Lipid Raft, Regulator of Plasmodesmal Callose Homeostasis
Abstract
:1. Lipid Raft Components
2. The Action of Plasmodesmata Callose
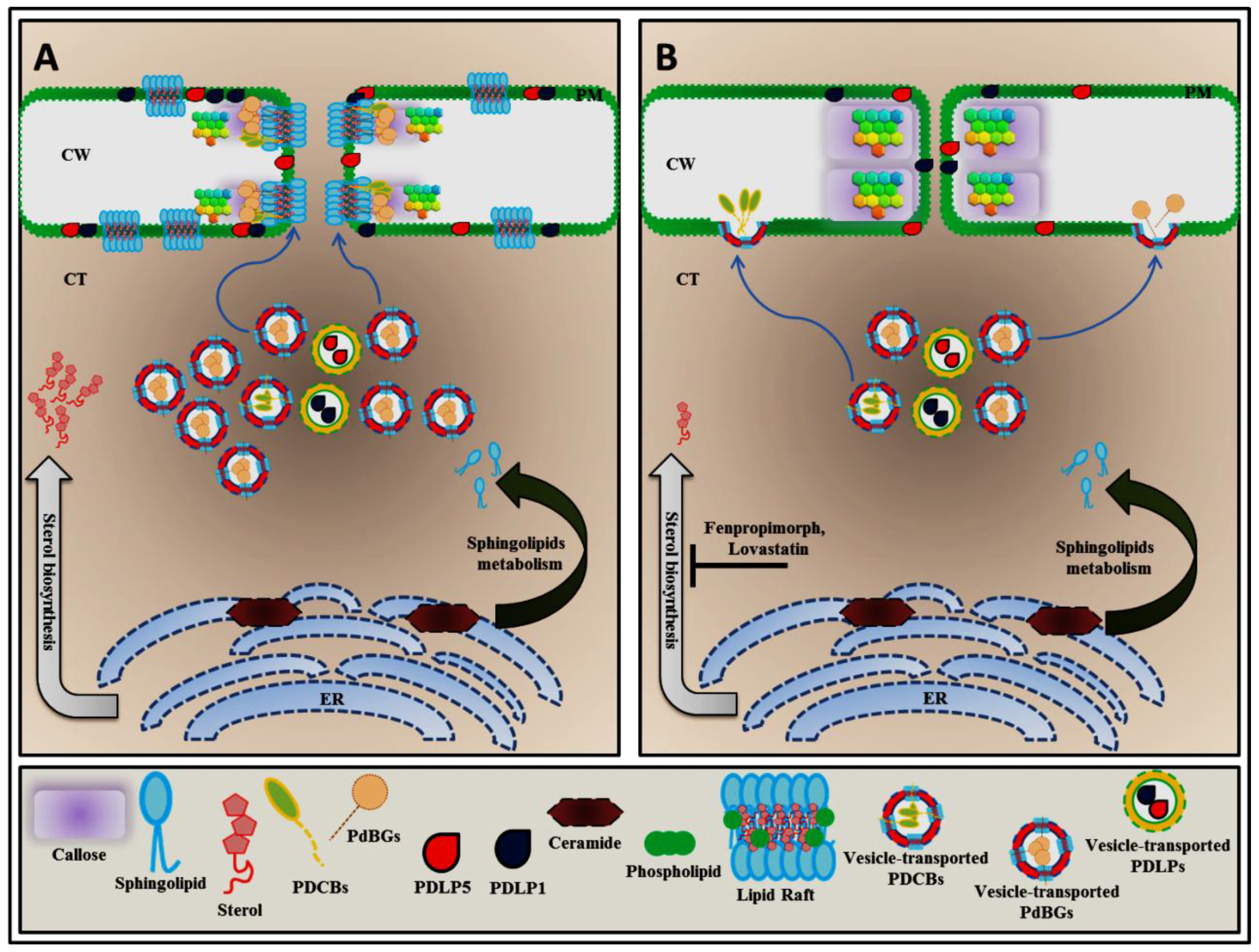
3. Sphingolipid and Sterol Biosynthesis Pathway Involved in Plasmodesmata Callose Maintenance
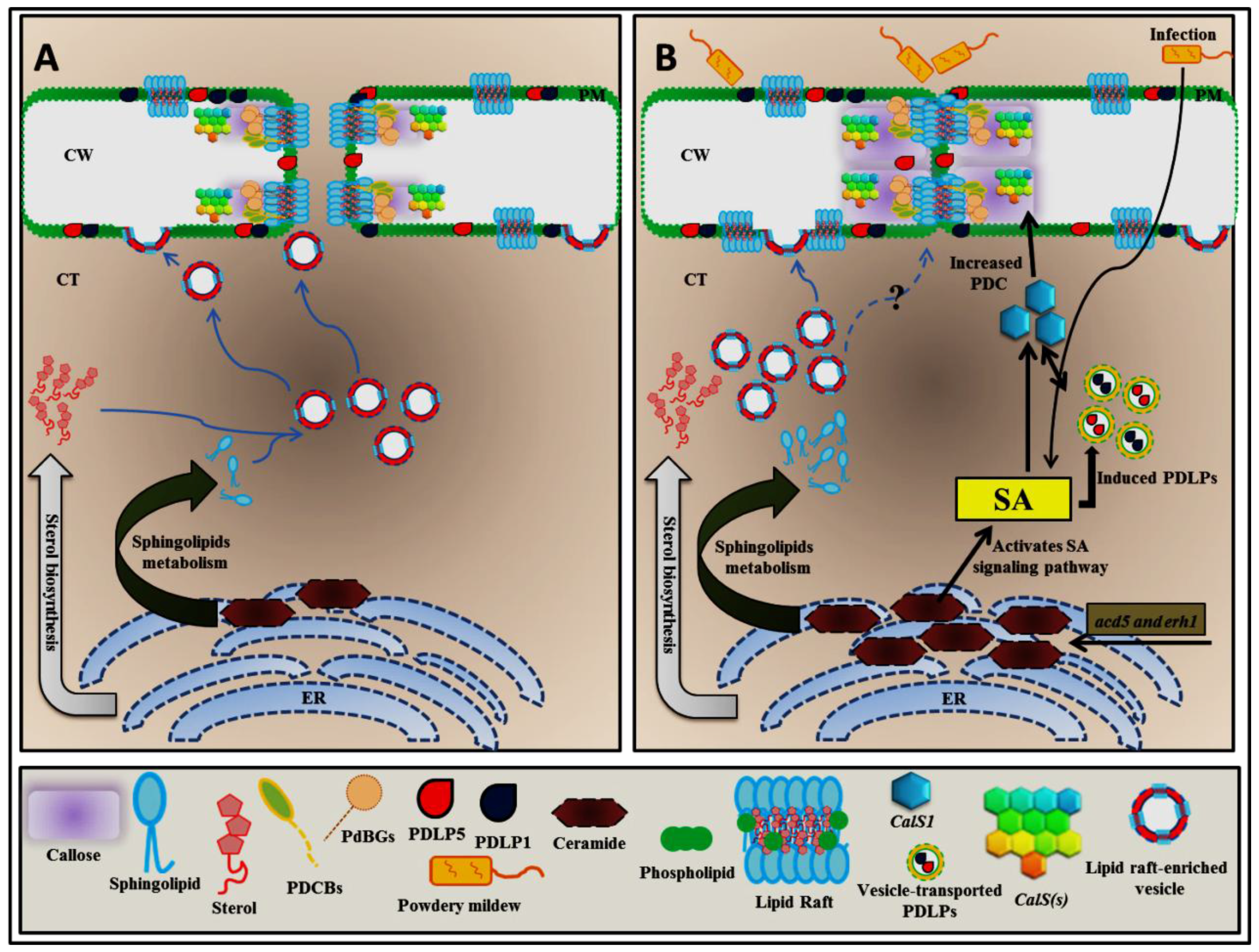
4. Alteration of Sphingolipid Homeostasis Controls PDLP5 (PLASMODESMATA-LOCATED PROTEIN 5) Expression through Salicylic Acid (SA)-Dependent Pathway
5. Plasmodesmal Localization of GPI-Anchored Plasmodesmata Proteins is Regulated by Lipid Rafts
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Korn, E.D. Structure and function of the plasma membrane. A biochemical perspective. J. Gen. Physiol. 1968, 52, 257s–278s. [Google Scholar] [CrossRef]
- Frohlich, F.; Christiano, R.; Olson, D.K.; Alcazar-Roman, A.; DeCamilli, P.; Walther, T.C. A role for eisosomes in maintenance of plasma membrane phosphoinositide levels. Mol. Biol. Cell 2014, 25, 2797–2806. [Google Scholar] [CrossRef] [PubMed]
- Yeagle, P.L. Lipid regulation of cell membrane structure and function. FASEB J. 1989, 3, 1833–1842. [Google Scholar] [PubMed]
- Karnovsky, M.J.; Kleinfeld, A.M.; Hoover, R.L.; Klausner, R.D. The concept of lipid domains in membranes. J. Cell Biol. 1982, 94, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, K.; Mongrand, S.; Beney, L.; Simon-Plas, F.; Gerbeau-Pissot, P. Differential effect of plant lipids on membrane organization: Specificities of phytosphingolipids and phytosterols. J. Biol. Chem. 2015, 290, 5810–5825. [Google Scholar] [CrossRef] [PubMed]
- Grison, M.S.; Brocard, L.; Fouillen, L.; Nicolas, W.; Wewer, V.; Dormann, P.; Nacir, H.; Benitez-Alfonso, Y.; Claverol, S.; Germain, V.; et al. Specific membrane lipid composition is important for plasmodesmata function in arabidopsis. Plant Cell 2015, 27, 1228–1250. [Google Scholar] [CrossRef] [PubMed]
- Mongrand, S.; Stanislas, T.; Bayer, E.M.; Lherminier, J.; Simon-Plas, F. Membrane rafts in plant cells. Trends Plant Sci. 2010, 15, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Laude, A.J.; Prior, I.A. Plasma membrane microdomains: Organization, function and trafficking. Mol. Membr. Biol. 2004, 21, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Duggan, J.; Jamal, G.; Tilley, M.; Davis, B.; McKenzie, G.; Vere, K.; Somekh, M.G.; O’Shea, P.; Harris, H. Functional imaging of microdomains in cell membranes. Eur. Biophys. J. 2008, 37, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Ando, J.; Kinoshita, M.; Cui, J.; Yamakoshi, H.; Dodo, K.; Fujita, K.; Murata, M.; Sodeoka, M. Sphingomyelin distribution in lipid rafts of artificial monolayer membranes visualized by raman microscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 4558–4563. [Google Scholar] [CrossRef] [PubMed]
