Taking the Occam’s Razor Approach to Hedgehog Lipidation and Its Role in Development
Abstract
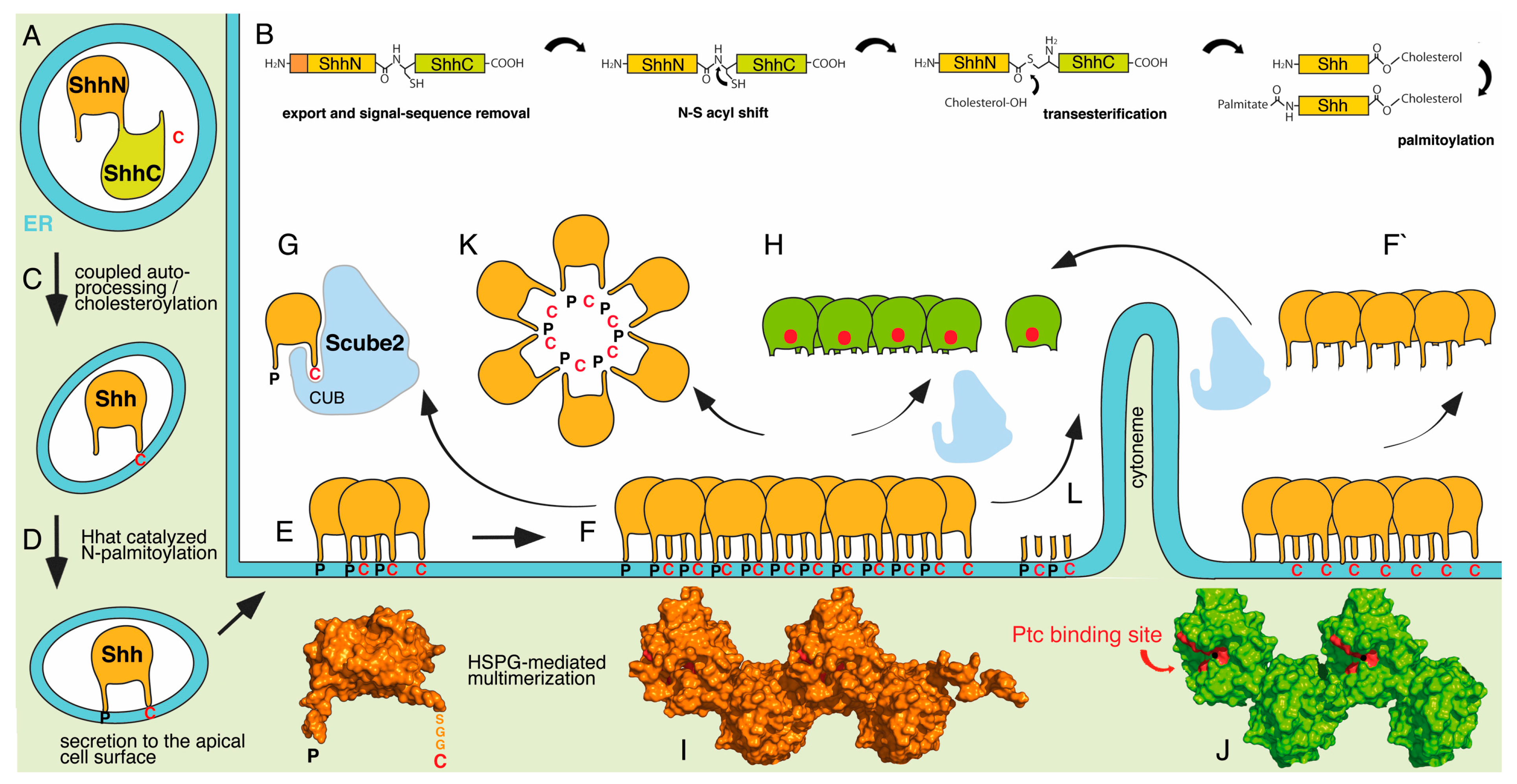
:1. Hedgehog (Hh) Proteins: Extensively Post-Translationally Modified Signals
2. Conserved Post-Translational Hh Lipidation
3. Hh Lipidation as a Prerequisite for Cell-Autonomous Hh Interactions at the Cell Surface
4. Hh Lipidation as a Prerequisite for Cell-Non-Autonomous Hh Interactions with Soluble Molecules
5. Proposed Model of Direct Disp- and Scube2-Mediated Shh Extraction from the Producing Cell
6. Proposed Model of Indirect Scube2 Effects as Enhancers of Proteolytic Shh Processing from the Surface of Producing Cells
7. Proposed Models of Micelle-, Lipophorin- and Membrane-Linked Hh Transport
8. Conclusion and Outlook
Conflicts of Interest
Abbreviations
| Hh | Hedgehog |
| Shh | Sonic hedgehog |
| HhC | C-terminal autoprocessing/cholesterol transferase domain |
| Hh/Shh | fully bioactive, dual-lipidated proteins |
| HhN/ShhN | non-cholesteroylated artificial forms of invertebrate Hh/vertebrate Shh due to deletion of HhC/ShhC ShhC25S represent C-cholesteroylated, N-terminally unpalmitoylated protein |
| PTM | posttranslational protein modification |
| ECM | extracellular matrix |
| Ptc | Patched |
| GPI | glycosylphosphatidyl-inositol |
| ER | endoplasmic reticulum |
| Disp | Dispatched |
| Hhat | Hedgehog acyltransferase, also called Ski (Skinny hedgehog) |
| EGF | epidermal growth factor |
| Scube | Signal sequence, cubulin domain, EGF-like growth factor domain |
| HS | heparan sulfate |
| HSPG | HS proteoglycan |
| PCPE | procollagen C-proteinase enhancer |
| MO | morpholino |
| GFP | green fluorescent protein |
| ESCRT | endosomal sorting complex required for transport |
References
- Ramsbottom, S.A.; Pownall, M.E. Regulation of hedgehog signalling inside and outside the cell. J. Dev. Biol. 2016, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.E. Smoothened translates hedgehog levels into distinct responses. Development 2003, 130, 3951–3963. [Google Scholar] [CrossRef] [PubMed]
- Strigini, M.; Cohen, S.M. A hedgehog activity gradient contributes to ap axial patterning of the drosophila wing. Development 1997, 124, 4697–4705. [Google Scholar] [PubMed]
- Ericson, J.; Morton, S.; Kawakami, A.; Roelink, H.; Jessell, T.M. Two critical periods of sonic hedgehog signaling required for the specification of motor neuron identity. Cell 1996, 87, 661–673. [Google Scholar] [CrossRef]
- Ryan, K.E.; Chiang, C. Hedgehog secretion and signal transduction in vertebrates. J. Biol. Chem. 2012, 287, 17905–17913. [Google Scholar] [CrossRef] [PubMed]
- Nusslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Joyner, A.L. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 2004, 118, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Roelink, H.; Porter, J.A.; Chiang, C.; Tanabe, Y.; Chang, D.T.; Beachy, P.A.; Jessell, T.M. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 1995, 81, 445–455. [Google Scholar] [CrossRef]
- Outram, S.V.; Varas, A.; Pepicelli, C.V.; Crompton, T. Hedgehog signaling regulates differentiation from double-negative to double-positive thymocyte. Immunity 2000, 13, 187–197. [Google Scholar] [CrossRef]
- Rowbotham, N.J.; Hager-Theodorides, A.L.; Cebecauer, M.; Shah, D.K.; Drakopoulou, E.; Dyson, J.; Outram, S.V.; Crompton, T. Activation of the hedgehog signaling pathway in t-lineage cells inhibits tcr repertoire selection in the thymus and peripheral t-cell activation. Blood 2007, 109, 3757–3766. [Google Scholar] [CrossRef] [PubMed]
- Solanki, A.; Lau, C.I.; Saldana, J.I.; Ross, S.; Crompton, T. The transcription factor gli3 promotes b cell development in fetal liver through repression of shh. J. Exp. Med. 2017, 214, 2041–2058. [Google Scholar] [CrossRef] [PubMed]
