Tumor Uptake of Triazine Dendrimers Decorated with Four, Sixteen, and Sixty-Four PSMA-Targeted Ligands: Passive versus Active Tumor Targeting
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Procedures
2.2. Synthesis
2.3. Cell Lines and Animal Models
2.4. Preparation of 64Cu-Labeld G1-(DUPA)4, G3-(DUPA)16, and G5-(DUPA)64
2.5. Serum Stability Assay
2.6. Competition Assay
2.7. Cell Uptake and Internalization
2.8. Small Animal PET/CT Imaging
3. Results and Discussion
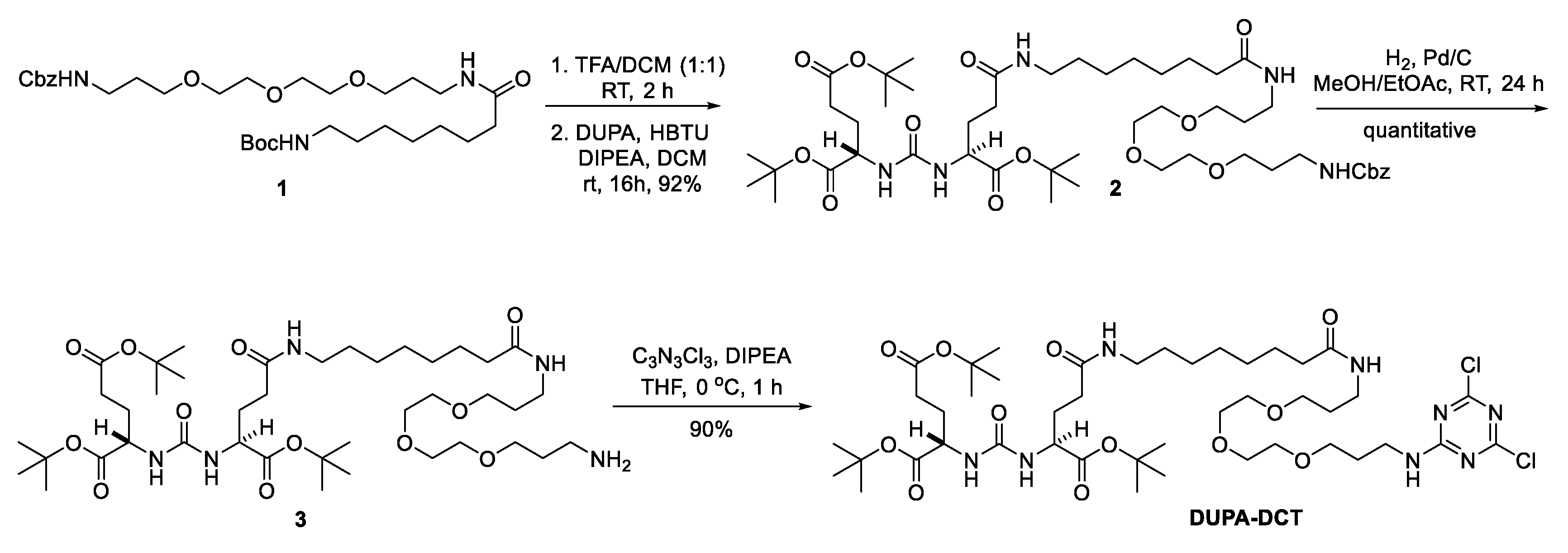
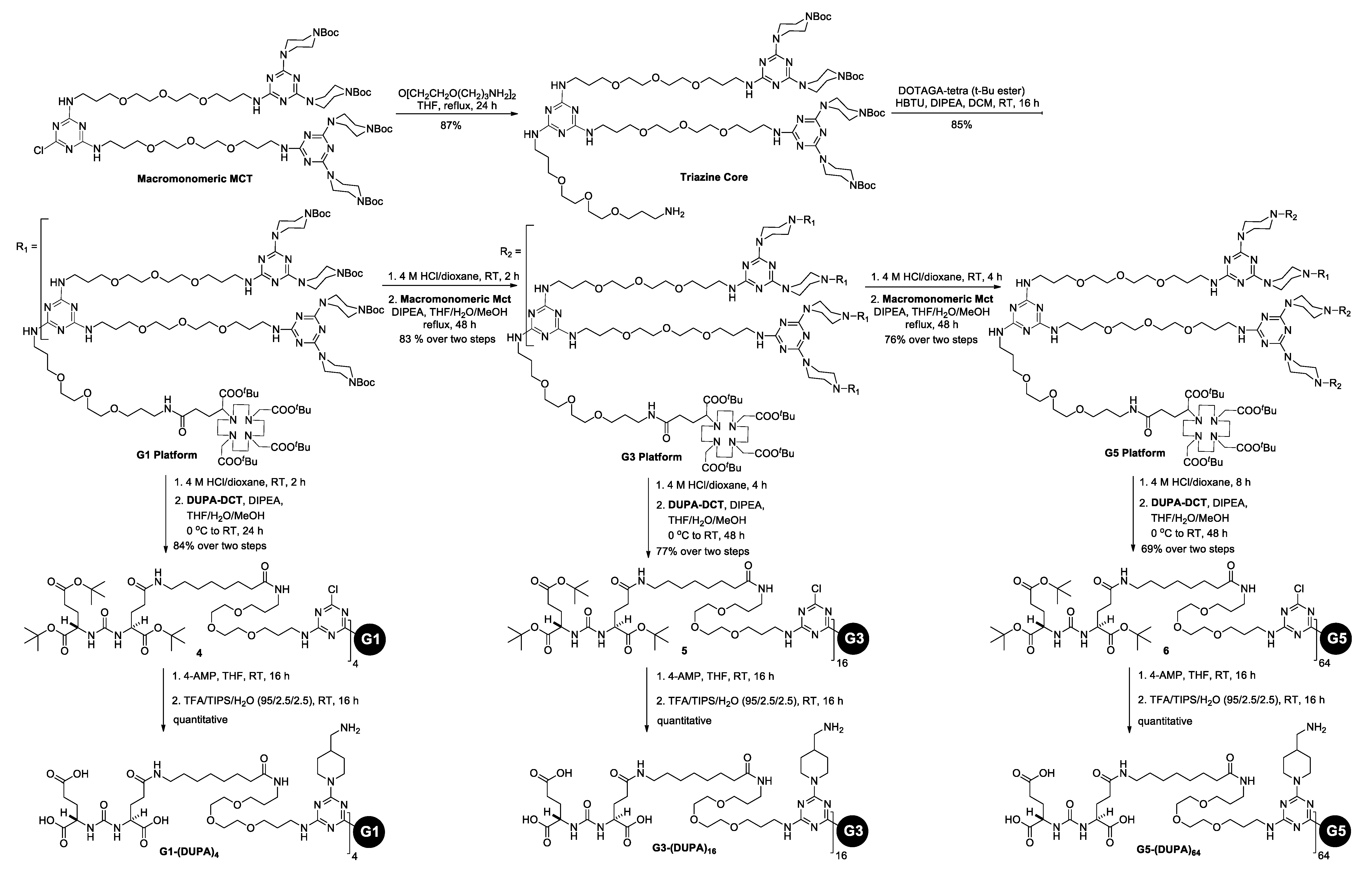
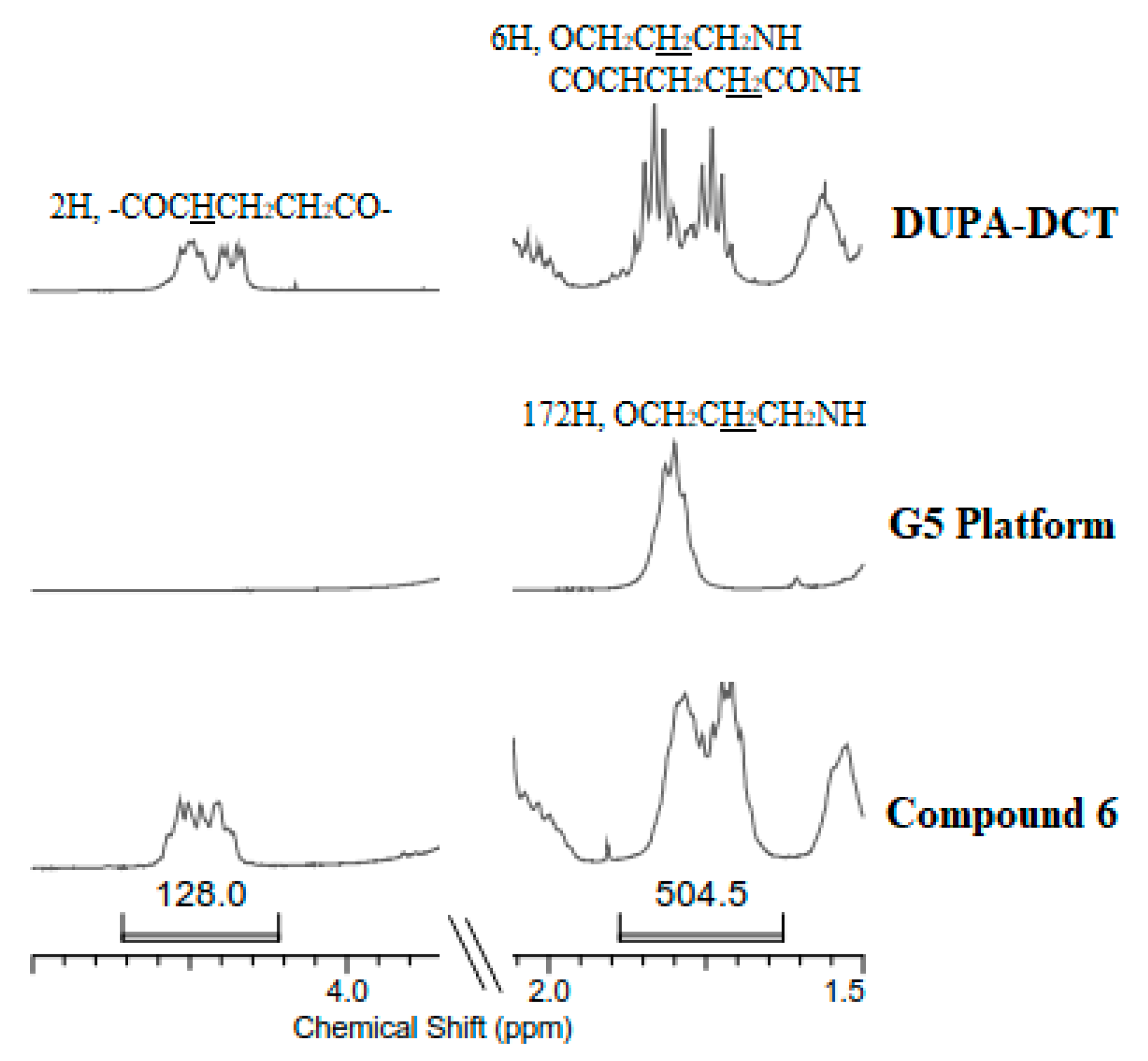
3.1. Synthesis and Characterization of G1-(DUPA)4, G3-(DUPA)16, and G5-(DUPA)64
3.2. Radiolabeling and Serum Stability
3.3. In Vitro Cell Assays
3.4. PET/CT Imaging in Mouse Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Srinivasarao, M.; Low, P.S. Ligand-Targeted Drug Delivery. Chem. Rev. 2017, 117, 12133–12164. [Google Scholar] [CrossRef] [PubMed]
- Srinivasarao, M.; Galliford, C.V.; Low, P.S. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nat. Rev. Drug Discov. 2015, 14, 203. [Google Scholar] [CrossRef] [PubMed]
- Byrne, J.D.; Betancourt, T.; Brannon-Peppas, L. Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv. Drug Deliv. Rev. 2008, 60, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H. The enhanced permeability and retention (EPR) effect in tumor vasculature: The key role of tumor-selective macromolecular drug targeting. Adv. Enzym. Regul. 2001, 41, 189–207. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Madan, R.A.; Mohebtash, M.; Arlen, P.M.; Vergati, M.; Rauckhorst, M.; Steinberg, S.M.; Tsang, K.Y.; Poole, D.J.; Parnes, H.L.; Wright, J.J. Ipilimumab and a poxviral vaccine targeting prostate-specific antigen in metastatic castration-resistant prostate cancer: A phase 1 dose-escalation trial. Lancet Oncol. 2012, 13, 501–508. [Google Scholar] [CrossRef]