- Frisz, J.F.; Klitzing, H.A.; Lou, K.; Hutcheon, I.D.; Weber, P.K.; Zimmerberg, J.; Kraft, M.L. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J. Biol. Chem. 2013, 288, 16855–16861. [Google Scholar] [CrossRef] [PubMed]
- Peskan, T.; Oelmuller, R. Heterotrimeric G-protein β-subunit is localized in the plasma membrane and nuclei of tobacco leaves. Plant Mol. Biol. 2000, 42, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Peskan, T.; Westermann, M.; Oelmuller, R. Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. FEBS J. 2000, 267, 6989–6995. [Google Scholar] [CrossRef]
- Borner, G.H.; Sherrier, D.J.; Weimar, T.; Michaelson, L.V.; Hawkins, N.D.; Macaskill, A.; Napier, J.A.; Beale, M.H.; Lilley, K.S.; Dupree, P. Analysis of detergent-resistant membranes in arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005, 137, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Garner, A.E.; Smith, D.A.; Hooper, N.M. Visualization of detergent solubilization of membranes: Implications for the isolation of rafts. Biophys. J. 2008, 94, 1326–1340. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Gerl, M.J. Revitalizing membrane rafts: New tools and insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.; Mouritsen, O.G.; Anderson, R.G. Lipid rafts: At a crossroad between cell biology and physics. Nat. Cell Biol. 2007, 9, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, F.J.; Douglas, L.M.; Konopka, J.B. Sterol-rich plasma membrane domains in fungi. Eukaryot. Cell 2007, 6, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Sampaio, J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.A.; Rose, J.K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 1992, 68, 533–544. [Google Scholar] [CrossRef]
- Brown, D.A. Interactions between GPI-anchored proteins and membrane lipids. Trends Cell Biol. 1992, 2, 338–343. [Google Scholar] [CrossRef]
- Grennan, A.K. Lipid rafts in plants. Plant Physiol. 2007, 143, 1083–1085. [Google Scholar] [CrossRef] [PubMed]
- Korlach, J.; Schwille, P.; Webb, W.W.; Feigenson, G.W. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA 1999, 96, 8461–8466. [Google Scholar] [CrossRef] [PubMed]
- Klymchenko, A.S.; Kreder, R. Fluorescent probes for lipid rafts: From model membranes to living cells. Chem. Biol. 2014, 21, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Silvius, J.R. Role of cholesterol in lipid raft formation: Lessons from lipid model systems. Biochim. Biophys. Acta 2003, 1610, 174–183. [Google Scholar] [CrossRef]