- Crompton, T.; Outram, S.V.; Hager-Theodorides, A.L. Sonic hedgehog signalling in t-cell development and activation. Nat. Rev. Immunol. 2007, 7, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Vortkamp, A.; Lee, K.; Lanske, B.; Segre, G.V.; Kronenberg, H.M.; Tabin, C.J. Regulation of rate of cartilage differentiation by indian hedgehog and pth-related protein. Science 1996, 273, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Outram, S.V.; Hager-Theodorides, A.L.; Shah, D.K.; Rowbotham, N.J.; Drakopoulou, E.; Ross, S.E.; Lanske, B.; Dessens, J.T.; Crompton, T. Indian hedgehog (ihh) both promotes and restricts thymocyte differentiation. Blood 2009, 113, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- De la Roche, M.; Ritter, A.T.; Angus, K.L.; Dinsmore, C.; Earnshaw, C.H.; Reiter, J.F.; Griffiths, G.M. Hedgehog signaling controls t cell killing at the immunological synapse. Science 2013, 342, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.I.; Outram, S.V.; Saldana, J.I.; Furmanski, A.L.; Dessens, J.T.; Crompton, T. Regulation of murine normal and stress-induced erythropoiesis by desert hedgehog. Blood 2012, 119, 4741–4751. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef] [PubMed]
- Pettigrew, C.A.; Asp, E.; Emerson, C.P., Jr. A new role for hedgehogs in juxtacrine signaling. Mech. Dev. 2014, 131, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Riddle, R.D.; Johnson, R.L.; Laufer, E.; Tabin, C. Sonic hedgehog mediates the polarizing activity of the zpa. Cell 1993, 75, 1401–1416. [Google Scholar] [CrossRef]
- Heemskerk, J.; DiNardo, S. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell 1994, 76, 449–460. [Google Scholar] [CrossRef]
- Li, S.; Wang, M.; Chen, Y.; Wang, W.; Wu, J.; Yu, C.; Zheng, Y.; Pan, Z. Role of the hedgehog signaling pathway in regulating the behavior of germline stem cells. Stem Cells Int. 2017, 2017, 5714608. [Google Scholar] [CrossRef] [PubMed]
- Petrova, R.; Joyner, A.L. Roles for hedgehog signaling in adult organ homeostasis and repair. Development 2014, 141, 3445–3457. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, Y.; Sun, B.; McMahon, A.P.; Wang, Y. Hedgehog signaling: From basic biology to cancer therapy. Cell Chem. Biol. 2017, 24, 252–280. [Google Scholar] [CrossRef] [PubMed]
- Xavier, G.M.; Seppala, M.; Barrell, W.; Birjandi, A.A.; Geoghegan, F.; Cobourne, M.T. Hedgehog receptor function during craniofacial development. Dev. Biol. 2016, 415, 198–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, C.; Litingtung, Y.; Lee, E.; Young, K.E.; Corden, J.L.; Westphal, H.; Beachy, P.A. Cyclopia and defective axial patterning in mice lacking sonic hedgehog gene function. Nature 1996, 383, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Maity, T.; Fuse, N.; Beachy, P.A. Molecular mechanisms of sonic hedgehog mutant effects in holoprosencephaly. Proc. Natl. Acad. Sci. USA 2005, 102, 17026–17031. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Helms, J.A. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development 1999, 126, 4873–4884. [Google Scholar] [PubMed]
- Porter, J.A.; Young, K.E.; Beachy, P.A. Cholesterol modification of hedgehog signaling proteins in animal development. Science 1996, 274, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Bumcrot, D.A.; Takada, R.; McMahon, A.P. Proteolytic processing yields two secreted forms of sonic hedgehog. Mol. Cell. Biol. 1995, 15, 2294–2303. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. A protein splice-junction motif in hedgehog family proteins. Trends Biochem. Sci. 1995, 20, 141–142. [Google Scholar] [CrossRef]
- Perler, F.B. Inbase: The intein database. Nucleic Acids Res. 2002, 30, 383–384. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.A.; Ekker, S.C.; Park, W.J.; von Kessler, D.P.; Young, K.E.; Chen, C.H.; Ma, Y.; Woods, A.S.; Cotter, R.J.; Koonin, E.V.; et al. Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 1996, 86, 21–34. [Google Scholar] [CrossRef]
- Pepinsky, R.B.; Zeng, C.; Wen, D.; Rayhorn, P.; Baker, D.P.; Williams, K.P.; Bixler, S.A.; Ambrose, C.M.; Garber, E.A.; Miatkowski, K.; et al. Identification of a palmitic acid-modified form of human sonic hedgehog. J. Biol. Chem. 1998, 273, 14037–14045. [Google Scholar] [CrossRef] [PubMed]
- Buglino, J.A.; Resh, M.D. Hhat is a palmitoylacyltransferase with specificity for n-palmitoylation of sonic hedgehog. J. Biol. Chem. 2008, 283, 22076–22088. [Google Scholar] [CrossRef] [PubMed]
- Konitsiotis, A.D.; Jovanovic, B.; Ciepla, P.; Spitaler, M.; Lanyon-Hogg, T.; Tate, E.W.; Magee, A.I. Topological analysis of hedgehog acyltransferase, a multi-palmitoylated transmembrane protein. J. Biol. Chem. 2015, 290, 3293–3307. [Google Scholar] [CrossRef] [PubMed]
