Genome, Metabolism, or Immunity: Which Is the Primary Decider of Pancreatic Cancer Fate through Non-Apoptotic Cell Death?
Abstract
:1. Introduction
2. Ferroptosis
2.1. Signaling Pathway
2.2. Immunogenetics
2.2.1. Ferroptosis-Related Genes (FRGs) as Risk Models in PDAC Prognosis
2.2.2. Interactions among Genomes, Immune System, Metabolism, and Signaling Pathways in PDAC
2.3. Treatment
2.3.1. Drugs That Modulate Signaling Pathways through Ferroptosis
2.3.2. Drugs That Modulate Organelle Functions through Ferroptosis
2.3.3. Combination Therapy
3. Necroptosis
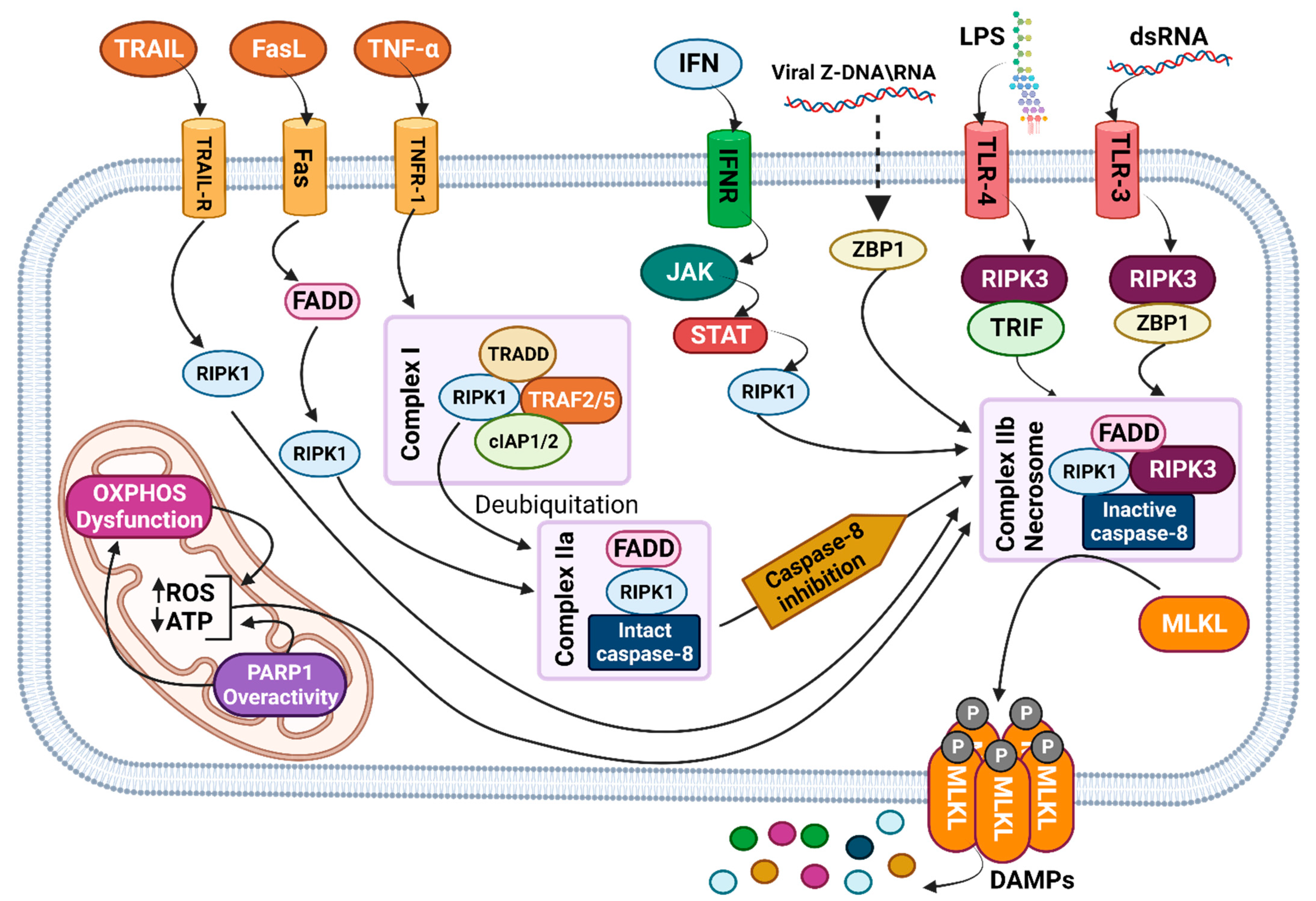
3.1. Signaling Pathway
3.2. Immunogenetics
3.2.1. Necroptosis-Related Genes (NRGs) as a Risk Model in PDAC Prognosis
3.2.2. Correlation between NRGs, Immune System, Metabolism, and Signaling Pathways in PDAC
3.3. Treatment
3.3.1. Drugs That Modulate Necroptosis Signaling Pathways
3.3.2. Drugs That Modulate Organelle Function
3.3.3. Combination Therapy
4. Pyroptosis
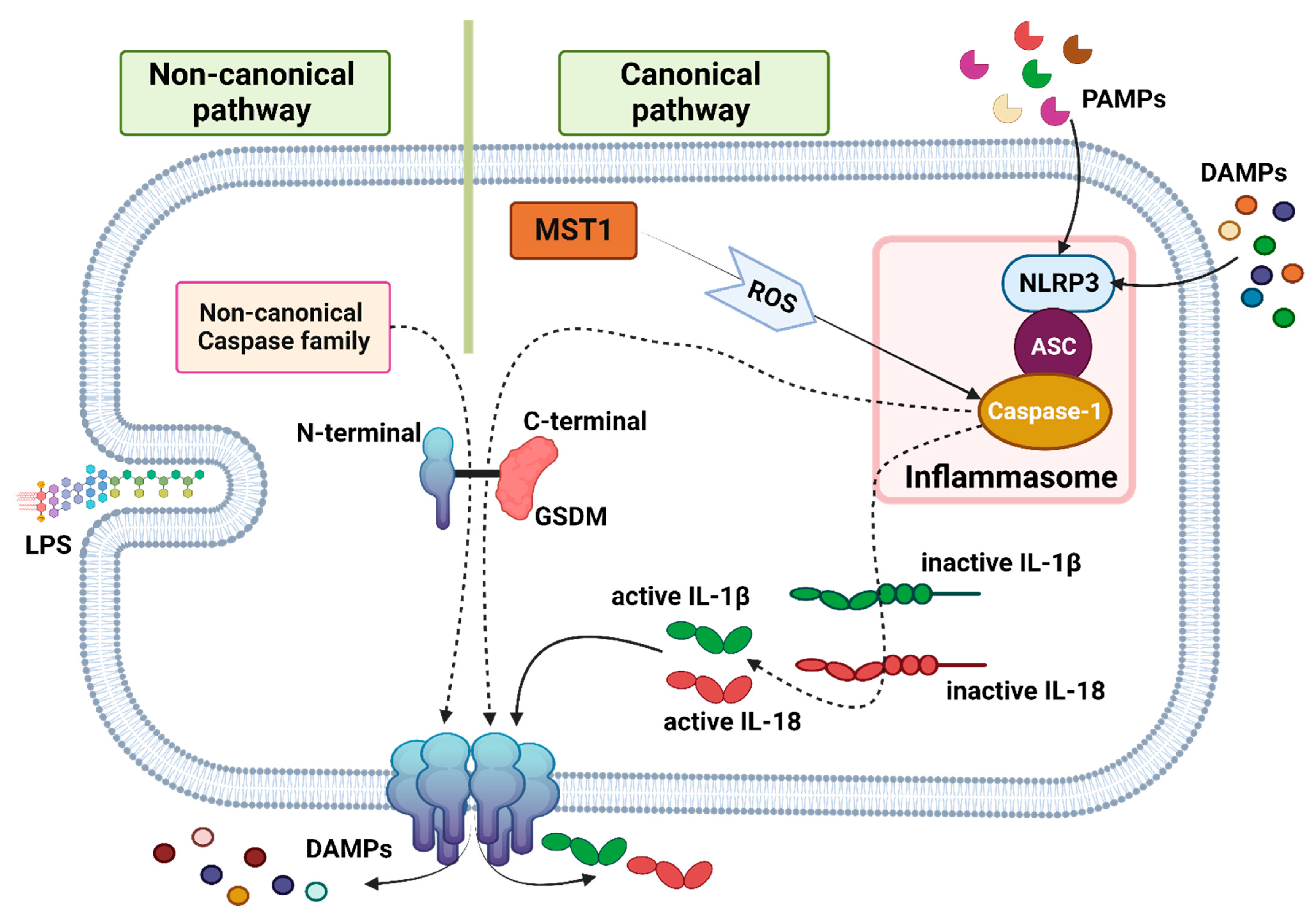
4.1. Signaling Pathway
4.2. Immunogenetics
4.2.1. Pyroptosis-Related Genes (PRGs) Risk Model in Pancreatic Cancer Prognosis
4.2.2. Correlation among PRGs, Immune System, Metabolism, and Signaling Pathways in Pancreatic Cancer
4.3. Treatment
4.3.1. Drugs That Modulate Signaling Pathways and Other Cell Death through Pyroptosis
4.3.2. Combination Therapy
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J. Oncol. 2019, 10, 10. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Comish, P.B.; Tang, D.; Kang, R. Characteristics and biomarkers of ferroptosis. Front. Cell Dev. Biol. 2021, 9, 637162. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, A.A.; Philip, P.A. Adjuvant treatment of surgically resectable pancreatic ductal adenocarcinoma. Clin. Adv. Hematol. Oncol. 2019, 17, 54–63. [Google Scholar] [PubMed]
- Singh, R.R.; O’Reilly, E.M. New treatment strategies for metastatic pancreatic ductal adenocarcinoma. Drugs 2020, 80, 647–669. [Google Scholar] [CrossRef]
- Vivarelli, M.; Mocchegiani, F.; Nicolini, D.; Vecchi, A.; Conte, G.; Dalla Bona, E.; Rossi, R.; Benedetti Cacciaguerra, A. Neoadjuvant treatment in resectable pancreatic cancer. Is it time for pushing on it? Front. Oncol. 2022, 12, 914203. [Google Scholar] [CrossRef]
- Tran, N.H.; Bekaii-Saab, T. Optimizing Chemotherapy Choice in the Treatment of Advanced Pancreatic Cancer—It Is Complicated. JAMA Netw. Open 2021, 4, e2134458. [Google Scholar] [CrossRef]
- Quiñonero, F.; Mesas, C.; Doello, K.; Cabeza, L.; Perazzoli, G.; Jimenez-Luna, C.; Rama, A.R.; Melguizo, C.; Prados, J. The challenge of drug resistance in pancreatic ductal adenocarcinoma: A current overview. Cancer Biol. Med. 2019, 16, 688. [Google Scholar] [CrossRef]
- Pajewska, M.; Partyka, O.; Czerw, A.; Deptała, A.; Cipora, E.; Gąska, I.; Wojtaszek, M.; Sygit, K.; Sygit, M.; Krzych-Fałta, E.; et al. Management of Metastatic Pancreatic Cancer-Comparison of Global Guidelines over the Last 5 Years. Cancers 2023, 15, 4400. [Google Scholar] [CrossRef]
- Ischenko, I.; D’Amico, S.; Rao, M.; Li, J.; Hayman, M.J.; Powers, S.; Petrenko, O.; Reich, N.C. KRAS drives immune evasion in a genetic model of pancreatic cancer. Nat. Commun. 2021, 12, 1482. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lu, X.; Dong, X.; Hao, F.; Liu, Z.; Ni, G.; Chen, D. pERK1/2 silencing sensitizes pancreatic cancer BXPC-3 cell to gemcitabine-induced apoptosis via regulating Bax and Bcl-2 expression. World J. Surg. Oncol. 2015, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, A.; De Castro Silva, I.; Deshpande, N.U.; Singh, S.; Mehra, S.; Garrido, V.T.; Guo, X.; Nivelo, L.A.; Kolonias, D.S.; Saigh, S.J. Cell-autonomous Cxcl1 sustains tolerogenic circuitries and stromal inflammation via neutrophil-derived TNF in pancreatic cancer. Cancer Discov. 2023, 13, 1428–1453. [Google Scholar] [CrossRef]
- Adamska, A.; Elaskalani, O.; Emmanouilidi, A.; Kim, M.; Razak, N.B.A.; Metharom, P.; Falasca, M. Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv. Biol. Regul. 2018, 68, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Kabacaoglu, D.; Ruess, D.A.; Ai, J.; Algül, H. NF-κB/Rel transcription factors in pancreatic cancer: Focusing on RelA, c-Rel, and RelB. Cancers 2019, 11, 937. [Google Scholar] [CrossRef]
- Silke, J.; O’Reilly, L.A. NF-κB and pancreatic cancer; chapter and verse. Cancers 2021, 13, 4510. [Google Scholar] [CrossRef]
- Rubin, S.J.; Sojwal, R.S.; Gubatan, J.; Rogalla, S. The tumor immune microenvironment in pancreatic ductal adenocarcinoma: Neither hot nor cold. Cancers 2022, 14, 4236. [Google Scholar] [CrossRef]
- Ligorio, M.; Sil, S.; Malagon-Lopez, J.; Nieman, L.T.; Misale, S.; Di Pilato, M.; Ebright, R.Y.; Karabacak, M.N.; Kulkarni, A.S.; Liu, A. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell 2019, 178, 160–175.e27. [Google Scholar] [CrossRef]
- Prabhu, L.; Mundade, R.; Korc, M.; Loehrer, P.; Lu, T. Critical role of NF-kappaB in pancreatic cancer. Oncotarget 2014, 5, 10969–10975. [Google Scholar] [CrossRef]
- Pramanik, K.C.; Makena, M.R.; Bhowmick, K.; Pandey, M.K. Advancement of NF-κB signaling pathway: A novel target in pancreatic cancer. Int. J. Mol. Sci. 2018, 19, 3890. [Google Scholar] [CrossRef]
- Bulle, A.; Lim, K.-H. Beyond just a tight fortress: Contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Signal Transduct. Target. Ther. 2020, 5, 249. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Wang, S.; Chen, B.; Zhang, Z.; Ma, Z.; Li, Z.; Liu, C.; Zhou, Z.; Gong, Y.; Huang, S. Prognostic Stratification Based on HIF-1 Signaling for Evaluating Hypoxic Status and Immune Infiltration in Pancreatic Ductal Adenocarcinomas. Front. Immunol. 2021, 12, 790661. [Google Scholar] [CrossRef] [PubMed]
- Sebestyén, A.; Kopper, L.; Dankó, T.; Tímár, J. Hypoxia signaling in cancer: From basics to clinical practice. Pathol. Oncol. Res. 2021, 27, 1609802. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Dong, N.; Lu, D.; Jiang, X.; Xu, J.; Wu, Z.; Zheng, D.; Wechsler, D.S. A positive feedback loop between ROS and Mxi1-0 promotes hypoxia-induced VEGF expression in human hepatocellular carcinoma cells. Cell. Signal. 2017, 31, 79–86. [Google Scholar] [CrossRef]
- Chen, D.; Huang, H.; Zang, L.; Gao, W.; Zhu, H.; Yu, X. Development and verification of the hypoxia-and immune-associated prognostic signature for pancreatic ductal adenocarcinoma. Front. Immunol. 2021, 12, 728062. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, Y.; Xu, H.; Zhang, X.; Mao, T.; Cui, J.; Yao, J.; Wang, Y.; Jiao, F.; Xiao, X. Single-cell analysis of pancreatic ductal adenocarcinoma identifies a novel fibroblast subtype associated with poor prognosis but better immunotherapy response. Cell Discov. 2021, 7, 36. [Google Scholar] [CrossRef]