- Dufourc, E.J. Sterols and membrane dynamics. J. Chem. Biol. 2008, 1, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Wewer, V.; Dombrink, I.; vom Dorp, K.; Dormann, P. Quantification of sterol lipids in plants by quadrupole time-of-flight mass spectrometry. J. Lipid Res. 2011, 52, 1039–1054. [Google Scholar] [CrossRef] [PubMed]
- Coursol, S.; Le Stunff, H.; Lynch, D.V.; Gilroy, S.; Assmann, S.M.; Spiegel, S. Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol. 2005, 137, 724–737. [Google Scholar] [CrossRef] [PubMed]
- Cacas, J.L.; Bure, C.; Grosjean, K.; Gerbeau-Pissot, P.; Lherminier, J.; Rombouts, Y.; Maes, E.; Bossard, C.; Gronnier, J.; Furt, F.; et al. Revisiting plant plasma membrane lipids in tobacco: A focus on sphingolipids. Plant Physiol. 2016, 170, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Baluska, F.; Hlavacka, A.; Volkmann, D.; Menzel, D. Getting connected: Actin-based cell-to-cell channels in plants and animals. Trends Cell Biol. 2004, 14, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Kragler, F. Plasmodesmata: Intercellular tunnels facilitating transport of macromolecules in plants. Cell Tissue Res. 2013, 352, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Thyssen, G.; Svab, Z.; Maliga, P. Cell-to-cell movement of plastids in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 2439–2443. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Zambryski, P.C. Cell-to-cell communication via plasmodesmata during arabidopsis embryogenesis. Curr. Opin. Plant Biol. 2005, 8, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.O.; Runkel, A.M.; Zambryski, P.C. Plasmodesmata dynamics are coordinated by intracellular signaling pathways. Curr. Opin. Plant Biol. 2013, 16, 614–620. [Google Scholar] [CrossRef] [PubMed]
- De Storme, N.; Geelen, D. Callose homeostasis at plasmodesmata: Molecular regulators and developmental relevance. Front. Plant Sci. 2014, 5, 138. [Google Scholar] [CrossRef] [PubMed]
- Lucas, W.J.; Bouche-Pillon, S.; Jackson, D.P.; Nguyen, L.; Baker, L.; Ding, B.; Hake, S. Selective trafficking of knotted1 homeodomain protein and its mrna through plasmodesmata. Science 1995, 270, 1980–1983. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Rim, Y.; Wang, J.; Jackson, D. A novel cell-to-cell trafficking assay indicates that the knox homeodomain is necessary and sufficient for intercellular protein and mrna trafficking. Genes Dev. 2005, 19, 788–793. [Google Scholar] [CrossRef] [PubMed]