- Rietveld, A.; Neutz, S.; Simons, K.; Eaton, S. Association of sterol- and glycosylphosphatidylinositol-linked proteins with drosophila raft lipid microdomains. J. Biol. Chem. 1999, 274, 12049–12054. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.; Wolf, A.; Wagner, M.; Kuhlmann, J.; Waldmann, H. The cholesterol membrane anchor of the hedgehog protein confers stable membrane association to lipid-modified proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 8531–8536. [Google Scholar] [CrossRef] [PubMed]
- Levental, I.; Lingwood, D.; Grzybek, M.; Coskun, U.; Simons, K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 22050–22054. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2006, 2, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Goetz, J.A.; Suber, L.M.; Scott, W.J., Jr.; Schreiner, C.M.; Robbins, D.J. A freely diffusible form of sonic hedgehog mediates long-range signalling. Nature 2001, 411, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.M.; Dunn, M.P.; McMahon, J.A.; Logan, M.; Martin, J.F.; St-Jacques, B.; McMahon, A.P. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by ptc1. Cell 2001, 105, 599–612. [Google Scholar] [CrossRef]
- Dawber, R.J.; Hebbes, S.; Herpers, B.; Docquier, F.; van den Heuvel, M. Differential range and activity of various forms of the hedgehog protein. BMC Dev. Biol. 2005, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Goetz, J.A.; Singh, S.; Suber, L.M.; Kull, F.J.; Robbins, D.J. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J. Biol. Chem. 2006, 281, 4087–4093. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Litingtung, Y.; Chiang, C. Region-specific requirement for cholesterol modification of sonic hedgehog in patterning the telencephalon and spinal cord. Development 2007, 134, 2095–2105. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Litingtung, Y.; Chiang, C. Cholesterol modification restricts the spread of shh gradient in the limb bud. Proc. Natl. Acad. Sci. USA 2006, 103, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Li, Y.J.; Kawakami, T.; Xu, S.M.; Chuang, P.T. Palmitoylation is required for the production of a soluble multimeric hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004, 18, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Kohtz, J.D.; Lee, H.Y.; Gaiano, N.; Segal, J.; Ng, E.; Larson, T.; Baker, D.P.; Garber, E.A.; Williams, K.P.; Fishell, G. N-terminal fatty-acylation of sonic hedgehog enhances the induction of rodent ventral forebrain neurons. Development 2001, 128, 2351–2363. [Google Scholar] [PubMed]
- Taylor, F.R.; Wen, D.; Garber, E.A.; Carmillo, A.N.; Baker, D.P.; Arduini, R.M.; Williams, K.P.; Weinreb, P.H.; Rayhorn, P.; Hronowski, X.; et al. Enhanced potency of human sonic hedgehog by hydrophobic modification. Biochemistry 2001, 40, 4359–4371. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.P.; Rayhorn, P.; Chi-Rosso, G.; Garber, E.A.; Strauch, K.L.; Horan, G.S.; Reilly, J.O.; Baker, D.P.; Taylor, F.R.; Koteliansky, V.; et al. Functional antagonists of sonic hedgehog reveal the importance of the n terminus for activity. J. Cell Sci. 1999, 112, 4405–4414. [Google Scholar] [PubMed]
- Lee, J.D.; Kraus, P.; Gaiano, N.; Nery, S.; Kohtz, J.; Fishell, G.; Loomis, C.A.; Treisman, J.E. An acylatable residue of hedgehog is differentially required in drosophila and mouse limb development. Dev. Biol. 2001, 233, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Burke, R.; Nellen, D.; Bellotto, M.; Hafen, E.; Senti, K.A.; Dickson, B.J.; Basler, K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 1999, 99, 803–815. [Google Scholar] [CrossRef]
- Ma, Y.; Erkner, A.; Gong, R.; Yao, S.; Taipale, J.; Basler, K.; Beachy, P.A. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 2002, 111, 63–75. [Google Scholar] [CrossRef]
- Tukachinsky, H.; Kuzmickas, R.P.; Jao, C.Y.; Liu, J.; Salic, A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2012, 2, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Belenkaya, T.Y.; Wang, B.; Lin, X. Drosophila glypicans control the cell-to-cell movement of hedgehog by a dynamin-independent process. Development 2004, 131, 601–611. [Google Scholar] [CrossRef] [PubMed]
- The, I.; Bellaiche, Y.; Perrimon, N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol. Cell 1999, 4, 633–639. [Google Scholar] [CrossRef]
