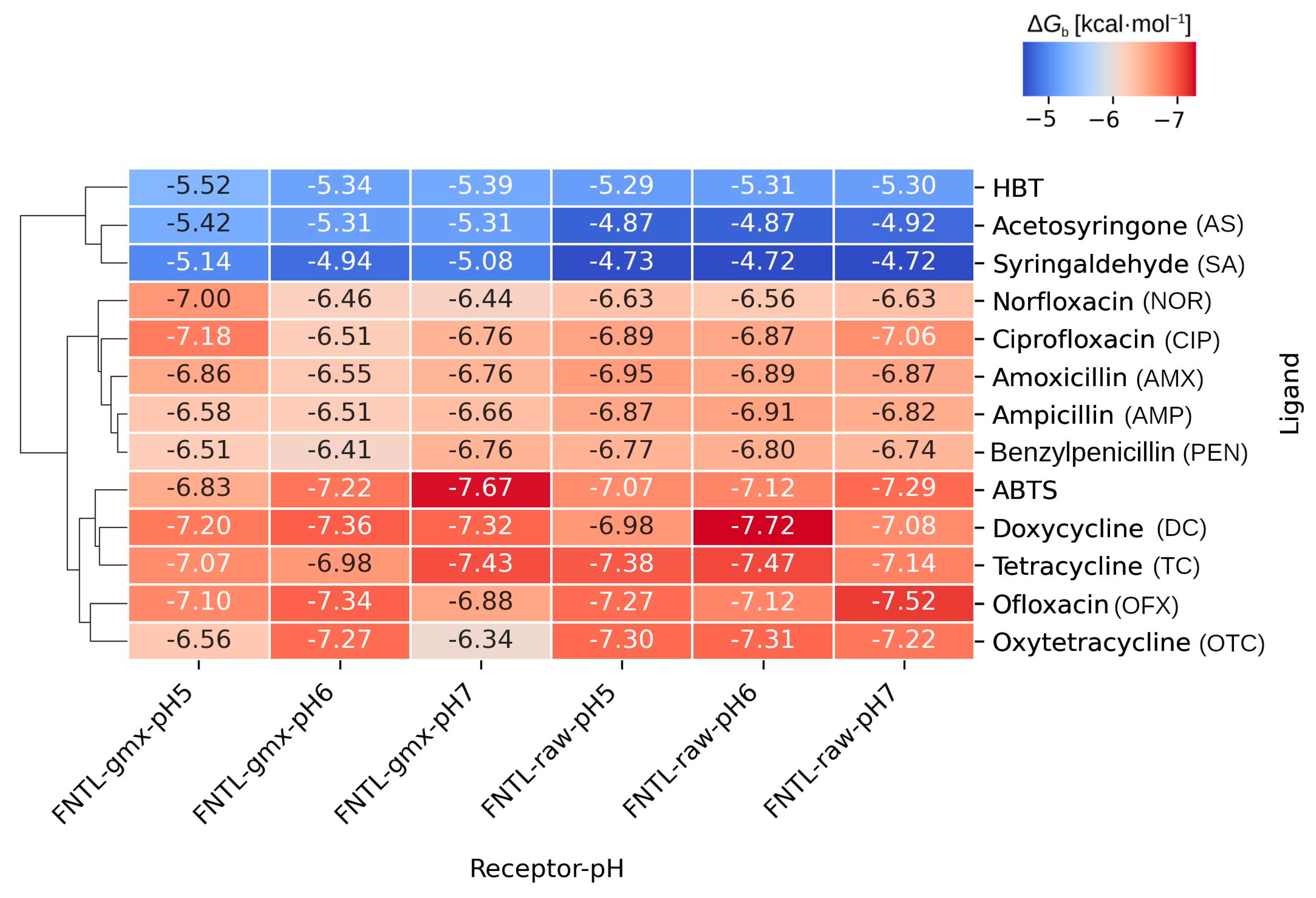



| Type | Molecule | Abbreviation |
|---|---|---|
| Artificial redox mediator | 1-hydroxybenzotriazole | HBT |
| 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) | ABTS | |
| Natural redox mediator | Acetosyringone | AS |
| Syringaldehyde | SA | |
| Tetracycline antibiotics | Tetracycline | TC |
| Oxytetracycline | OTC | |
| Doxycycline | DC | |
| β-lactams antibiotics | Amoxicillin | AMX |
| Ampicillin | AMP | |
| Benzylpenicillin | PEN | |
| Fluoroquinolone antibiotics | Ciprofloxacin | CIP |
| Norfloxacin | NOR | |
| Ofloxacin | OFX |
| Antibiotics | |||||
|---|---|---|---|---|---|
| Residue | Interaction | TC | OTC | AMX | CIP |
| PRO224 | Hydrophobic | False | False | True | False |
| PRO224 | VdWContact | False | False | True | False |
| THR258 | Hydrophobic | True | True | True | False |
| THR258 | VdWContact | True | False | False | False |
| ARG259 | VdWContact | True | False | True | True |
| ILE260 | Hydrophobic | True | True | True | True |
| ILE260 | HBAcceptor | True | False | True | True |
| ILE260 | HBDonor | False | False | True | False |
| ILE260 | VdWContact | True | True | True | True |
| ILE318 | Hydrophobic | True | True | False | True |
| ILE318 | VdWContact | True | True | False | True |
| GLY319 | HBDonor | True | False | False | False |
| GLY319 | VdWContact | True | False | True | True |
| CYS320 | VdWContact | True | False | True | True |
| GLY321 | VdWContact | True | False | False | False |
| GLY322 | VdWContact | True | True | False | True |
| ARG415 | HBDonor | False | False | False | True |
| ARG415 | VdWContact | False | True | False | True |
| ALA416 | VdWContact | False | True | False | True |
| ILE417 | Hydrophobic | True | True | False | True |
| ILE417 | HBAcceptor | False | True | True | False |
| ILE417 | HBDonor | False | False | True | False |
| ILE417 | VdWContact | True | True | True | False |
| PRO470 | Hydrophobic | False | True | False | False |
| PRO470 | VdWContact | False | False | False | True |
| HIS496 | Hydrophobic | False | False | True | False |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-SanMartín, J.; Márquez, S.L.; Espina, G.; Cortés-Antiquera, R.; Sun, J.; Blamey, J.M. Evaluation of Antibiotic Biodegradation by a Versatile and Highly Active Recombinant Laccase from the Thermoalkaliphilic Bacterium Bacillus sp. FNT. Biomolecules 2024, 14, 369. https://doi.org/10.3390/biom14030369
Sánchez-SanMartín J, Márquez SL, Espina G, Cortés-Antiquera R, Sun J, Blamey JM. Evaluation of Antibiotic Biodegradation by a Versatile and Highly Active Recombinant Laccase from the Thermoalkaliphilic Bacterium Bacillus sp. FNT. Biomolecules. 2024; 14(3):369. https://doi.org/10.3390/biom14030369
Chicago/Turabian StyleSánchez-SanMartín, Jorge, Sebastián L. Márquez, Giannina Espina, Rodrigo Cortés-Antiquera, Junsong Sun, and Jenny M. Blamey. 2024. "Evaluation of Antibiotic Biodegradation by a Versatile and Highly Active Recombinant Laccase from the Thermoalkaliphilic Bacterium Bacillus sp. FNT" Biomolecules 14, no. 3: 369. https://doi.org/10.3390/biom14030369