- Chen, X.; Zeh, H.J.; Kang, R.; Kroemer, G.; Tang, D. Cell death in pancreatic cancer: From pathogenesis to therapy. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 804–823. [Google Scholar] [CrossRef]
- Hänggi, K.; Ruffell, B. Cell death, therapeutics, and the immune response in cancer. Trends Cancer 2023, 9, 381–396. [Google Scholar] [CrossRef]
- Hsu, S.-K.; Chu, Y.-H.; Syue, W.-J.; Lin, H.Y.-H.; Chang, W.-T.; Chen, J.Y.-F.; Wu, C.-Y.; Yen, C.-H.; Cheng, K.-C.; Chiu, C.-C. The Role of Nonapoptotic Programmed Cell Death—Ferroptosis, Necroptosis, and Pyroptosis—In Pancreatic Ductal Adenocarcinoma Treatment. Front. Oncol. 2022, 12, 872883. [Google Scholar] [CrossRef]
- Bedoui, S.; Herold, M.J.; Strasser, A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat. Rev. Mol. Cell Biol. 2020, 21, 678–695. [Google Scholar] [CrossRef]
- Demarco, B.; Chen, K.W.; Broz, P. Cross talk between intracellular pathogens and cell death. Immunol. Rev. 2020, 297, 174–193. [Google Scholar] [CrossRef] [PubMed]
- Karlowitz, R.; van Wijk, S.J. Surviving death: Emerging concepts of RIPK3 and MLKL ubiquitination in the regulation of necroptosis. FEBS J. 2023, 290, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.; Vince, J.E. Pyroptosis versus necroptosis: Similarities, differences, and crosstalk. Cell Death Differ. 2019, 26, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, X.; Comish, P.B.; Kang, R. The dual role of ferroptosis in pancreatic cancer: A narrative review. J. Pancreatol. 2021, 4, 76–81. [Google Scholar] [CrossRef]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296. [Google Scholar] [CrossRef]
- Dai, E.; Han, L.; Liu, J.; Xie, Y.; Kroemer, G.; Klionsky, D.J.; Zeh, H.J.; Kang, R.; Wang, J.; Tang, D. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy 2020, 16, 2069–2083. [Google Scholar] [CrossRef]
- Seibt, T.M.; Proneth, B.; Conrad, M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 2019, 133, 144–152. [Google Scholar] [CrossRef]
- Ingold, I.; Berndt, C.; Schmitt, S.; Doll, S.; Poschmann, G.; Buday, K.; Roveri, A.; Peng, X.; Freitas, F.P.; Seibt, T. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell 2018, 172, 409–422.e21. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Badgley, M.A.; Kremer, D.M.; Maurer, H.C.; DelGiorno, K.E.; Lee, H.-J.; Purohit, V.; Sagalovskiy, I.R.; Ma, A.; Kapilian, J.; Firl, C.E. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 2020, 368, 85–89. [Google Scholar] [CrossRef]
- Chen, X.; Yu, C.; Kang, R.; Kroemer, G.; Tang, D. Cellular degradation systems in ferroptosis. Cell Death Differ. 2021, 28, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Du, J.; Zhang, Y.; Wang, Y.; Wang, B.; Zhang, T. GPX4-independent ferroptosis—A new strategy in disease’s therapy. Cell Death Discov. 2022, 8, 434. [Google Scholar] [CrossRef] [PubMed]
- Latunde-Dada, G.O. Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy. Biochim. Et. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 1893–1900. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Xie, Y.; Song, X.; Sun, X.; Lotze, M.T.; Zeh III, H.J.; Kang, R.; Tang, D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 2016, 12, 1425–1428. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, J.; Xu, Y.; Wu, R.; Chen, X.; Song, X.; Zeh, H.; Kang, R.; Klionsky, D.J.; Wang, X. Tumor heterogeneity in autophagy-dependent ferroptosis. Autophagy 2021, 17, 3361–3374. [Google Scholar] [CrossRef]
- Liu, J.; Kang, R.; Tang, D. The art of war: Ferroptosis and pancreatic cancer. Front. Pharmacol. 2021, 12, 773909. [Google Scholar] [CrossRef]
- Görgülü, K.; Diakopoulos, K.N.; Kaya-Aksoy, E.; Ciecielski, K.J.; Ai, J.; Lesina, M.; Algül, H. The role of autophagy in pancreatic cancer: From bench to the dark bedside. Cells 2020, 9, 1063. [Google Scholar] [CrossRef]
- Zhu, S.; Zhang, Q.; Sun, X.; Zeh III, H.J.; Lotze, M.T.; Kang, R.; Tang, D. HSPA5 regulates ferroptotic cell death in cancer cells. Cancer Res. 2017, 77, 2064–2077. [Google Scholar] [CrossRef]
- Chen, G.; Guo, G.; Zhou, X.; Chen, H. Potential mechanism of ferroptosis in pancreatic cancer. Oncol. Lett. 2020, 19, 579–587. [Google Scholar] [CrossRef]
- Ye, Z.; Hu, Q.; Zhuo, Q.; Zhu, Y.; Fan, G.; Liu, M.; Sun, Q.; Zhang, Z.; Liu, W.; Xu, W. Abrogation of ARF6 promotes RSL3-induced ferroptosis and mitigates gemcitabine resistance in pancreatic cancer cells. Am. J. Cancer Res. 2020, 10, 1182. [Google Scholar]
- Song, X.; Liu, J.; Kuang, F.; Chen, X.; Zeh, H.J.; Kang, R.; Kroemer, G.; Xie, Y.; Tang, D. PDK4 dictates metabolic resistance to ferroptosis by suppressing pyruvate oxidation and fatty acid synthesis. Cell Rep. 2021, 34, 108767. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Ricciardiello, F.; Yang, G.; Qiu, J.; Huang, H.; Xiao, J.; Cao, Z.; Zhao, F.; Liu, Y.; Luo, W. The role of mitochondria in the chemoresistance of pancreatic cancer cells. Cells 2021, 10, 497. [Google Scholar] [CrossRef] [PubMed]
- Masoud, R.; Lac, S.; Garcia, J.; Reyes-Castellanos, G.; Iovanna, J.; Carrier, A. Targeting mitochondrial energy metabolism in PDAC is a promising strategy to overcome resistance to chemotherapy. Pancreatology 2018, 18, S154. [Google Scholar] [CrossRef]
- Yang, H.; Villani, R.M.; Wang, H.; Simpson, M.J.; Roberts, M.S.; Tang, M.; Liang, X. The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Cancer Res. 2018, 37, 266. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Qian, H.; Li, K.; Lou, J.; Wu, Y.; Peng, C. Development and validation of a 7-gene prognostic signature to improve survival prediction in pancreatic ductal adenocarcinoma. Front. Mol. Biosci. 2021, 8, 676291. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Chen, P.; Li, K.; Lou, J.; Wu, Y.; Li, T.; Peng, C. A Novel Ferroptosis-Related Gene Signature Predicts Recurrence in Patients With Pancreatic Ductal Adenocarcinoma. Front. Mol. Biosci. 2021, 8, 650264. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Yang, F.; Zou, C.; Bao, T.; Wu, M.; Yang, D.; Bu, S. The construction and analysis of a ferroptosis-related gene prognostic signature for pancreatic cancer. Aging 2021, 13, 10396. [Google Scholar] [CrossRef]
- Wu, M.; Li, X.; Liu, R.; Yuan, H.; Liu, W.; Liu, Z. Development and validation of a metastasis-related gene signature for predicting the overall survival in patients with pancreatic ductal adenocarcinoma. J. Cancer 2020, 11, 6299. [Google Scholar] [CrossRef]
- Qiu, C.-J.; Wang, X.-B.; Zheng, Z.-R.; Yang, C.-Z.; Lin, K.; Zhang, K.; Tu, M.; Jiang, K.-R.; Gao, W.-T. Development and validation of a ferroptosis-related prognostic model in pancreatic cancer. Investig. New Drugs 2021, 39, 1507–1522. [Google Scholar] [CrossRef]