- Bellaiche, Y.; The, I.; Perrimon, N. Tout-velu is a drosophila homologue of the putative tumour suppressor ext-1 and is needed for hh diffusion. Nature 1998, 394, 85–88. [Google Scholar] [PubMed]
- Ortmann, C.; Pickhinke, U.; Exner, S.; Ohlig, S.; Lawrence, R.; Jboor, H.; Dreier, R.; Grobe, K. Sonic hedgehog processing and release are regulated by glypican heparan sulfate proteoglycans. J. Cell Sci. 2015, 128, 2374–2385. [Google Scholar] [CrossRef] [PubMed]
- Vyas, N.; Goswami, D.; Manonmani, A.; Sharma, P.; Ranganath, H.A.; VijayRaghavan, K.; Shashidhara, L.S.; Sowdhamini, R.; Mayor, S. Nanoscale organization of hedgehog is essential for long-range signaling. Cell 2008, 133, 1214–1227. [Google Scholar] [CrossRef] [PubMed]
- Wojcinski, A.; Nakato, H.; Soula, C.; Glise, B. Dsulfatase-1 fine-tunes hedgehog patterning activity through a novel regulatory feedback loop. Dev. Biol. 2011, 358, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Al Oustah, A.; Danesin, C.; Khouri-Farah, N.; Farreny, M.A.; Escalas, N.; Cochard, P.; Glise, B.; Soula, C. Dynamics of sonic hedgehog signaling in the ventral spinal cord are controlled by intrinsic changes in source cells requiring sulfatase 1. Development 2014, 141, 1392–1403. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.F.; Yan, Y.T.; Wu, S.Y.; Djoko, B.; Tsai, M.T.; Cheng, C.J.; Yang, R.B. Domain and functional analysis of a novel platelet-endothelial cell surface protein, scube1. J. Biol. Chem. 2008, 283, 12478–12488. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.B.; Ng, C.K.; Wasserman, S.M.; Colman, S.D.; Shenoy, S.; Mehraban, F.; Komuves, L.G.; Tomlinson, J.E.; Topper, J.N. Identification of a novel family of cell-surface proteins expressed in human vascular endothelium. J. Biol. Chem. 2002, 277, 46364–46373. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.T.; Su, Y.H.; Tsai, M.T.; Wasserman, S.M.; Topper, J.N.; Yang, R.B. A novel secreted, cell-surface glycoprotein containing multiple epidermal growth factor-like repeats and one cub domain is highly expressed in primary osteoblasts and bones. J. Biol. Chem. 2004, 279, 37485–37490. [Google Scholar] [CrossRef] [PubMed]
- Hollway, G.E.; Maule, J.; Gautier, P.; Evans, T.M.; Keenan, D.G.; Lohs, C.; Fischer, D.; Wicking, C.; Currie, P.D. Scube2 mediates hedgehog signalling in the zebrafish embryo. Dev. Biol. 2006, 294, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, A.; Nojima, Y.; Toyoda, A.; Takahoko, M.; Satoh, M.; Tanaka, H.; Wada, H.; Masai, I.; Terasaki, H.; Sakaki, Y.; et al. The zebrafish-secreted matrix protein you/scube2 is implicated in long-range regulation of hedgehog signaling. Curr. Biol. 2005, 15, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Woods, I.G.; Talbot, W.S. The you gene encodes an egf-cub protein essential for hedgehog signaling in zebrafish. PLoS Biol. 2005, 3, e66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauerte, H.E.; van Eeden, F.J.; Fricke, C.; Odenthal, J.; Strahle, U.; Haffter, P. Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development 1998, 125, 2983–2993. [Google Scholar] [PubMed]
- Johnson, J.L.; Hall, T.E.; Dyson, J.M.; Sonntag, C.; Ayers, K.; Berger, S.; Gautier, P.; Mitchell, C.; Hollway, G.E.; Currie, P.D. Scube activity is necessary for hedgehog signal transduction in vivo. Dev. Biol. 2012, 368, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.T.; Cheng, C.J.; Lin, Y.C.; Chen, C.C.; Wu, A.R.; Wu, M.T.; Hsu, C.C.; Yang, R.B. Isolation and characterization of a secreted, cell-surface glycoprotein scube2 from humans. Biochem. J. 2009, 422, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Creanga, A.; Glenn, T.D.; Mann, R.K.; Saunders, A.M.; Talbot, W.S.; Beachy, P.A. Scube/you activity mediates release of dually lipid-modified hedgehog signal in soluble form. Genes Dev. 2012, 26, 1312–1325. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, P.; Exner, S.; Schurmann, S.; Pickhinke, U.; Bandari, S.; Ortmann, C.; Kupich, S.; Schulz, P.; Hansen, U.; Seidler, D.G.; et al. Scube2 enhances proteolytic shh processing from the surface of shh-producing cells. J. Cell Sci. 2014, 127, 1726–1737. [Google Scholar] [CrossRef] [PubMed]