- Veeramani, S.; Yuan, T.-C.; Chen, S.-J.; Lin, F.-F.; Petersen, J.E.; Shaheduzzaman, S.; Srivastava, S.; MacDonald, R.G.; Lin, M.-F. Cellular prostatic acid phosphatase: A protein tyrosine phosphatase involved in androgen-independent proliferation of prostate cancer. Endocr. Relat. Cancer 2005, 12, 805–822. [Google Scholar] [CrossRef]
- Reiter, R.E.; Gu, Z.; Watabe, T.; Thomas, G.; Szigeti, K.; Davis, E.; Wahl, M.; Nisitani, S.; Yamashiro, J.; Le Beau, M.M. Prostate stem cell antigen: A cell surface marker overexpressed in prostate cancer. Proc. Natl. Acad. Sci. USA 1998, 95, 1735–1740. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, T.; Mitsunaga, M.; Bander, N.H.; Heston, W.D.; Choyke, P.L.; Kobayashi, H. Targeted, activatable, in vivo fluorescence imaging of prostate-specific membrane antigen (PSMA) positive tumors using the quenched humanized J591 antibody–indocyanine green (ICG) conjugate. Bioconjug. Chem. 2011, 22, 1700–1705. [Google Scholar] [CrossRef]
- Haberkorn, U.; Eder, M.; Kopka, K.; Babich, J.W.; Eisenhut, M. New strategies in prostate cancer: Prostate-specific membrane antigen (PSMA) ligands for diagnosis and therapy. Clin. Cancer Res. 2016, 22, 9–15. [Google Scholar] [CrossRef]
- Sokoloff, R.; Norton, K.; Gasior, C.; Marker, K.; Grauer, L. A dual-monoclonal sandwich assay for prostate-specific membrane antigen: Levels in tissues, seminal fluid and urine. Prostate 2000, 43, 150–157. [Google Scholar] [CrossRef]
- Maurer, T.; Eiber, M.; Schwaiger, M.; Gschwend, J.E. Current use of PSMA–PET in prostate cancer management. Nat. Rev. Urol. 2016, 13, 226. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Rajasekaran, A.K.; Moy, P.; Xia, Y.; Kim, S.; Navarro, V.; Rahmati, R.; Bander, N.H. Constitutive and antibody-induced internalization of prostate-specific membrane antigen. Cancer Res. 1998, 58, 4055–4060. [Google Scholar] [PubMed]