- Chen, D.; Gao, W.; Zang, L.; Zhang, X.; Li, Z.; Zhu, H.; Yu, X. Ferroptosis-related IncRNAs are prognostic biomarker of overall survival in pancreatic cancer patients. Front. Cell Dev. Biol. 2022, 10, 819724. [Google Scholar] [CrossRef]
- Xie, H.; Xu, J.; Xie, Z.; Xie, N.; Lu, J.; Yu, L.; Li, B.; Cheng, L. Identification and validation of prognostic model for pancreatic ductal adenocarcinoma based on necroptosis-related genes. Front. Genet. 2022, 13, 919638. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Wang, L.; Shang, Z.; Guo, Q.; Liu, Q.; Zhang, M.; Li, T.; Wang, Y.; Zhang, Y.; Zhang, Y. A panel of necroptosis-related genes predicts the prognosis of pancreatic adenocarcinoma. Transl. Oncol. 2022, 22, 101462. [Google Scholar] [CrossRef] [PubMed]
- Widmann, S.; Srivastava, S.; Lin, C.-Y. A Novel Liver X Receptor Inverse Agonist Impairs Cholesterol and Phospholipid Metabolism and Induces Apoptosis and Necroptosis in Pancreatic Ductal Adenocarcinoma Cells. Receptors 2023, 2, 34–46. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, X.; Cai, M.; Huang, P.; Guan, Z. Novel necroptosis-related gene signature for predicting the prognosis of pancreatic adenocarcinoma. Aging 2022, 14, 869. [Google Scholar] [CrossRef]
- Fang, K.; Tang, D.-S.; Yan, C.-S.; Ma, J.; Cheng, L.; Li, Y.; Wang, G. Comprehensive analysis of necroptosis in pancreatic cancer for appealing its implications in prognosis, immunotherapy, and chemotherapy responses. Front. Pharmacol. 2022, 13, 862502. [Google Scholar] [CrossRef]
- Lu, S.-Y.; Hua, J.; Liu, J.; Wei, M.-Y.; Liang, C.; Meng, Q.-C.; Zhang, B.; Yu, X.-J.; Wang, W.; Xu, J. A new approach: Evaluation of necroptosis and immune status enables prediction of the tumor microenvironment and treatment targets in pancreatic cancer. Comput. Struct. Biotechnol. J. 2023, 21, 2419–2433. [Google Scholar] [CrossRef]
- Song, W.; Liu, Z.; Wang, K.; Tan, K.; Zhao, A.; Li, X.; Yuan, Y.; Yang, Z. Pyroptosis-related genes regulate proliferation and invasion of pancreatic cancer and serve as the prognostic signature for modeling patient survival. Discov. Oncol. 2022, 13, 39. [Google Scholar] [CrossRef]
- Cui, J.; Zhou, Z.; Yang, H.; Jiao, F.; Li, N.; Gao, Y.; Wang, L.; Chen, J.; Quan, M. MST1 suppresses pancreatic cancer progression via ROS-induced pyroptosis. Mol. Cancer Res. 2019, 17, 1316–1325. [Google Scholar] [CrossRef]
- Li, S.; Yue, M.; Xu, H.; Zhang, X.; Mao, T.; Quan, M.; Ma, J.; Wang, Y.; Ge, W.; Wang, Y. Chemotherapeutic drugs-induced pyroptosis mediated by gasdermin E promotes the progression and chemoresistance of pancreatic cancer. Cancer Lett. 2023, 564, 216206. [Google Scholar] [CrossRef]
- Xia, X.-H.; Yin, W.-J.; Mao, J.-F.; Liu, P.; Qin, C.-D.; Hu, J.-J.; Liu, S.-Y.; Wang, C.-M.; Zou, D.-H.; Yang, H.-J. The expression profile of Gasdermin C-related genes predicts the prognosis and immunotherapy response of pancreatic adenocarcinoma. Am. J. Cancer Res. 2023, 13, 1240. [Google Scholar]
- Li, C.; Wang, M.; Wei, J.; Zhang, W.; Liu, H.; Zhao, D. Construction of a pyroptosis-related genes signature to improve the prognostic prediction and therapeutic drugs selection in patients with pancreatic cancer. Int. J. Gen. Med. 2022, 15, 6387–6403. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Tian, L.; Wang, X.; Shou, Y. A pyroptosis-related gene signature for prognosis and immune microenvironment of pancreatic cancer. Front. Genet. 2022, 13, 817919. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Shen, K.; Wang, K.; Ge, W.; Mao, T.; Li, S.; Zhang, X.; Xu, H.; Wang, Y.; Yao, J. Prediction of Survival and Tumor Microenvironment Infiltration Based on Pyroptosis-Related lncRNAs in Pancreatic Cancer. Dis. Markers 2022, 2022, 5634887. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liang, J.-H.; Li, J.-H.; Xu, Q.-C.; Yin, X.-Y. Values of a novel pyroptosis-related genetic signature in predicting outcome and immune status of pancreatic ductal adenocarcinoma. Gastroenterol. Rep. 2022, 10, goac051. [Google Scholar] [CrossRef]
- Xie, W.; Li, X.; Yang, C.; Li, J.; Shen, G.; Chen, H.; Xiao, S.-Y.; Li, Y. The pyroptosis-related gene prognostic index associated with tumor immune infiltration for pancreatic cancer. Int. J. Mol. Sci. 2022, 23, 6178. [Google Scholar] [CrossRef]
- Li, L.; Deng, Z.; Xiao, Z.; Zou, W.; Liu, R. Analysis of pyroptosis-related signature for predicting prognosis and tumor immune microenvironment in pancreatic cancer. Front. Oncol. 2022, 12, 770005. [Google Scholar] [CrossRef]
- Zhang, J.; You, X.; Kang, D.; Zhou, G. Exploring the potential of pyroptosis-related genes in predicting prognosis and immunological characteristics of pancreatic cancer from the perspective of genome and transcriptome. Front. Oncol. 2022, 12, 932786. [Google Scholar] [CrossRef]
- Bai, Z.; Xu, F.; Feng, X.; Wu, Y.; Lv, J.; Shi, Y.; Pei, H. Pyroptosis regulators exert crucial functions in prognosis, progression and immune microenvironment of pancreatic adenocarcinoma: A bioinformatic and in vitro research. Bioengineered 2022, 13, 1717–1735. [Google Scholar] [CrossRef]
- Xu, Z.; Yu, W.; Li, L.; Wang, G. Identification of pyroptosis-related gene signature for predicting prognosis of patients with pancreatic cancer using bioinformatics. Medicine 2022, 101, e31043. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef]
- Shang, M.; Weng, L.; Xu, G.; Wu, S.; Liu, B.; Yin, X.; Mao, A.; Zou, X.; Wang, Z. TRIM11 suppresses ferritinophagy and gemcitabine sensitivity through UBE2N/TAX1BP1 signaling in pancreatic ductal adenocarcinoma. J. Cell. Physiol. 2021, 236, 6868–6883. [Google Scholar] [CrossRef]
- Hu, T.; Shukla, S.K.; Vernucci, E.; He, C.; Wang, D.; King, R.J.; Jha, K.; Siddhanta, K.; Mullen, N.J.; Attri, K.S. Metabolic rewiring by loss of Sirt5 promotes Kras-induced pancreatic cancer progression. Gastroenterology 2021, 161, 1584–1600. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Yamasaki, S.; Kaneko, O.; Taoka, N.; Tomimoto, Y.; Namatame, I.; Yahata, T.; Kuromitsu, S.; Cantley, L.C.; Lyssiotis, C.A. A covalent small molecule inhibitor of glutamate-oxaloacetate transaminase 1 impairs pancreatic cancer growth. Biochem. Biophys. Res. Commun. 2020, 522, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Magtanong, L.; Ko, P.-J.; To, M.; Cao, J.Y.; Forcina, G.C.; Tarangelo, A.; Ward, C.C.; Cho, K.; Patti, G.J.; Nomura, D.K. Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem. Biol. 2019, 26, 420–432.e29. [Google Scholar] [CrossRef] [PubMed]
- Kerk, S.A.; Papagiannakopoulos, T.; Shah, Y.M.; Lyssiotis, C.A. Metabolic networks in mutant KRAS-driven tumours: Tissue specificities and the microenvironment. Nat. Rev. Cancer 2021, 21, 510–525. [Google Scholar] [CrossRef]