- van Eeden, F.J.; Granato, M.; Schach, U.; Brand, M.; Furutani-Seiki, M.; Haffter, P.; Hammerschmidt, M.; Heisenberg, C.P.; Jiang, Y.J.; Kane, D.A.; et al. Mutations affecting somite formation and patterning in the zebrafish, danio rerio. Development 1996, 123, 153–164. [Google Scholar] [PubMed]
- Liao, W.J.; Tsao, K.C.; Yang, R.B. Electrostatics and n-glycan-mediated membrane tethering of scube1 is critical for promoting bone morphogenetic protein signalling. Biochem. J. 2016, 473, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, P.; Schulz, P.; Ortmann, C.; Schurmann, S.; Exner, S.; Rebollido-Rios, R.; Dreier, R.; Seidler, D.G.; Grobe, K. Bridging the gap: Heparan sulfate and scube2 assemble sonic hedgehog release complexes at the surface of producing cells. Sci. Rep. 2016, 6, 26435. [Google Scholar] [CrossRef] [PubMed]
- Tukachinsky, H.; Petrov, K.; Watanabe, M.; Salic, A. Mechanism of inhibition of the tumor suppressor patched by sonic hedgehog. Proc. Natl. Acad. Sci. USA 2016, 113, E5866–E5875. [Google Scholar] [CrossRef] [PubMed]
- Lopez, C.A.; de Vries, A.H.; Marrink, S.J. Computational microscopy of cyclodextrin mediated cholesterol extraction from lipid model membranes. Sci. Rep. 2013, 3, 2071. [Google Scholar] [CrossRef] [PubMed]
- Lopez, C.A.; de Vries, A.H.; Marrink, S.J. Molecular mechanism of cyclodextrin mediated cholesterol extraction. PLoS Comput. Biol. 2011, 7, e1002020. [Google Scholar] [CrossRef] [PubMed]
- Chamoun, Z.; Mann, R.K.; Nellen, D.; von Kessler, D.P.; Bellotto, M.; Beachy, P.A.; Basler, K. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 2001, 293, 2080–2084. [Google Scholar] [CrossRef] [PubMed]
- Konitsiotis, A.D.; Chang, S.C.; Jovanovic, B.; Ciepla, P.; Masumoto, N.; Palmer, C.P.; Tate, E.W.; Couchman, J.R.; Magee, A.I. Attenuation of hedgehog acyltransferase-catalyzed sonic hedgehog palmitoylation causes reduced signaling, proliferation and invasiveness of human carcinoma cells. PLoS ONE 2014, 9, e89899. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE 2006, 2006, re14. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Tokhunts, R.; Old, W.M.; Houel, S.; Rodgriguez-Blanco, J.; Singh, S.; Schilling, N.; Anthony, J.C.; Ahn, N.G.; Robbins, D.J. Identification of a family of fatty-acid-speciated sonic hedgehog proteins, whose members display differential biological properties. Cell Rep. 2015, 10, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Font, B.; Eichenberger, D.; Moreau, C.; Ricard-Blum, S.; Hulmes, D.J.; Moali, C. Insights into how cub domains can exert specific functions while sharing a common fold: Conserved and specific features of the cub1 domain contribute to the molecular basis of procollagen c-proteinase enhancer-1 activity. J. Biol. Chem. 2007, 282, 16924–16933. [Google Scholar] [CrossRef] [PubMed]
- Gaboriaud, C.; Teillet, F.; Gregory, L.A.; Thielens, N.M.; Arlaud, G.J. Assembly of c1 and the mbl- and ficolin-masp complexes: Structural insights. Immunobiology 2007, 212, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Takahara, K.; Kessler, E.; Biniaminov, L.; Brusel, M.; Eddy, R.L.; Jani-Sait, S.; Shows, T.B.; Greenspan, D.S. Type i procollagen cooh-terminal proteinase enhancer protein: Identification, primary structure, and chromosomal localization of the cognate human gene (pcolce). J. Biol. Chem. 1994, 269, 26280–26285. [Google Scholar] [PubMed]
- Bourhis, J.M.; Vadon-Le Goff, S.; Afrache, H.; Mariano, N.; Kronenberg, D.; Thielens, N.; Moali, C.; Hulmes, D.J. Procollagen c-proteinase enhancer grasps the stalk of the c-propeptide trimer to boost collagen precursor maturation. Proc. Natl. Acad. Sci. USA 2013, 110, 6394–6399. [Google Scholar] [CrossRef] [PubMed]
- Hulmes, D.J.; Mould, A.P.; Kessler, E. The cub domains of procollagen c-proteinase enhancer control collagen assembly solely by their effect on procollagen c-proteinase/bone morphogenetic protein-1. Matrix Biol. 1997, 16, 41–45. [Google Scholar] [CrossRef]
- Weiss, T.; Brusel, M.; Rousselle, P.; Kessler, E. The ntr domain of procollagen c-proteinase enhancer-1 (pcpe-1) mediates pcpe-1 binding to syndecans-1, -2 and -4 as well as fibronectin. Int. J. Biochem. Cell Biol. 2014, 57, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Bekhouche, M.; Kronenberg, D.; Vadon-Le Goff, S.; Bijakowski, C.; Lim, N.H.; Font, B.; Kessler, E.; Colige, A.; Nagase, H.; Murphy, G.; et al. Role of the netrin-like domain of procollagen c-proteinase enhancer-1 in the control of metalloproteinase activity. J. Biol. Chem. 2010, 285, 15950–15959. [Google Scholar] [CrossRef] [PubMed]