- Min, K.; Jo, H.; Song, K.; Cho, M.; Chun, Y.-S.; Jon, S.; Kim, W.J.; Ban, C. Dual-aptamer-based delivery vehicle of doxorubicin to both PSMA (+) and PSMA (−) prostate cancers. Biomaterials 2011, 32, 2124–2132. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, S.T.; Milowsky, M.I.; Morris, M.; Vallabhajosula, S.; Christos, P.; Akhtar, N.H.; Osborne, J.; Goldsmith, S.J.; Larson, S.; Taskar, N.P. Phase II study of lutetium-177–labeled anti-prostate-specific membrane antigen monoclonal antibody J591 for metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2013, 19, 5182–5191. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Singh, P.; Topaloglu, O.; Isaacs, J.T.; Denmeade, S.R. A dimeric peptide that binds selectively to prostate-specific membrane antigen and inhibits its enzymatic activity. Cancer Res. 2006, 66, 9171–9177. [Google Scholar] [CrossRef] [PubMed]
- Kularatne, S.A.; Wang, K.; Santhapuram, H.-K.R.; Low, P.S. Prostate-specific membrane antigen targeted imaging and therapy of prostate cancer using a PSMA inhibitor as a homing ligand. Mol. Pharm. 2009, 6, 780–789. [Google Scholar] [CrossRef]
- Zechmann, C.M.; Afshar-Oromieh, A.; Armor, T.; Stubbs, J.B.; Mier, W.; Hadaschik, B.; Joyal, J.; Kopka, K.; Debus, J.; Babich, J.W. Radiation dosimetry and first therapy results with a 124 I/131 I-labeled small molecule (MIP-1095) targeting PSMA for prostate cancer therapy. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 1280–1292. [Google Scholar] [CrossRef]
- Kratochwil, C.; Giesel, F.L.; Stefanova, M.; Benešová, M.; Bronzel, M.; Afshar-Oromieh, A.; Mier, W.; Eder, M.; Kopka, K.; Haberkorn, U. PSMA-targeted radionuclide therapy of metastatic castration-resistant prostate cancer with 177Lu-labeled PSMA-617. J. Nucl. Med. 2016, 57, 1170–1176. [Google Scholar] [CrossRef]
- Kumar, A.; Mastren, T.; Wang, B.; Hsieh, J.-T.; Hao, G.; Sun, X. Design of a small-molecule drug conjugate for prostate cancer targeted theranostics. Bioconjug. Chem. 2016, 27, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Hrkach, J.; Von Hoff, D.; Ali, M.M.; Andrianova, E.; Auer, J.; Campbell, T.; De Witt, D.; Figa, M.; Figueiredo, M.; Horhota, A. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci. Transl. Med. 2012, 4, 128ra39. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Gu, F.X.; Langer, R.; Farokhzad, O.C.; Lippard, S.J. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt (IV) prodrug-PLGA–PEG nanoparticles. Proc. Natl. Acad. Sci. USA 2008, 105, 17356–17361. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Penet, M.-F.; Nimmagadda, S.; Li, C.; Banerjee, S.R.; Winnard, P.T., Jr.; Artemov, D.; Glunde, K.; Pomper, M.G.; Bhujwalla, Z.M. PSMA-targeted theranostic nanoplex for prostate cancer therapy. ACS Nano 2012, 6, 7752–7762. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Siddiqui, I.A.; Nihal, M.; Pilla, S.; Rosenthal, K.; Mukhtar, H.; Gong, S. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials 2013, 34, 5244–5253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barinka, C.; Byun, Y.; Dusich, C.L.; Banerjee, S.R.; Chen, Y.; Castanares, M.; Kozikowski, A.P.; Mease, R.C.; Pomper, M.G.; Lubkowski, J. Interactions between human glutamate carboxypeptidase II and urea-based inhibitors: Structural characterization. J. Med. Chem. 2008, 51, 7737–7743. [Google Scholar] [CrossRef] [PubMed]
- Kozikowski, A.P.; Zhang, J.; Nan, F.; Petukhov, P.A.; Grajkowska, E.; Wroblewski, J.T.; Yamamoto, T.; Bzdega, T.; Wroblewska, B.; Neale, J.H. Synthesis of urea-based inhibitors as active site probes of glutamate carboxypeptidase II: Efficacy as analgesic agents. J. Med. Chem. 2004, 47, 1729–1738. [Google Scholar] [CrossRef]
- Roy, J.; Nguyen, T.X.; Kanduluru, A.K.; Venkatesh, C.; Lv, W.; Reddy, P.N.; Low, P.S.; Cushman, M. DUPA conjugation of a cytotoxic indenoisoquinoline topoisomerase i inhibitor for selective prostate cancer cell targeting. J. Med. Chem. 2015, 58, 3094–3103. [Google Scholar] [CrossRef]
- Eder, M.; Schäfer, M.; Bauder-Wüst, U.; Hull, W.-E.; Wängler, C.; Mier, W.; Haberkorn, U.; Eisenhut, M. 68Ga-complex lipophilicity and the targeting property of a urea-based PSMA inhibitor for PET imaging. Bioconjug. Chem. 2012, 23, 688–697. [Google Scholar] [CrossRef]