- Li, J.; Lama, R.; Galster, S.L.; Inigo, J.R.; Wu, J.; Chandra, D.; Chemler, S.R.; Wang, X. Small-molecule MMRi62 induces ferroptosis and inhibits metastasis in pancreatic cancer via degradation of ferritin heavy chain and mutant p53. Mol. Cancer Ther. 2022, 21, 535–545. [Google Scholar] [CrossRef]
- Bansod, S.; Dodhiawala, P.B.; Lim, K.-H. Oncogenic KRAS-induced feedback inflammatory signaling in pancreatic cancer: An overview and new therapeutic opportunities. Cancers 2021, 13, 5481. [Google Scholar] [CrossRef]
- Santana-Codina, N.; Del Rey, M.Q.; Kapner, K.S.; Zhang, H.; Gikandi, A.; Malcolm, C.; Poupault, C.; Kuljanin, M.; John, K.M.; Biancur, D.E.; et al. NCOA4-Mediated Ferritinophagy Is a Pancreatic Cancer Dependency via Maintenance of Iron Bioavailability for Iron-Sulfur Cluster Proteins. Cancer Discov. 2022, 12, 2180–2197. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Encarnación-Rosado, J.; Lin, E.Y.; Sohn, A.S.W.; Zhang, H.; Mancias, J.D.; Kimmelman, A.C. Autophagy supports mitochondrial metabolism through the regulation of iron homeostasis in pancreatic cancer. Sci. Adv. 2023, 9, eadf9284. [Google Scholar] [CrossRef]
- Liu, P.; Zu, F.; Chen, H.; Yin, X.; Tan, X. Exosomal DNAJB11 promotes the development of pancreatic cancer by modulating the EGFR/MAPK pathway. Cell. Mol. Biol. Lett. 2022, 27, 87. [Google Scholar] [CrossRef]
- Robinson, C.M.; Talty, A.; Logue, S.E.; Mnich, K.; Gorman, A.M.; Samali, A. An emerging role for the unfolded protein response in pancreatic cancer. Cancers 2021, 13, 261. [Google Scholar] [CrossRef]
- Wu, Z.; Geng, Y.; Lu, X.; Shi, Y.; Wu, G.; Zhang, M.; Shan, B.; Pan, H.; Yuan, J. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc. Natl. Acad. Sci. USA 2019, 116, 2996–3005. [Google Scholar] [CrossRef]
- Zhang, Y.; Ware, M.B.; Zaidi, M.Y.; Ruggieri, A.N.; Olson, B.M.; Komar, H.; Farren, M.R.; Nagaraju, G.P.; Zhang, C.; Chen, Z. Heat shock protein-90 inhibition alters activation of pancreatic stellate cells and enhances the efficacy of PD-1 blockade in pancreatic cancer. Mol. Cancer Ther. 2021, 20, 150–160. [Google Scholar] [CrossRef]
- Chen, D.; Chu, B.; Yang, X.; Liu, Z.; Jin, Y.; Kon, N.; Rabadan, R.; Jiang, X.; Stockwell, B.R.; Gu, W. iPLA2β-mediated lipid detoxification controls p53-driven ferroptosis independent of GPX4. Nat. Commun. 2021, 12, 3644. [Google Scholar] [CrossRef]
- Kremer, D.M.; Nelson, B.S.; Lin, L.; Yarosz, E.L.; Halbrook, C.J.; Kerk, S.A.; Sajjakulnukit, P.; Myers, A.; Thurston, G.; Hou, S.W. GOT1 inhibition promotes pancreatic cancer cell death by ferroptosis. Nat. Commun. 2021, 12, 4860. [Google Scholar] [CrossRef]
- Sharbeen, G.; McCarroll, J.A.; Akerman, A.; Kopecky, C.; Youkhana, J.; Kokkinos, J.; Holst, J.; Boyer, C.; Erkan, M.; Goldstein, D. Cancer-associated fibroblasts in pancreatic ductal adenocarcinoma determine response to SLC7A11 inhibition. Cancer Res. 2021, 81, 3461–3479. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Liu, J.; Kang, R.; Tang, D. Interplay between MTOR and GPX4 signaling modulates autophagy-dependent ferroptotic cancer cell death. Cancer Gene Ther. 2021, 28, 55–63. [Google Scholar] [CrossRef]
- Eling, N.; Reuter, L.; Hazin, J.; Hamacher-Brady, A.; Brady, N.R. Identification of artesunate as a specific activator of ferroptosis in pancreatic cancer cells. Oncoscience 2015, 2, 517. [Google Scholar] [CrossRef]
- Song, Z.; Xiang, X.; Li, J.; Deng, J.; Fang, Z.; Zhang, L.; Xiong, J. Ruscogenin induces ferroptosis in pancreatic cancer cells. Oncol. Rep. 2020, 43, 516–524. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Kasukabe, T.; Kumakura, S. Piperlongumine rapidly induces the death of human pancreatic cancer cells mainly through the induction of ferroptosis. Int. J. Oncol. 2018, 52, 1011–1022. [Google Scholar] [CrossRef]
- Santofimia-Castaño, P.; Xia, Y.; Peng, L.; Velázquez-Campoy, A.; Abián, O.; Lan, W.; Lomberk, G.; Urrutia, R.; Rizzuti, B.; Soubeyran, P. Targeting the stress-induced protein NUPR1 to treat pancreatic adenocarcinoma. Cells 2019, 8, 1453. [Google Scholar] [CrossRef] [PubMed]
- Santofimia-Castaño, P.; Lan, W.; Bintz, J.; Gayet, O.; Carrier, A.; Lomberk, G.; Neira, J.L.; González, A.; Urrutia, R.; Soubeyran, P. Inactivation of NUPR1 promotes cell death by coupling ER-stress responses with necrosis. Sci. Rep. 2018, 8, 16999. [Google Scholar] [CrossRef] [PubMed]
- Rizzuti, B.; Lan, W.; Santofimia-Castaño, P.; Zhou, Z.; Velázquez-Campoy, A.; Abián, O.; Peng, L.; Neira, J.L.; Xia, Y.; Iovanna, J.L. Design of inhibitors of the intrinsically disordered protein NUPR1: Balance between drug affinity and target function. Biomolecules 2021, 11, 1453. [Google Scholar] [CrossRef] [PubMed]
- Ruoxi, Z.; Kang, R.; Daolin, T. The STING1 network regulates autophagy and cell death. Signal Transduct. Target. Ther. 2021, 6, 208. [Google Scholar]
- Li, C.; Zhang, Y.; Liu, J.; Kang, R.; Klionsky, D.J.; Tang, D. Mitochondrial DNA stress triggers autophagy-dependent ferroptotic death. Autophagy 2021, 17, 948–960. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, C.; Zhong, W.; Wang, Q.; Zhang, D.; Zhang, J.; Xie, S.; Xu, M. Chrysin induces autophagy-dependent ferroptosis to increase chemosensitivity to gemcitabine by targeting CBR1 in pancreatic cancer cells. Biochem. Pharmacol. 2021, 193, 114813. [Google Scholar] [CrossRef]
- Nayak, D.; Weadick, B.; Persaud, A.K.; Raj, R.; Shakya, R.; Li, J.; Campbell, M.J.; Govindarajan, R. EMT alterations in the solute carrier landscape uncover SLC22A10/A15 imposed vulnerabilities in pancreatic cancer. Iscience 2022, 25, 104193. [Google Scholar] [CrossRef]
- Hung, W.-C.; Lee, D.-Y.; Chiang, E.-P.I.; Syu, J.-N.; Chao, C.-Y.; Yang, M.-D.; Tsai, S.-Y.; Tang, F.-Y. Docosahexaenoic acid inhibits the proliferation of Kras/TP53 double mutant pancreatic ductal adenocarcinoma cells through modulation of glutathione level and suppression of nucleotide synthesis. PLoS ONE 2020, 15, e0241186. [Google Scholar] [CrossRef]
- Du, J.; Wang, X.; Li, Y.; Ren, X.; Zhou, Y.; Hu, W.; Zhou, C.; Jing, Q.; Yang, C.; Wang, L. DHA exhibits synergistic therapeutic efficacy with cisplatin to induce ferroptosis in pancreatic ductal adenocarcinoma via modulation of iron metabolism. Cell Death Dis. 2021, 12, 705. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Z.; Cheng, J.; Pan, H.; Lin, T.; Shen, X.; Chen, W.; Chen, Q.; Gu, C.; Mao, Q. Platelet-Vesicles-Encapsulated RSL-3 Enable Anti-Angiogenesis and Induce Ferroptosis to Inhibit Pancreatic Cancer Progress. Front. Endocrinol. 2022, 13, 865655. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.-L.; Li, J.; Ye, Z.-P.; Du, T.; Li, L.-C.; Guo, Y.-Q.; Yang, D.; Li, Z.-L.; Cao, J.-H. Tubastatin A potently inhibits GPX4 activity to potentiate cancer radiotherapy through boosting ferroptosis. Redox Biol. 2023, 62, 102677. [Google Scholar] [CrossRef] [PubMed]