- Dierker, T.; Dreier, R.; Petersen, A.; Bordych, C.; Grobe, K. Heparan sulfate-modulated, metalloprotease-mediated sonic hedgehog release from producing cells. J. Biol. Chem. 2009, 284, 8013–8022. [Google Scholar] [CrossRef] [PubMed]
- Ohlig, S.; Farshi, P.; Pickhinke, U.; van den Boom, J.; Hoing, S.; Jakuschev, S.; Hoffmann, D.; Dreier, R.; Scholer, H.R.; Dierker, T.; et al. Sonic hedgehog shedding results in functional activation of the solubilized protein. Dev. Cell 2011, 20, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, P.; Schulz, P.; Schurmann, S.; Niland, S.; Exner, S.; Rebollido-Rios, R.; Manikowski, D.; Hoffmann, D.; Seidler, D.G.; Grobe, K. Calcium coordination controls sonic hedgehog structure and scube2-cubulin domain regulated release. J. Cell Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Damhofer, H.; Veenstra, V.L.; Tol, J.A.; van Laarhoven, H.W.; Medema, J.P.; Bijlsma, M.F. Blocking hedgehog release from pancreatic cancer cells increases paracrine signaling potency. J. Cell Sci. 2015, 128, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Miura, G.I.; Buglino, J.; Alvarado, D.; Lemmon, M.A.; Resh, M.D.; Treisman, J.E. Palmitoylation of the egfr ligand spitz by rasp increases spitz activity by restricting its diffusion. Dev. Cell 2006, 10, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Hardy, R.Y.; Resh, M.D. Identification of N-terminal residues of sonic hedgehog important for palmitoylation by hedgehog acyltransferase. J. Biol. Chem. 2012, 287, 42881–42889. [Google Scholar] [CrossRef] [PubMed]
- Kakugawa, S.; Langton, P.F.; Zebisch, M.; Howell, S.A.; Chang, T.H.; Liu, Y.; Feizi, T.; Bineva, G.; O’Reilly, N.; Snijders, A.P.; et al. Notum deacylates wnt proteins to suppress signalling activity. Nature 2015, 519, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Buglino, J.A.; Resh, M.D. Palmitoylation of hedgehog proteins. Vitam. Horm. 2012, 88, 229–252. [Google Scholar] [PubMed]
- Peschon, J.J.; Slack, J.L.; Reddy, P.; Stocking, K.L.; Sunnarborg, S.W.; Lee, D.C.; Russell, W.E.; Castner, B.J.; Johnson, R.S.; Fitzner, J.N.; et al. An essential role for ectodomain shedding in mammalian development. Science 1998, 282, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; Swierczynska, M.M.; Kumari, V.; Ehrhart-Bornstein, M.; Bornstein, S.R.; Eaton, S. Secretion and signaling activities of lipoprotein-associated hedgehog and non-sterol-modified hedgehog in flies and mammals. PLoS Biol. 2013, 11, e1001505. [Google Scholar] [CrossRef] [PubMed]
- Pepinsky, R.B.; Rayhorn, P.; Day, E.S.; Dergay, A.; Williams, K.P.; Galdes, A.; Taylor, F.R.; Boriack-Sjodin, P.A.; Garber, E.A. Mapping sonic hedgehog-receptor interactions by steric interference. J. Biol. Chem. 2000, 275, 10995–11001. [Google Scholar] [CrossRef] [PubMed]
- Bishop, B.; Aricescu, A.R.; Harlos, K.; O’Callaghan, C.A.; Jones, E.Y.; Siebold, C. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein hhip. Nat. Struct. Mol. Biol. 2009, 16, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Bosanac, I.; Maun, H.R.; Scales, S.J.; Wen, X.; Lingel, A.; Bazan, J.F.; de Sauvage, F.J.; Hymowitz, S.G.; Lazarus, R.A. The structure of shh in complex with hhip reveals a recognition role for the shh pseudo active site in signaling. Nat. Struct. Mol. Biol. 2009, 16, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Maun, H.R.; Wen, X.; Lingel, A.; de Sauvage, F.J.; Lazarus, R.A.; Scales, S.J.; Hymowitz, S.G. The hedgehog pathway antagonist 5e1 binds hedgehog at the pseudo-active site. J. Biol. Chem. 2010, 285, 26570–26580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Abreu, J.G.; Yokota, C.; MacDonald, B.T.; Singh, S.; Coburn, K.L.; Cheong, S.M.; Zhang, M.M.; Ye, Q.Z.; Hang, H.C.; et al. Tiki1 is required for head formation via wnt cleavage-oxidation and inactivation. Cell 2012, 149, 1565–1577. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, C.E.; Jeong, J.; Guo, C.; Allen, B.L.; McMahon, A.P. Notochord-derived shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. Development 2008, 135, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Matthews, V.; Schuster, B.; Schutze, S.; Bussmeyer, I.; Ludwig, A.; Hundhausen, C.; Sadowski, T.; Saftig, P.; Hartmann, D.; Kallen, K.J.; et al. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by adam10 and adam17 (tace). J. Biol. Chem. 2003, 278, 38829–38839. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Huang, M.; Sun, S.; Wu, Y.; Lin, X. Epithelial heparan sulfate regulates sonic hedgehog signaling in lung development. PLoS Genet. 2017, 13, e1006992. [Google Scholar] [CrossRef] [PubMed]