- Oparka, K.J.; Cruz, S.S. The great escape: Phloem transport and unloading of macromolecules1. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 323–347. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Erlanger, M.; Rosenthal, M.; Epel, B.L. A plasmodesmata-associated β-1,3-glucanase in arabidopsis. Plant J. 2007, 49, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Vaten, A.; Dettmer, J.; Wu, S.; Stierhof, Y.D.; Miyashima, S.; Yadav, S.R.; Roberts, C.J.; Campilho, A.; Bulone, V.; Lichtenberger, R.; et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 2011, 21, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Guseman, J.M.; Lee, J.S.; Bogenschutz, N.L.; Peterson, K.M.; Virata, R.E.; Xie, B.; Kanaoka, M.M.; Hong, Z.; Torii, K.U. Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in arabidopsis chorus (glucan synthase-like 8). Development 2010, 137, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, K.N.; Voitsekhovskaja, O.V.; Pawlowski, K. Plasmodesmata without callose and calreticulin in higher plants—Open channels for fast symplastic transport. Front. Plant Sci. 2014, 5, 74. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Kim, J.Y. Callose synthesis in higher plants. Plant Signal. Behav. 2009, 4, 489–492. [Google Scholar] [CrossRef]
- Galatis, B.; Apostolakos, P. A new callose function: Involvement in differentiation and function of fern stomatal complexes. Plant Signal. Behav. 2010, 5, 1359–1364. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose deposition: A multifaceted plant defense response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Liu, L.; Lee, E.; Han, X.; Rim, Y.; Chu, H.; Kim, S.W.; Sack, F.; Kim, J.Y. The arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol. 2009, 150, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Bachman, E.S.; McClay, D.R. Molecular cloning of the first metazoan beta-1,3 glucanase from eggs of the sea urchin strongylocentrotus purpuratus. Proc. Natl. Acad. Sci. USA 1996, 93, 6808–6813. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Diaz-Moreno, S.M.; Davis, D.J.; Wilkop, T.E.; Bulone, V.; Drakakaki, G. Endosidin 7 specifically arrests late cytokinesis and inhibits callose biosynthesis, revealing distinct trafficking events during cell plate maturation. Plant Physiol. 2014, 165, 1019–1034. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Hong, Z.; Sivaramakrishnan, M.; Mahfouz, M.; Verma, D.P. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in arabidopsis. Plant J. 2005, 42, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Delauney, A.J.; Verma, D.P. A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 2001, 13, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Zhang, Z.; Olson, J.M.; Verma, D.P. A novel udp-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 2001, 13, 769–779. [Google Scholar] [CrossRef]