- Dai, E.; Han, L.; Liu, J.; Xie, Y.; Zeh, H.J.; Kang, R.; Bai, L.; Tang, D. Ferroptotic damage promotes pancreatic tumorigenesis through a TMEM173/STING-dependent DNA sensor pathway. Nat. Commun. 2020, 11, 6339. [Google Scholar] [CrossRef] [PubMed]
- Dagorn, J.-C.; Vasseur, S.; Iovanna, J.L. p8 Is a New Target of Gemcitabine in Pancreatic Cancer Cells. Clin. Cancer Res. 2006, 12, 235–241. [Google Scholar]
- Li, C.; Yin, X.; Liu, Z.; Wang, J. Emerging Potential Mechanism and Therapeutic Target of Ferroptosis in PDAC: A Promising Future. Int. J. Mol. Sci. 2022, 23, 15031. [Google Scholar] [CrossRef]
- Zhu, F.; Zhang, W.; Yang, T.; He, S.-D. Complex roles of necroptosis in cancer. J. Zhejiang Univ. Sci. B 2019, 20, 399. [Google Scholar] [CrossRef]
- Qin, X.; Ma, D.; Tan, Y.-X.; Wang, H.-Y.; Cai, Z. The role of necroptosis in cancer: A double-edged sword? Biochim. Et. Biophys. Acta (BBA) Rev. Cancer 2019, 1871, 259–266. [Google Scholar] [CrossRef]
- Yuan, J.; Amin, P.; Ofengeim, D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 2019, 20, 19–33. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Seo, J.; Nam, Y.W.; Kim, S.; Oh, D.-B.; Song, J. Necroptosis molecular mechanisms: Recent findings regarding novel necroptosis regulators. Exp. Mol. Med. 2021, 53, 1007–1017. [Google Scholar] [CrossRef]
- Liu, H.; Li, Z.; Zhang, L.; Zhang, M.; Liu, S.; Wang, J.; Yang, C.; Peng, Q.; Du, C.; Jiang, N. Necroptosis-Related Prognostic Model for Pancreatic Carcinoma Reveals Its Invasion and Metastasis Potential through Hybrid EMT and Immune Escape. Biomedicines 2023, 11, 1738. [Google Scholar] [CrossRef]
- Vandenabeele, P. Necroptosis and its role in inflammation. Nature 2015, 517, 311–320. [Google Scholar]
- Hsu, S.-K.; Chang, W.-T.; Lin, I.-L.; Chen, Y.-F.; Padalwar, N.B.; Cheng, K.-C.; Teng, Y.-N.; Wang, C.-H.; Chiu, C.-C. The role of necroptosis in ROS-mediated cancer therapies and its promising applications. Cancers 2020, 12, 2185. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Yousif, A.; Chesnokov, M.; Hong, L.; Chefetz, I. A decade of cell death studies: Breathing new life into necroptosis. Pharmacol. Ther. 2021, 220, 107717. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Rowe, D.C.; Barnes, B.J.; Caffrey, D.R.; Visintin, A.; Latz, E.; Monks, B.; Pitha, P.M.; Golenbock, D.T. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the toll adapters TRAM and TRIF. J. Exp. Med. 2003, 198, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.E.; Price, D.R.; Ryter, S.W.; Choi, A.M. Necroptosis: A crucial pathogenic mediator of human disease. JCI Insight 2019, 4, e128834. [Google Scholar] [CrossRef]
- Fulda, S. The mechanism of necroptosis in normal and cancer cells. Cancer Biol. Ther. 2013, 14, 999–1004. [Google Scholar] [CrossRef]
- Wang, L.; Chang, X.; Feng, J.; Yu, J.; Chen, G. TRADD mediates RIPK1-independent necroptosis induced by tumor necrosis factor. Front. Cell Dev. Biol. 2020, 7, 393. [Google Scholar] [CrossRef]
- Chen, H.; Li, Y.; Wu, J.; Li, G.; Tao, X.; Lai, K.; Yuan, Y.; Zhang, X.; Zou, Z.; Xu, Y. RIPK3 collaborates with GSDMD to drive tissue injury in lethal polymicrobial sepsis. Cell Death Differ. 2020, 27, 2568–2585. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, N.; Su, W.; Zhuo, Y. Necroptosis: A novel pathway in neuroinflammation. Front. Pharmacol. 2021, 12, 701564. [Google Scholar] [CrossRef]
- Yang, D.; Liang, Y.; Zhao, S.; Ding, Y.; Zhuang, Q.; Shi, Q.; Ai, T.; Wu, S.-Q.; Han, J. ZBP1 mediates interferon-induced necroptosis. Cell. Mol. Immunol. 2020, 17, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Wachsmuth, L.; Kumari, S.; Schwarzer, R.; Lin, J.; Eren, R.O.; Fisher, A.; Lane, R.; Young, G.R.; Kassiotis, G. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 2020, 580, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, D.A.; Quarato, G.; Liedmann, S.; Tummers, B.; Zhang, T.; Guy, C.; Crawford, J.C.; Palacios, G.; Pelletier, S.; Kalkavan, H. Caspase-8 and FADD prevent spontaneous ZBP1 expression and necroptosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2207240119. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.J.; Nogusa, S.; Chen, P.; Maki, J.L.; Lerro, A.; Andrake, M.; Rall, G.F.; Degterev, A.; Balachandran, S. Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc. Natl. Acad. Sci. USA 2013, 110, E3109–E3118. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Zhou, Y.; Feng, Z.; Ma, M.; Yao, Y.; Wang, Y.; Deng, Y.; Wu, Y. Caspase-independent regulated necrosis pathways as potential targets in cancer management. Front. Oncol. 2021, 10, 616952. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Y.; Zhang, Y.; He, X.; Zhong, C.-Q.; Ni, H.; Chen, X.; Liang, Y.; Wu, J.; Zhao, S. RIP3 targets pyruvate dehydrogenase complex to increase aerobic respiration in TNF-induced necroptosis. Nat. Cell Biol. 2018, 20, 186–197. [Google Scholar] [CrossRef]
- Fulda, S. Regulation of necroptosis signaling and cell death by reactive oxygen species. Biol. Chem. 2016, 397, 657–660. [Google Scholar] [CrossRef]
- Huang, C.; Lan, W.; Fraunhoffer, N.; Meilerman, A.; Iovanna, J.; Santofimia-Castaño, P. Dissecting the anticancer mechanism of trifluoperazine on pancreatic ductal adenocarcinoma. Cancers 2019, 11, 1869. [Google Scholar] [CrossRef]
- Brillo, V.; Chieregato, L.; Leanza, L.; Muccioli, S.; Costa, R. Mitochondrial dynamics, ROS, and cell signaling: A blended overview. Life 2021, 11, 332. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP inhibitors: Clinical relevance, mechanisms of action and tumor resistance. Front. Cell Dev. Biol. 2020, 8, 564601. [Google Scholar] [CrossRef]
- Dias, M.P.; Moser, S.C.; Ganesan, S.; Jonkers, J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2021, 18, 773–791. [Google Scholar] [CrossRef]
- Spiegel, J.O.; Van Houten, B.; Durrant, J.D. PARP1: Structural insights and pharmacological targets for inhibition. DNA Repair 2021, 103, 103125. [Google Scholar] [CrossRef]
- Looi, C.-K.; Gan, L.-L.; Sim, W.; Hii, L.-W.; Chung, F.F.-L.; Leong, C.-O.; Lim, W.-M.; Mai, C.-W. Histone deacetylase inhibitors restore cancer cell sensitivity towards T lymphocytes mediated cytotoxicity in pancreatic cancer. Cancers 2022, 14, 3709. [Google Scholar] [CrossRef] [PubMed]
- Messex, J.K.; Adams, K.L.; Hawkins, W.G.; DeNardo, D.; Bardeesy, N.; Billadeau, D.D.; Liou, G.-Y. Oncogenic Kras-mediated cytokine CCL15 regulates pancreatic cancer cell migration and invasion through ROS. Cancers 2022, 14, 2153. [Google Scholar] [CrossRef]
- Binker-Cosen, M.J.; Richards, D.; Oliver, B.; Gaisano, H.Y.; Binker, M.G.; Cosen-Binker, L.I. Palmitic acid increases invasiveness of pancreatic cancer cells AsPC-1 through TLR4/ROS/NF-κB/MMP-9 signaling pathway. Biochem. Biophys. Res. Commun. 2017, 484, 152–158. [Google Scholar] [CrossRef]