- Xavier, G.M.; Panousopoulos, L.; Cobourne, M.T. Scube3 is expressed in multiple tissues during development but is dispensable for embryonic survival in the mouse. PLoS ONE 2013, 8, e55274. [Google Scholar] [CrossRef]
- Muller, P.; Schier, A.F. Extracellular movement of signaling molecules. Dev. Cell 2011, 21, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.; Kondev, J.; Theriot, J. Physical Biology of the Cell, 2nd ed.; Garland Science, Taylor and Francis Group LLC: New York, NY, USA, 2009; pp. 312–314. ISBN 978-0815344506. [Google Scholar]
- Berg, H.C. Random Walks in Biology; Princeton University Press: Princeton, NJ, USA, 1993. [Google Scholar]
- Gallet, A.; Therond, P.P. Temporal modulation of the hedgehog morphogen gradient by a patched-dependent targeting to lysosomal compartment. Dev. Biol. 2005, 277, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Panakova, D.; Sprong, H.; Marois, E.; Thiele, C.; Eaton, S. Lipoprotein particles are required for hedgehog and wingless signalling. Nature 2005, 435, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Eugster, C.; Panakova, D.; Mahmoud, A.; Eaton, S. Lipoprotein-heparan sulfate interactions in the hh pathway. Dev. Cell 2007, 13, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Gradilla, A.C.; Gonzalez, E.; Seijo, I.; Andres, G.; Bischoff, M.; Gonzalez-Mendez, L.; Sanchez, V.; Callejo, A.; Ibanez, C.; Guerra, M.; et al. Exosomes as hedgehog carriers in cytoneme-mediated transport and secretion. Nat. Commun. 2014, 5, 5649. [Google Scholar] [CrossRef] [PubMed]
- Parchure, A.; Vyas, N.; Ferguson, C.; Parton, R.G.; Mayor, S. Oligomerization and endocytosis of hedgehog is necessary for its efficient exovesicular secretion. Mol. Biol. Cell 2015, 26, 4700–4717. [Google Scholar] [CrossRef] [PubMed]
- Matusek, T.; Wendler, F.; Poles, S.; Pizette, S.; D’Angelo, G.; Furthauer, M.; Therond, P.P. The escrt machinery regulates the secretion and long-range activity of hedgehog. Nature 2014, 516, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Gallet, A.; Ruel, L.; Staccini-Lavenant, L.; Therond, P.P. Cholesterol modification is necessary for controlled planar long-range activity of hedgehog in drosophila epithelia. Development 2006, 133, 407–418. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G.; Matusek, T.; Pizette, S.; Therond, P.P. Endocytosis of hedgehog through dispatched regulates long-range signaling. Dev. Cell 2015, 32, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Ayers, K.L.; Gallet, A.; Staccini-Lavenant, L.; Therond, P.P. The long-range activity of hedgehog is regulated in the apical extracellular space by the glypican dally and the hydrolase notum. Dev. Cell 2010, 18, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Callejo, A.; Bilioni, A.; Mollica, E.; Gorfinkiel, N.; Andres, G.; Ibanez, C.; Torroja, C.; Doglio, L.; Sierra, J.; Guerrero, I. Dispatched mediates hedgehog basolateral release to form the long-range morphogenetic gradient in the drosophila wing disk epithelium. Proc. Natl. Acad. Sci. USA 2011, 108, 12591–12598. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Hannoush, R.N. Method for cellular imaging of palmitoylated proteins with clickable probes and proximity ligation applied to hedgehog, tubulin, and ras. J. Am. Chem. Soc. 2014, 136, 4544–4550. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, P.; Nabi, I.R. Lipid rafts, caveolae, and their endocytosis. Int. Rev. Cell Mol. Biol. 2010, 282, 135–163. [Google Scholar] [PubMed]