- Mamun, E.A.; Alfred, S.; Cantrill, L.C.; Overall, R.L.; Sutton, B.G. Effects of chilling on male gametophyte development in rice. Cell Biol. Int. 2006, 30, 583–591. [Google Scholar] [CrossRef] [PubMed]
- De Storme, N.; de Schrijver, J.; van Criekinge, W.; Wewer, V.; Dormann, P.; Geelen, D. Glucan SYNTHASE-LIKE8 and STEROL METHYLTRANSFERASE2 are required for ploidy consistency of the sexual reproduction system in arabidopsis. Plant Cell 2013, 25, 387–403. [Google Scholar] [CrossRef] [PubMed]
- De Storme, N.; Geelen, D. The impact of environmental stress on male reproductive development in plants: Biological processes and molecular mechanisms. Plant Cell Environ. 2014, 37, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Benitez-Alfonso, Y.; Faulkner, C.; Pendle, A.; Miyashima, S.; Helariutta, Y.; Maule, A. Symplastic intercellular connectivity regulates lateral root patterning. Dev. Cell 2013, 26, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Hyun, T.K.; Zhang, M.; Kumar, R.; Koh, E.J.; Kang, B.H.; Lucas, W.J.; Kim, J.Y. Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev. Cell 2014, 28, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Barratt, D.H.; Kolling, K.; Graf, A.; Pike, M.; Calder, G.; Findlay, K.; Zeeman, S.C.; Smith, A.M. Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in arabidopsis. Plant Physiol. 2011, 155, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Wang, X.; Zhu, M.; Zhang, Z.; Hong, Z. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 2011, 65, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Toller, A.; Brownfield, L.; Neu, C.; Twell, D.; Schulze-Lefert, P. Dual function of arabidopsis glucan synthase-like genes gsl8 and gsl10 in male gametophyte development and plant growth. Plant J. 2008, 54, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Sevilem, I.; Miyashima, S.; Helariutta, Y. Cell-to-cell communication via plasmodesmata in vascular plants. Cell Adhes. Migr. 2013, 7, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; O’Lexy, R.; Xu, M.; Sang, Y.; Chen, X.; Yu, Q.; Gallagher, K.L. Symplastic signaling instructs cell division, cell expansion, and cell polarity in the ground tissue of arabidopsis thaliana roots. Proc. Natl. Acad. Sci. USA 2016, 113, 11621–11626. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Lee, J.Y. Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants 2016, 2, 16034. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.S. The role of auxin transport in plant patterning mechanisms. PLoS Biol. 2008, 6, e323. [Google Scholar] [CrossRef] [PubMed]
- Vieten, A.; Vanneste, S.; Wisniewska, J.; Benkova, E.; Benjamins, R.; Beeckman, T.; Luschnig, C.; Friml, J. Functional redundancy of pin proteins is accompanied by auxin-dependent cross-regulation of pin expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef] [PubMed]