- Massoumi, R.L.; Teper, Y.; Ako, S.; Ye, L.; Wang, E.; Hines, O.J.; Eibl, G. Direct effects of lipopolysaccharide on human pancreatic cancer cells. Pancreas 2021, 50, 524. [Google Scholar] [CrossRef]
- Yin, H.; Pu, N.; Chen, Q.; Zhang, J.; Zhao, G.; Xu, X.; Wang, D.; Kuang, T.; Jin, D.; Lou, W. Gut-derived lipopolysaccharide remodels tumoral microenvironment and synergizes with PD-L1 checkpoint blockade via TLR4/MyD88/AKT/NF-κB pathway in pancreatic cancer. Cell Death Dis. 2021, 12, 1033. [Google Scholar] [CrossRef]
- Tang, R.; Ji, J.; Ding, J.; Huang, J.; Gong, B.; Zhang, X.; Li, F. Overexpression of MYEOV predicting poor prognosis in patients with pancreatic ductal adenocarcinoma. Cell Cycle 2020, 19, 1602–1610. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, R.; Sun, W.; Wang, J.; Liu, J. Analysis of methylation-driven genes in pancreatic ductal adenocarcinoma for predicting prognosis. J. Cancer 2021, 12, 6507. [Google Scholar] [CrossRef]
- Yang, J.; Chheda, C.; Lim, A.; Hauptschein, D.; Zayou, L.; Tang, J.; Pandol, S.J.; Edderkaoui, M. HDAC4 mediates smoking-induced pancreatic cancer metastasis. Pancreas 2022, 51, 190–195. [Google Scholar] [CrossRef]
- Yuan, P.; Tang, C.; Chen, B.; Lei, P.; Song, J.; Xin, G.; Wang, Z.; Hui, Y.; Yao, W.; Wang, G. miR-32-5p suppresses the proliferation and migration of pancreatic adenocarcinoma cells by targeting TLDC1. Mol. Med. Rep. 2021, 24, 752. [Google Scholar] [CrossRef]
- Zuazo, M.; Arasanz, H.; Bocanegra, A.; Fernandez, G.; Chocarro, L.; Vera, R.; Kochan, G.; Escors, D. Systemic CD4 immunity as a key contributor to PD-L1/PD-1 blockade immunotherapy efficacy. Front. Immunol. 2020, 11, 586907. [Google Scholar] [CrossRef]
- Scott, E.N.; Gocher, A.M.; Workman, C.J.; Vignali, D.A. Regulatory T cells: Barriers of immune infiltration into the tumor microenvironment. Front. Immunol. 2021, 12, 702726. [Google Scholar] [CrossRef]
- Titov, A.; Zmievskaya, E.; Ganeeva, I.; Valiullina, A.; Petukhov, A.; Rakhmatullina, A.; Miftakhova, R.; Fainshtein, M.; Rizvanov, A.; Bulatov, E. Adoptive immunotherapy beyond CAR T-cells. Cancers 2021, 13, 743. [Google Scholar] [CrossRef]
- Kosmidis, C.; Sapalidis, K.; Koletsa, T.; Kosmidou, M.; Efthimiadis, C.; Anthimidis, G.; Varsamis, N.; Michalopoulos, N.; Koulouris, C.; Atmatzidis, S. Interferon-γ and Colorectal Cancer: An up-to date. J. Cancer 2018, 9, 232. [Google Scholar] [CrossRef]
- Jin, L.; Kim, H.S.; Shi, J. Neutrophil in the pancreatic tumor microenvironment. Biomolecules 2021, 11, 1170. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Q.; Peng, J.; Wang, M.; Li, T.; Liu, J.; Cui, M.; Zhang, X.; Gao, X.; Liao, Q. CXCL5 overexpression predicts a poor prognosis in pancreatic ductal adenocarcinoma and is correlated with immune cell infiltration. J. Cancer 2020, 11, 2371. [Google Scholar] [CrossRef]
- Ando, Y.; Ohuchida, K.; Otsubo, Y.; Kibe, S.; Takesue, S.; Abe, T.; Iwamoto, C.; Shindo, K.; Moriyama, T.; Nakata, K. Necroptosis in pancreatic cancer promotes cancer cell migration and invasion by release of CXCL5. PLoS ONE 2020, 15, e0228015. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, H.; Zeng, Z.; Hu, Y.; Fan, T.; Zheng, L.; Wang, Q.; Xiao, S.; Wang, T.; Xiao, W. Single-cell RNA sequencing and influences of necroptosis-associated myeloid lineages in immune microenvironment of PDAC. J. Clin. Oncol. 2023, 41, e16261. [Google Scholar] [CrossRef]
- Agostinis, P.; Berg, K.; Cengel, K.A.; Foster, T.H.; Girotti, A.W.; Gollnick, S.O.; Hahn, S.M.; Hamblin, M.R.; Juzeniene, A.; Kessel, D. Photodynamic therapy of cancer: An update. CA A Cancer J. Clin. 2011, 61, 250–281. [Google Scholar] [CrossRef] [PubMed]
- Karimnia, V.; Slack, F.J.; Celli, J.P. Photodynamic therapy for pancreatic ductal adenocarcinoma. Cancers 2021, 13, 4354. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, D.R.; Dos Santos, A.F.; Wailemann, R.A.; Terra, L.F.; Gomes, V.M.; Arini, G.S.; Bertoldi, E.R.; Reis, E.M.; Baptista, M.S.; Labriola, L. Necroptosis activation is associated with greater methylene blue-photodynamic therapy-induced cytotoxicity in human pancreatic ductal adenocarcinoma cells. Photochem. Photobiol. Sci. 2023, 22, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Tatiana, M.; Irina, B.; Anastasia, G.; Vedunova, M.; Krysko, D.V. Which cell death modality wins the contest for photodynamic therapy of cancer? Cell Death Dis. 2022, 13, 455. [Google Scholar]
- Zielinska, E.; Zauszkiewicz-Pawlak, A.; Wojcik, M.; Inkielewicz-Stepniak, I. Silver nanoparticles of different sizes induce a mixed type of programmed cell death in human pancreatic ductal adenocarcinoma. Oncotarget 2018, 9, 4675. [Google Scholar] [CrossRef]
- Liu, L.; An, X.; Schaefer, M.; Yan, B.; de la Torre, C.; Hillmer, S.; Gladkich, J.; Herr, I. Nanosilver inhibits the progression of pancreatic cancer by inducing a paraptosis-like mixed type of cell death. Biomed. Pharmacother. 2022, 153, 113511. [Google Scholar] [CrossRef]
- Barcińska, E.; Wierzbicka, J.; Zauszkiewicz-Pawlak, A.; Jacewicz, D.; Dabrowska, A.; Inkielewicz-Stepniak, I. Role of oxidative and nitro-oxidative damage in silver nanoparticles cytotoxic effect against human pancreatic ductal adenocarcinoma cells. Oxidative Med. Cell. Longev. 2018, 2018, 8251961. [Google Scholar] [CrossRef]
- Zhao, Q.; Zheng, Y.; Lv, X.; Gong, J.; Yang, L. IMB5036 inhibits human pancreatic cancer growth primarily through activating necroptosis. Basic. Clin. Pharmacol. Toxicol. 2022, 130, 375–384. [Google Scholar] [CrossRef]
- Gomes-Filho, S.M.; Dos Santos, E.O.; Bertoldi, E.R.M.; Scalabrini, L.C.; Heidrich, V.; Dazzani, B.; Levantini, E.; Reis, E.M.; Bassères, D.S. Aurora A kinase and its activator TPX2 are potential therapeutic targets in KRAS-induced pancreatic cancer. Cell. Oncol. 2020, 43, 445–460. [Google Scholar] [CrossRef]
- Xu, X.; Yu, Y.; Yang, L.; Wang, B.; Fan, Y.; Ruan, B.; Zhang, X.; Dai, H.; Mei, W.; Jie, W. Integrated analysis of Dendrobium nobile extract Dendrobin A against pancreatic ductal adenocarcinoma based on network pharmacology, bioinformatics, and validation experiments. Front. Pharmacol. 2023, 14, 1079539. [Google Scholar] [CrossRef]
- Xie, Y.; Zhu, S.; Zhong, M.; Yang, M.; Sun, X.; Liu, J.; Kroemer, G.; Lotze, M.; Zeh III, H.J.; Kang, R. Inhibition of aurora kinase A induces necroptosis in pancreatic carcinoma. Gastroenterology 2017, 153, 1429–1443.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Nikhil, K.; Viccaro, K.; Chang, L.; White, J.; Shah, K. Phosphorylation-dependent regulation of ALDH1A1 by Aurora kinase A: Insights on their synergistic relationship in pancreatic cancer. BMC Biol. 2017, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ma, Y.; Wang, Y.; Mukhopadhyay, D.; Bi, Y.; Ji, B. Aurora kinase a inhibitor MLN8237 suppresses pancreatic cancer growth. Pancreatology 2022, 22, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, H.; Ding, Y.; Wan, M.; Xu, M. The Role of Adipokines in Pancreatic Cancer. Front. Oncol. 2022, 12, 926230. [Google Scholar] [CrossRef]