- Vincent, S.; Thomas, A.; Brasher, B.; Benson, J.D. Targeting of proteins to membranes through hedgehog auto-processing. Nat. Biotechnol. 2003, 21, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Hsiung, F.; Ramirez-Weber, F.A.; Iwaki, D.D.; Kornberg, T.B. Dependence of drosophila wing imaginal disc cytonemes on decapentaplegic. Nature 2005, 437, 560–563. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Hsiung, F.; Kornberg, T.B. Specificity of drosophila cytonemes for distinct signaling pathways. Science 2011, 332, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Huang, H.; Liu, S.; Kornberg, T.B. Cytoneme-mediated contact-dependent transport of the drosophila decapentaplegic signaling protein. Science 2014, 343, 1244624. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Huang, H.; Hatori, R.; Kornberg, T.B. Essential basal cytonemes take up hedgehog in the drosophila wing imaginal disc. Development 2017, 144, 3134–3144. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, M.; Gradilla, A.C.; Seijo, I.; Andres, G.; Rodriguez-Navas, C.; Gonzalez-Mendez, L.; Guerrero, I. Cytonemes are required for the establishment of a normal hedgehog morphogen gradient in drosophila epithelia. Nat. Cell Biol. 2013, 15, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Sanders, T.A.; Llagostera, E.; Barna, M. Specialized filopodia direct long-range transport of shh during vertebrate tissue patterning. Nature 2013, 497, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Mendez, L.; Seijo-Barandiaran, I.; Guerrero, I. Cytoneme-mediated cell-cell contacts for hedgehog reception. Elife 2017, 6, e24045. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, I.; Kornberg, T.B. Hedgehog and its circuitous journey from producing to target cells. Semin. Cell Dev. Biol. 2014, 33, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Vyas, N.; Walvekar, A.; Tate, D.; Lakshmanan, V.; Bansal, D.; Lo Cicero, A.; Raposo, G.; Palakodeti, D.; Dhawan, J. Vertebrate hedgehog is secreted on two types of extracellular vesicles with different signaling properties. Sci. Rep. 2014, 4, 7357. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Struhl, G. Dual roles for patched in sequestering and transducing hedgehog. Cell 1996, 87, 553–563. [Google Scholar] [CrossRef]
| Hh Cholesterol Extraction by Scube2 | Scube2-Regulated Hh Shedding | Micelle, Lipoprotein, and Exosome Transport | Model of Hh Transport Via Cytonemes | |
|---|---|---|---|---|
| Dual Hh membrane association and multimerization | − Problem: Energy required for Hh release from membrane | + No energy required, general release mechanism in ECM | − Problem: Energy required for Hh release from membrane | − Problem: required Hh transfer at the cytoneme synapse |
| Established CUB-domain functions, exergonic Hh release | ? CUB binding/extraction of cholesterol endergonic | + CUBs are established protease regulators | ? Not addressed | ? Not addressed |
| Variable role of palmitate and functionality of other hydrophobic Hh modifications | ? Not addressed | + Processing of lipidated termini activates Hh, lipids function indirectly as membrane tethers | ? Not addressed | ? Not addressed |
| Short-range and long-range Hh transport | − Problem: Subsequent diffusion-based transport is not sufficient | − Problem: Subsequent diffusion-based transport is not sufficient | − Problem: Subsequent diffusion-based transport is not sufficient | + Regulated transport, no long-range diffusion required |
| HSPGs in Hh release and transport | ? Not addressed | + Recruit Scube2 and generate release hubs (in producing compartment) | + Permissive factors for Hh transfer and transport | + Permissive factors for cytoneme extension (in receiving compartment) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manikowski, D.; Kastl, P.; Grobe, K. Taking the Occam’s Razor Approach to Hedgehog Lipidation and Its Role in Development. J. Dev. Biol. 2018, 6, 3. https://doi.org/10.3390/jdb6010003
Manikowski D, Kastl P, Grobe K. Taking the Occam’s Razor Approach to Hedgehog Lipidation and Its Role in Development. Journal of Developmental Biology. 2018; 6(1):3. https://doi.org/10.3390/jdb6010003
Chicago/Turabian StyleManikowski, Dominique, Philipp Kastl, and Kay Grobe. 2018. "Taking the Occam’s Razor Approach to Hedgehog Lipidation and Its Role in Development" Journal of Developmental Biology 6, no. 1: 3. https://doi.org/10.3390/jdb6010003
APA StyleManikowski, D., Kastl, P., & Grobe, K. (2018). Taking the Occam’s Razor Approach to Hedgehog Lipidation and Its Role in Development. Journal of Developmental Biology, 6(1), 3. https://doi.org/10.3390/jdb6010003