- Krecek, P.; Skupa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zazimalova, E. The pin-formed (pin) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Bi, F.C.; Liu, Z.; Wu, J.X.; Liang, H.; Xi, X.L.; Fang, C.; Sun, T.J.; Yin, J.; Dai, G.Y.; Rong, C.; et al. Loss of ceramide kinase in arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 2014, 26, 3449–3467. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.X.; Li, J.; Liu, Z.; Yin, J.; Chang, Z.Y.; Rong, C.; Wu, J.L.; Bi, F.C.; Yao, N. The arabidopsis ceramidase atacer functions in disease resistance and salt tolerance. Plant J. 2015, 81, 767–780. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.J.; Chen, Q.F.; Chen, M.X.; Yu, L.J.; Huang, L.; Chen, L.; Wang, F.Z.; Xia, F.N.; Zhu, T.R.; Wu, J.X.; et al. Unsaturation of very-long-chain ceramides protects plant from hypoxia-induced damages by modulating ethylene signaling in arabidopsis. PLoS Genet. 2015, 11, e1005143. [Google Scholar] [CrossRef] [PubMed]
- Luttgeharm, K.D.; Chen, M.; Mehra, A.; Cahoon, R.E.; Markham, J.E.; Cahoon, E.B. Overexpression of arabidopsis ceramide synthases differentially affects growth, sphingolipid metabolism, programmed cell death, and mycotoxin resistance. Plant Physiol. 2015, 169, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, X.; Tangchaiburana, S.; Ndeh, R.; Markham, J.E.; Tsegaye, Y.; Dunn, T.M.; Wang, G.L.; Bellizzi, M.; Parsons, J.F.; et al. An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in arabidopsis. Plant Cell 2008, 20, 3163–3179. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An arabidopsis gpi-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Hosogaya, N.; Miyazaki, T.; Nagi, M.; Tanabe, K.; Minematsu, A.; Nagayoshi, Y.; Yamauchi, S.; Nakamura, S.; Imamura, Y.; Izumikawa, K.; et al. The heme-binding protein Dap1 links iron homeostasis to azole resistance via the P450 protein Erg11 in candida glabrata. FEMS Yeast Res. 2013, 13, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Magnin-Robert, M.; Le Bourse, D.; Markham, J.; Dorey, S.; Clement, C.; Baillieul, F.; Dhondt-Cordelier, S. Modifications of sphingolipid content affect tolerance to hemibiotrophic and necrotrophic pathogens by modulating plant defense responses in arabidopsis. Plant Physiol. 2015, 169, 2255–2274. [Google Scholar] [CrossRef] [PubMed]