- Takenaga, K.; Akimoto, M.; Koshikawa, N.; Nagase, H. Obesity reduces the anticancer effect of AdipoRon against orthotopic pancreatic cancer in diet-induced obese mice. Sci. Rep. 2021, 11, 2923. [Google Scholar] [CrossRef]
- Kowalski, S.; Hać, S.; Wyrzykowski, D.; Zauszkiewicz-Pawlak, A.; Inkielewicz-Stępniak, I. Selective cytotoxicity of vanadium complexes on human pancreatic ductal adenocarcinoma cell line by inducing necroptosis, apoptosis and mitotic catastrophe process. Oncotarget 2017, 8, 60324. [Google Scholar] [CrossRef]
- Chen, J.A.; Huynh, J.C.; Wu, C.-Y.; Yu, A.-M.; Matsukuma, K.; Semrad, T.J.; Gandara, D.R.; Li, T.; Riess, J.W.; Tam, K. A phase I dose escalation, dose expansion and pharmacokinetic trial of gemcitabine and alisertib in advanced solid tumors and pancreatic cancer. Cancer Chemother. Pharmacol. 2022, 90, 217–228. [Google Scholar] [CrossRef]
- Chen, C.; Xiao, W.; Huang, L.; Yu, G.; Ni, J.; Yang, L.; Wan, R.; Hu, G. Shikonin induces apoptosis and necroptosis in pancreatic cancer via regulating the expression of RIP1/RIP3 and synergizes the activity of gemcitabine. Am. J. Transl. Res. 2017, 9, 5507. [Google Scholar]
- Hannes, S.; Karlowitz, R.; van Wijk, S.J. The Smac mimetic BV6 cooperates with STING to induce necroptosis in apoptosis-resistant pancreatic carcinoma cells. Cell Death Dis. 2021, 12, 816. [Google Scholar] [CrossRef]
- Oldfield, L.; Evans, A.; Rao, R.G.; Jenkinson, C.; Purewal, T.; Psarelli, E.E.; Menon, U.; Timms, J.F.; Pereira, S.P.; Ghaneh, P. Blood levels of adiponectin and IL-1Ra distinguish type 3c from type 2 diabetes: Implications for earlier pancreatic cancer detection in new-onset diabetes. EBioMedicine 2022, 75, 103802. [Google Scholar] [CrossRef]
- Fernandes, P.; O’Donovan, T.R.; McKenna, S.L.; Forde, P.F. Electrochemotherapy causes caspase-independent necrotic-like death in pancreatic cancer cells. Cancers 2019, 11, 1177. [Google Scholar] [CrossRef]
- Bosnjak, M.; Jesenko, T.; Markelc, B.; Cerovsek, A.; Sersa, G.; Cemazar, M. Sunitinib potentiates the cytotoxic effect of electrochemotherapy in pancreatic carcinoma cells. Radiol. Oncol. 2022, 56, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, J.; Wang, P.; Peng, Q.; Kang, Z.; Deng, Y.; Li, J.; Yan, D.; Ge, F.; Chen, Y. FERMT1 Is a Prognostic Marker Involved in Immune Infiltration of Pancreatic Adenocarcinoma Correlating with m6A Modification and Necroptosis. Genes 2023, 14, 734. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhang, X.; Liu, N.; Tang, L.; Peng, C.; Chen, X. Pyroptosis: Mechanisms and diseases. Signal Transduct. Target. Ther. 2021, 6, 128. [Google Scholar] [CrossRef]
- Shi, J.; Gao, W.; Shao, F. Pyroptosis: Gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 2017, 42, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Guo, T.; Zhang, X. Pyroptosis in cancer: Friend or foe? Cancers 2021, 13, 3620. [Google Scholar] [CrossRef]
- Xia, X.; Wang, X.; Cheng, Z.; Qin, W.; Lei, L.; Jiang, J.; Hu, J. The role of pyroptosis in cancer: Pro-cancer or pro-“host”? Cell Death Dis. 2019, 10, 650. [Google Scholar] [CrossRef]
- Marandi, Y.; Hashemzade, S.; Tayebinia, H.; Karimi, J.; Zamani, A.; Khodadadi, I. NLRP3-inflammasome activation is associated with epithelial-mesenchymal transition and progression of colorectal cancer. Iran. J. Basic. Med. Sci. 2021, 24, 483. [Google Scholar]
- Zhang, Y.; Yang, H.; Sun, M.; He, T.; Liu, Y.; Yang, X.; Shi, X.; Liu, X. Alpinumisoflavone suppresses hepatocellular carcinoma cell growth and metastasis via NLRP3 inflammasome-mediated pyroptosis. Pharmacol. Rep. 2020, 72, 1370–1382. [Google Scholar] [CrossRef]
- Gao, J.; Qiu, X.; Xi, G.; Liu, H.; Zhang, F.; Lv, T.; Song, Y. Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/Akt signaling and predicts a good prognosis in non-small cell lung cancer. Oncol. Rep. 2018, 40, 1971–1984. [Google Scholar] [CrossRef]
- Wang, W.J.; Chen, D.; Jiang, M.Z.; Xu, B.; Li, X.W.; Chu, Y.; Zhang, Y.J.; Mao, R.; Liang, J.; Fan, D.M. Downregulation of gasdermin D promotes gastric cancer proliferation by regulating cell cycle-related proteins. J. Dig. Dis. 2018, 19, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, H.; Kouadir, M.; Song, H.; Shi, F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019, 10, 128. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Wang, S.; Wang, J. Mechanism and regulation of pyroptosis-mediated in cancer cell death. Chem. Biol. Interact. 2020, 323, 109052. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.-C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Y.; Ding, J.; Wang, C.; Zhou, X.; Gao, W.; Huang, H.; Shao, F.; Liu, Z. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature 2020, 579, 421–426. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, Y.; Chen, X.; Wang, Z.; Liang, X.; Zhang, T.; Liu, M.; Zhou, N.; Lv, J.; Tang, K. Gasdermin E–mediated target cell pyroptosis by CAR T cells triggers cytokine release syndrome. Sci. Immunol. 2020, 5, eaax7969. [Google Scholar] [CrossRef]
- Fang, Y.; Tian, S.; Pan, Y.; Li, W.; Wang, Q.; Tang, Y.; Yu, T.; Wu, X.; Shi, Y.; Ma, P. Pyroptosis: A new frontier in cancer. Biomed. Pharmacother. 2020, 121, 109595. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, W.; Shi, X.; Ding, J.; Liu, W.; He, H.; Wang, K.; Shao, F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017, 547, 99–103. [Google Scholar] [CrossRef]
- Zuo, J.; Yi, C.; Chen, Z.; Zhou, B.; Yang, T.; Lin, J. A novel refined pyroptosis and inflammasome-related genes signature for predicting prognosis and immune microenvironment in pancreatic ductal adenocarcinoma. Sci. Rep. 2022, 12, 18384. [Google Scholar] [CrossRef]
- Yu, T.; Tan, H.; Liu, C.; Nie, W.; Wang, Y.; Zhou, K.; Shi, H. Integratively genomic analysis reveals the prognostic and immunological characteristics of pyroptosis and ferroptosis in pancreatic cancer for precision immunotherapy. Front. Cell Dev. Biol. 2022, 10, 826879. [Google Scholar] [CrossRef]
- Su, L.; Chen, Y.; Huang, C.; Wu, S.; Wang, X.; Zhao, X.; Xu, Q.; Sun, R.; Kong, X.; Jiang, X. Targeting Src reactivates pyroptosis to reverse chemoresistance in lung and pancreatic cancer models. Sci. Transl. Med. 2023, 15, eabl7895. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zhang, S.; Zhong, X.; Yuan, H.; Song, D.; Wang, X.; Yu, H.; Guo, Z. The induction of PANoptosis in KRAS-mutant pancreatic ductal adenocarcinoma cells by a multispecific platinum complex. Sci. China Chem. 2022, 65, 1978–1984. [Google Scholar] [CrossRef]