- Chen, Y.; Pullambhatla, M.; Foss, C.A.; Byun, Y.; Nimmagadda, S.; Senthamizhchelvan, S.; Sgouros, G.; Mease, R.C.; Pomper, M.G. 2-(3-{1-Carboxy-5-[(6-[18F] fluoro-pyridine-3-carbonyl)-amino]-pentyl}-ureido)-pentanedioic acid, [18F] DCFPyL, a PSMA-based PET imaging agent for prostate cancer. Clin. Cancer Res. 2011, 17, 7645–7653. [Google Scholar] [CrossRef]
- Kim, C.H.; Axup, J.Y.; Lawson, B.R.; Yun, H.; Tardif, V.; Choi, S.H.; Zhou, Q.; Dubrovska, A.; Biroc, S.L.; Marsden, R. Bispecific small molecule–antibody conjugate targeting prostate cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 17796–17801. [Google Scholar] [CrossRef] [PubMed]
- Wurzer, A.; Seidl, C.; Morgenstern, A.; Bruchertseifer, F.; Schwaiger, M.; Wester, H.-J.; Notnu, J. Dual-Nuclide Radiopharmaceuticals for Positron Emission Tomography Based Dosimetry in Radiotherapy. Chem. Eur. J. 2018, 24, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Wurzer, A.; Vagner, A.; Horvath, D.; Fellegi, F.; Wester, H.-J.; Kalma, F.K.; Notni, J. Synthesis of Symmetric Tetrameric Conjugates of the Radiolanthide Chelator DOTPI for Application in Endoradiotherapy by Means of Click Chemistry. Front. Chem. 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Reich, D.; Wurzer, A.; Wirtz, M.; Stiegler, V.; Spatz, P.; Pollman, J.; Wester, H.-J.; Notni, J. Dendritic poly-chelator frameworks for multimeric bioconjugation. Chem. Commun. 2017, 53, 2586–2589. [Google Scholar] [CrossRef] [PubMed]
- Wurzer, A.; Pollmann, J.; Schmidt, A.; Reich, D.; Wester, H.-J.; Notni, J. Molar Activity of Ga-68 Labeled PSMA Inhibitor Conjugates Determines PET Imaging Results. Mol. Pharm. 2018, 15, 4296–4302. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.-D.; Chung, H.-J.; Lee, S.J.; Lee, S.-H.; Jeong, B.-H.; Kim, H.-K. Synthesis of novel multivalent fluorescent inhibitors with high affinity to prostate cancer and their biological evaluation. Bioorg. Med. Chem. Lett. 2018, 28, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.R.; Pullambhatla, M.; Shallal, H.; Lisok, A.; Mease, R.C.; Pomper, M.G. A modular strategy to prepare multivalent inhibitors of prostate-specific membrane antigen (PSMA). Oncotarget 2011, 2, 1244. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Bauder-Wüst, U.; Leotta, K.; Zoller, F.; Mier, W.; Haberkorn, U.; Eisenhut, M.; Eder, M. A dimerized urea-based inhibitor of the prostate-specific membrane antigen for 68 Ga-PET imaging of prostate cancer. EJNMMI Res. 2012, 2, 23. [Google Scholar] [CrossRef]
- Humblet, V.; Misra, P.; Bhushan, K.R.; Nasr, K.; Ko, Y.-S.; Tsukamoto, T.; Pannier, N.; Frangioni, J.V.; Maison, W. Multivalent scaffolds for affinity maturation of small molecule cell surface binders and their application to prostate tumor targeting. J. Med. Chem. 2008, 52, 544–550. [Google Scholar] [CrossRef]
- Baranyai, Z.; Reich, D.; Vágner, A.; Weineisen, M.; Tóth, I.; Wester, H.-J.; Notni, J. A shortcut to high-affinity Ga-68 and Cu-64 radiopharmaceuticals: One-pot click chemistry trimerisation on the TRAP platform. Dalton Trans. 2015, 44, 11137–11146. [Google Scholar] [CrossRef]
- Huang, B.; Otis, J.; Joice, M.; Kotlyar, A.; Thomas, T.P. PSMA-targeted stably linked “dendrimer-glutamate urea-methotrexate” as a prostate cancer therapeutic. Biomacromolecules 2014, 15, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.M.; Wittrup, K.D. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol. Cancer Ther. 2009, 8, 2861–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Peng, X.; Wang, Y.; Wang, Y.; Shin, D.M.; El-Sayed, M.A.; Nie, S. A reexamination of active and passive tumor targeting by using rod-shaped gold nanocrystals and covalently conjugated peptide ligands. ACS Nano 2010, 4, 5887–5896. [Google Scholar] [CrossRef] [PubMed]