- Kimberlin, A.N.; Han, G.; Luttgeharm, K.D.; Chen, M.; Cahoon, R.E.; Stone, J.M.; Markham, J.E.; Dunn, T.M.; Cahoon, E.B. Orm expression alters sphingolipid homeostasis and differentially affects ceramide synthase activity. Plant Physiol. 2016, 172, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Ueki, S.; Citovsky, V. Plasmodesmata-associated proteins: Can we see the whole elephant? Plant Signal. Behav. 2014, 9, e27899. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Wang, X.; Cui, W.; Sager, R.; Modla, S.; Czymmek, K.; Zybaliov, B.; Van Wijk, K.; Zhang, C.; Lu, H.; et al. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in arabidopsis. Plant Cell 2011, 23, 3353–3373. [Google Scholar] [CrossRef] [PubMed]
- Lim, G.H.; Kachroo, A.; Kachroo, P. Role of plasmodesmata and plasmodesmata localizing proteins in systemic immunity. Plant Signal. Behav. 2016, 11, e1219829. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.L.; Bayer, E.M.; Ritzenthaler, C.; Fernandez-Calvino, L.; Maule, A.J. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 2008, 6, e7. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sager, R.; Cui, W.; Zhang, C.; Lu, H.; Lee, J.Y. Salicylic acid regulates plasmodesmata closure during innate immune responses in arabidopsis. Plant Cell 2013, 25, 2315–2329. [Google Scholar] [CrossRef] [PubMed]
- Mayor, S.; Riezman, H. Sorting gpi-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004, 5, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Zavaliev, R.; Dong, X.; Epel, B.L. Glycosylphosphatidylinositol (GPI) modification serves as a primary plasmodesmal sorting signal. Plant Physiol. 2016, 172, 1061–1073. [Google Scholar] [PubMed]
- Raffaele, S.; Bayer, E.; Lafarge, D.; Cluzet, S.; Retana, S.G.; Boubekeur, T.; Leborgne-Castel, N.; Carde, J.P.; Lherminier, J.; Noirot, E.; et al. Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus x movement. Plant Cell 2009, 21, 1541–1555. [Google Scholar] [CrossRef] [PubMed]
- Perraki, A.; Binaghi, M.; Mecchia, M.A.; Gronnier, J.; German-Retana, S.; Mongrand, S.; Bayer, E.; Zelada, A.M.; Germain, V. StRemorin1.3 hampers Potato virus X TGBP1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett. 2014, 588, 1699–1705. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Liu, C.; Shen, J.; Li, L. Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol. 2014, 166, 1463–1478. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iswanto, A.B.B.; Kim, J.-Y. Lipid Raft, Regulator of Plasmodesmal Callose Homeostasis. Plants 2017, 6, 15. https://doi.org/10.3390/plants6020015
Iswanto ABB, Kim J-Y. Lipid Raft, Regulator of Plasmodesmal Callose Homeostasis. Plants. 2017; 6(2):15. https://doi.org/10.3390/plants6020015
Chicago/Turabian StyleIswanto, Arya Bagus Boedi, and Jae-Yean Kim. 2017. "Lipid Raft, Regulator of Plasmodesmal Callose Homeostasis" Plants 6, no. 2: 15. https://doi.org/10.3390/plants6020015