- Kirpotin, D.B.; Drummond, D.C.; Shao, Y.; Shalaby, M.R.; Hong, K.; Nielsen, U.B.; Marks, J.D.; Benz, C.C.; Park, J.W. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006, 66, 6732–6740. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zheng, K.; Yuan, C.; Chen, Z.; Huang, M. Be Active or Not: The Relative Contribution of Active and Passive Tumor Targeting of Nanomaterials. Nanotheranostics 2017, 1, 346. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Mintzer, M.A.; Perez, L.M.; Simanek, E.E. Synthesis of odd generation triazine dendrimers using a divergent, macromonomer approach. Org. Lett. 2010, 12, 1148–1151. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Pavan, G.M.; Annunziata, O.; Simanek, E.E. Experimental and computational evidence for an inversion in guest capacity in high-generation triazine dendrimer hosts. J. Am. Chem. Soc. 2012, 134, 1942–1945. [Google Scholar] [CrossRef]
- Lim, J.; Simanek, E.E. Synthesis of water-soluble dendrimers based on melamine bearing 16 paclitaxel groups. Org. Lett. 2008, 10, 201–204. [Google Scholar] [CrossRef]
- Giri, J.; Diallo, M.S.; Simpson, A.J.; Liu, Y.; Goddard, W.A., III; Kumar, R.; Woods, G.C. Interactions of poly (amidoamine) dendrimers with human serum albumin: Binding constants and mechanisms. ACS Nano 2011, 5, 3456–3468. [Google Scholar] [CrossRef]
- Silver, D.A.; Pellicer, I.; Fair, W.R.; Heston, W.; Cordon-Cardo, C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin. Cancer Res. 1997, 3, 81–85. [Google Scholar]
- Lim, J.; Kostiainen, M.; Maly, J.; da Costa, V.C.P.; Annunziata, O.; Pavan, G.M.; Simanek, E.E. Synthesis of Large Dendrimers with the Dimensions of Small Viruses. J. Am. Chem. Soc. 2013, 135, 4660–4663. [Google Scholar] [CrossRef] [PubMed] [Green Version]

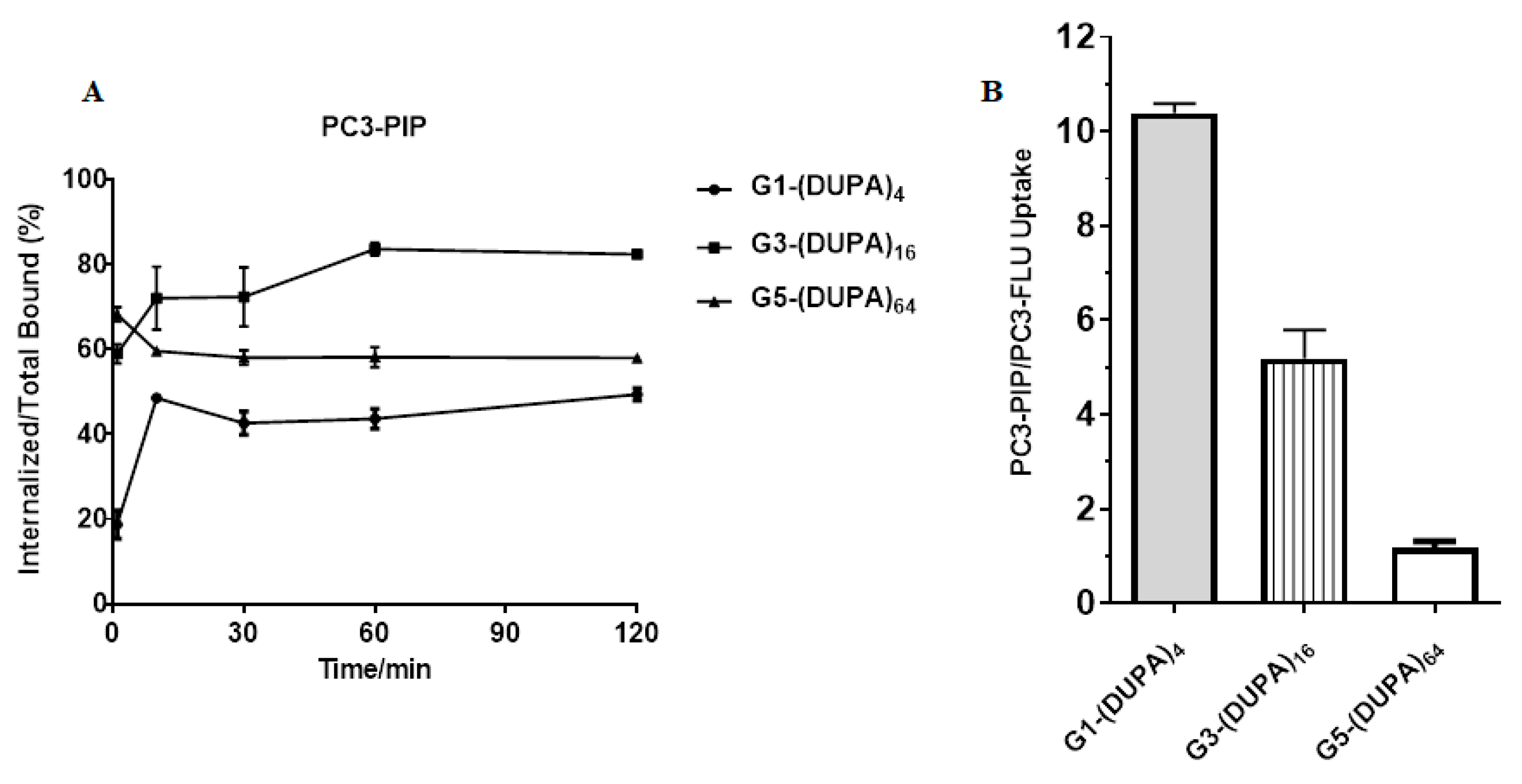
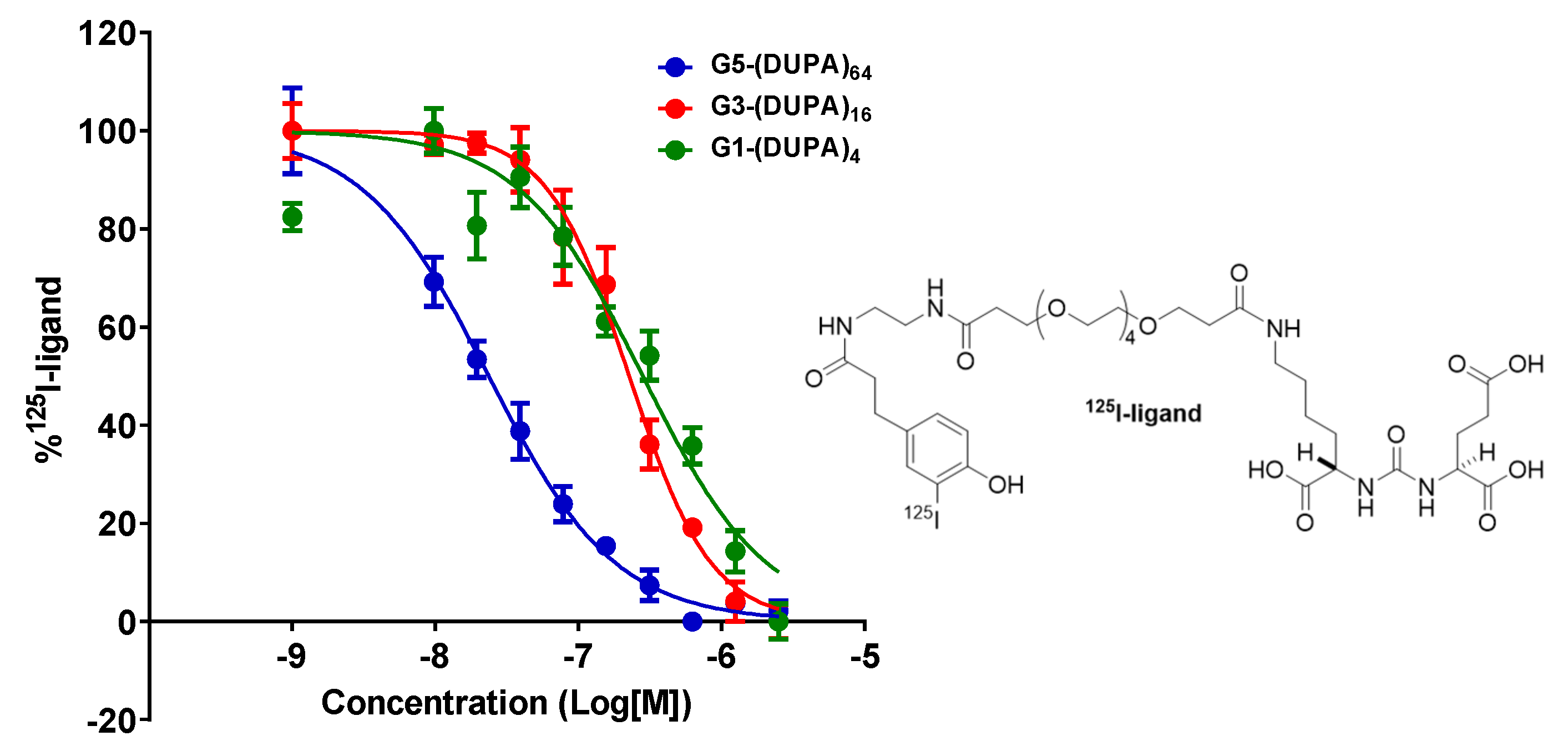


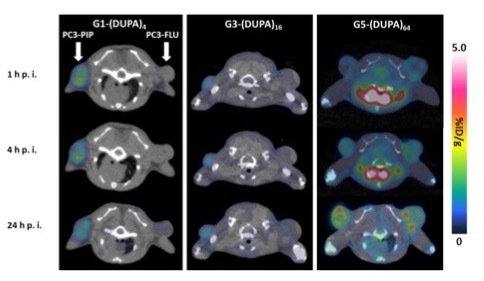
| G1-(DUPA)4 | G3-(DUPA)16 | G5-(DUPA)64 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 h p.i. | 4 h p.i. | 24 h p.i. | 1 h p.i. | 4 h. p.i. | 24 h. p.i. | 1 h p.i. | 4 h. p.i. | 24 h. p.i. | |
| Brain | 0.03 ± 0.01 | 0.02 ± 0.00 | 0.07 ± 0.00 | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.07 ± 0.03 | 0.94 ± 0.10 | 0.66 ± 0.08 | 0.20 ± 0.06 |
| Heart | 0.16 ± 0.05 | 0.10 ± 0.02 | 0.25 ± 0.05 | 0.20 ± 0.03 | 0.17 ± 0.03 | 0.29 ± 0.03 | 15.37 ± 1.46 | 10.77 ± 1.55 | 1.60 ± 0.10 |
| Liver | 1.03 ± 0.16 | 1.20 ± 0.20 | 1.93 ± 0.15 | 1.23 ± 0.21 | 1.45 ± 0.13 | 2,20 ± 0.27 | 11.43 ± 1.27 | 13.67 ± 0.97 | 18.90 ± 1.04 |
| Left Kidney | 53.27 ± 4.37 | 58.73 ± 4.98 | 54.00 ± 5.96 | 42.85 ± 3.43 | 45.00 ± 4.63 | 43.85 ± 5.13 | 6.10 ± 1.57 | 5.47 ± 0.76 | 2.93 ± 0.59 |
| Right Kidney | 56.83 ± 5.14 | 59.97 ± 3.27 | 55.03 ± 7.36 | 44.28 ± 4.79 | 48.55 ± 6.59 | 46.95 ± 6.35 | 6.23 ± 0.29 | 5.73 ± 0.51 | 3.17 ± 0.45 |
| Muscle | 0.06 ± 0.03 | 0.05 ± 0.01 | 0.13 ± 0.03 | 0.20 ± 0.04 | 0.18 ± 0.02 | 0.23 ± 0.03 | 0.49 ± 0.07 | 0.42 ± 0.06 | 0.29 ± 0.06 |
| Bone | 0.08 ± 0.03 | 0.07 ± 0.01 | 0.08 ± 0.02 | 0.27 ± 0.08 | 0.22 ± 0.03 | 0.27 ± 0.08 | 0.93 ± 0.04 | 0.90 ± 0.17 | 0.98 ± 0.20 |
| PC3-PIP | 0.99 ± 0.36 | 0.87 ± 0.40 | 0.81 ± 0.36 | 0.66 ± 0.15 | 0.64 ± 0.11 | 0.67 ± 0.08 | 0.97 ± 0.03 | 1.57 ± 0.15 | 1.83 ± 0.21 |
| PC3-FLU | 0.18 ± 0.05 | 0.13 ± 0.02 | 0.33 ± 0.06 | 0.45 ± 0.06 | 0.42 ± 0.05 | 0.54 ± 0.08 | 1.16 ± 0.21 | 1.83 ± 0.32 | 1.97 ± 0.31 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, J.; Guan, B.; Nham, K.; Hao, G.; Sun, X.; Simanek, E.E. Tumor Uptake of Triazine Dendrimers Decorated with Four, Sixteen, and Sixty-Four PSMA-Targeted Ligands: Passive versus Active Tumor Targeting. Biomolecules 2019, 9, 421. https://doi.org/10.3390/biom9090421
Lim J, Guan B, Nham K, Hao G, Sun X, Simanek EE. Tumor Uptake of Triazine Dendrimers Decorated with Four, Sixteen, and Sixty-Four PSMA-Targeted Ligands: Passive versus Active Tumor Targeting. Biomolecules. 2019; 9(9):421. https://doi.org/10.3390/biom9090421
Chicago/Turabian StyleLim, Jongdoo, Bing Guan, Kien Nham, Guiyang Hao, Xiankai Sun, and Eric E. Simanek. 2019. "Tumor Uptake of Triazine Dendrimers Decorated with Four, Sixteen, and Sixty-Four PSMA-Targeted Ligands: Passive versus Active Tumor Targeting" Biomolecules 9, no. 9: 421. https://doi.org/10.3390/biom9090421