- Malireddi, R.; Karki, R.; Sundaram, B.; Kancharana, B.; Lee, S.; Samir, P.; Kanneganti, T.-D. Inflammatory cell death, PANoptosis, mediated by cytokines in diverse cancer lineages inhibits tumor growth. Immunohorizons 2021, 5, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, W.; Zhou, H.; Chen, J. Steroidal saponins PPI/CCRIS/PSV induce cell death in pancreatic cancer cell through GSDME-dependent pyroptosis. Biochem. Biophys. Res. Commun. 2023, 173, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, W.; Yang, Z.; Luo, Y.; Qiao, C.; Xie, A.; Jia, Q.; Yang, P.; Wang, Z.; Zhang, R. Sonodynamic-immunomodulatory nanostimulators activate pyroptosis and remodel tumor microenvironment for enhanced tumor immunotherapy. Theranostics 2023, 13, 1571–1583. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Xu, H.; Liu, Q.; Liu, C.; Hu, J.; Liu, P.; Fang, T.; Bai, Y.; Zhu, J.; Xie, R. 5-Aminolevulinic acid hydrochloride loaded microbubbles-mediated sonodynamic therapy in pancreatic cancer cells. Artif. Cells Nanomed. Biotechnol. 2020, 48, 1178–1188. [Google Scholar] [CrossRef]
- Wang, M.; Wu, M.; Liu, X.; Shao, S.; Huang, J.; Liu, B.; Liang, T. Pyroptosis remodeling tumor microenvironment to enhance pancreatic cancer immunotherapy driven by membrane anchoring photosensitizer. Adv. Sci. 2022, 9, 2202914. [Google Scholar] [CrossRef]

| Cell Death | Immunogenic Feature | Lytic Feature | Major Morphological Change | Involved Organelles | Pore Executer | Caspase-Dependent | Main Signaling Pathway | Refs. |
|---|---|---|---|---|---|---|---|---|
Apoptosis  | Generally non-immunogenic. Apoptotic cells are typically eliminated by phagocytosis without initiating a strong immune response. | Non-lytic | Cell shrinkage, intact cell membrane, membrane bubbling | Mitochondria | _ | Dependent (caspase-3/-7/-8) | Apoptotic bodies are formed when caspase -3/7 is activated during extrinsic and intrinsic pathways. Extrinsic pathways activate caspase-3/-7 via activated caspase-8, and intrinsic pathways activate caspase-3/-7 via Bax/Bak-induced mitochondrial DNA fragmentation. | [27,28,29,30,31,32] |
Ferroptosis  | Moderately immunogenic. Can release DAMPs. | Not classically lytic, nanopores in the membrane may not disrupt the membrane integrity to the same extent as necroptosis or pyroptosis. | Cell swelling, nanopore | Mitochondria | Lipid peroxidation | Independent | Inhibition of XC-/GSH/GPX4 induces lipid peroxidation and iron accumulation. | [27,28,29,30,31] |
| ER | ||||||||
Necroptosis  | Highly immunogenic because of the production of abundant DAMPs. Activates immune cells and promotes inflammation. | Lytic, characterized by rupture of the cell membrane and release of intracellular contents. | Cell swelling, pore formation, plasma membrane bubbling | Mitochondria | P-MLKL | Independent | Activation of initiator factors by the death receptor family leads to necrosome formation, MLKL phosphorylation, and DAMP release. | [27,28,29,30,31,33] |
| ER | ||||||||
Pyroptosis  | Highly immunogenic. A strong inflammatory response is triggered due to the release of cytokines (IL-1β and IL-18). Promotes adaptive immune system response. | Lytic, the rupture of a cell membrane caused by GSDM releasing cytokines and cell contents. | Cell swelling, pore formation, plasma membrane bubbling | _ | N-terminal GSDM | Dependent | Canonical or non-canonical caspases cleave and activate GSDM to form pores. | [27,28,29,30,31,33] |
| (caspase-1 (canonical pathway) and caspase-4/-5/-11 (non-canonical pathway)) |
| Type of Cell-Death-Related Genes | Upregulated Genes Correlated with Better Prognosis | Upregulated Genes Correlated with Adverse Prognosis | Ref. |
|---|---|---|---|
| FRGs | CAV1, DDIT4, SRXN1, TFAP2C, MT1G, TUBE1, ATG4D, ENPP2, SETBP1-DT, ZNF93-AS1, SLC25A5-AS1, AC073896.2, LINC00242, PXN-AS1, AC036176.1 | SLC40A1, PTGS2, ATG4D, SLC16A1-AS1 | [56,57,59,60] |
| NRGs | BCL2, JAK3, PLA2G4C, STAT4, CAMK2B, PLA2G4C, STAT4, CASKIN2, TLE2, USP20, SPRN, ARSG, MIR106B, MIR98, PLA2G4C, STAT4, SLC25A6, SLC25A4, METTL14, METTL3 | CAPN, CHMP4C, PYGB, PLA2G4F, CHMP4C, TNFSF10, ACAT2, DHCR7, SQLE, FDPS, MSMO1, OSBPL5, PLBD1, PITPNM3, LPCAT2, LPCAT4, PNPLA3, CPNE3, SLC44A1, SLC2A1, PLA2R1, ALKBH5, HNRNPC, WTAP, YTHDC2, CAPN2, CHMP4C | [61,62,63,64,65,66,67] |
| PRGs | MST1, ELANE, NLRP1, AC090114.2, AC005332.6, PAN3-AS1, AC087501.4, APIP, CHMP6, PLCG1, SMAD4, CDKN2A | GSDME, GSDMC, IL-18, NLRP2, AC083841.1, LINC01133, AC015660.1, AIM2, CASP4, CASP6, CHMP4C, GSDMC, GZMB, CASP4, NLRP1, PLCG1, IL-18, CASP1, NLRP2, TLR3, BAK1, TP63, CHMP4C, PD-L1 | [68,69,70,71,72,73,74,75,76,77,78,79] |
| Agent | Mechanisms of Function | Development Stage | Ref. |
|---|---|---|---|
| Cysteinase | XC- system inhibitor | In vivo | [40] |
| IKE | XC- system inhibitor | In vitro | [40] |
| Rapamycin | GPX4 depletion | In vivo/in vitro | [97] |
| RSL3 | GPX4 depletion | In vivo/in vitro | [97] |
| ART | Increasing intracellular ROS and iron accumulation | In vitro | [98] |
| PL | Increasing intracellular ROS | In vitro | [100] |
| Ruscogenin | Increasing iron accumulation | In vitro | [99] |
| ZZW-115 | NUPR1 inhibitors, modulation of organelle function (ER and mitochondria), metabolic shifts to glycolysis, suppressing GPX4 and SLC7A11, increasing lipid peroxidation | In vivo/in vitro | [101,102,103] |
| Zalcitabine | Ferritinophagy | In vivo/in vitro | [104,105] |
| GEM + chrysin | Inhibition of CBR1, increasing the accumulation of ROS, ferritinophagy | In vivo/in vitro | [106] |
| GEM + lesinurad | Inhibitor of pan-SLC22A, reduces metastasis | In vivo | [107] |
| GEM + docosahexaenoic acid | Induced oxidative stress and cell death | In vivo/in vitro | [108] |
| GEM + EGCG | Inhibition of HSPA5, destabilizing GPX4 | In vivo/in vitro | [48] |
| GEM + SSZ | Inhibition of HSPA5, destabilizing GPX4 | In vivo/in vitro | [48] |
| SSZ + PL + cotylenin A | Accumulation of ROS | In vivo/in vitro | [100] |
| SSZ + docosahexaenoic acid | Inhibition of SLC7A11, modulating the GSH level, restricting nucleotide synthesis | In vivo/in vitro | [108] |
| DHA + DDP | Interruption of mitochondrial hemostasis, catastrophic accumulation of free iron, unrestricted lipid peroxidation, degradation of GPX4 and FTH | In vivo/in vitro | [109] |
| XL888 + anti-PD-1 | Inhibition of HSP90, promoting anti-PD-1 inhibitory | In vivo/in vitro | [93] |
| RSL-3@PVs | Tumor embolisms, inhibiting nutrient delivery, excessive lipid peroxidation, mitochondrial dysfunction | In vivo/in vitro | [110] |
| Agent | Mechanism of Function | Stage of Treatment | Ref. |
|---|---|---|---|
| SB225002 | Inhibition of CXCR2 | In vitro | [159] |
| PDT-MB | Increasing expression of RIPK1, RIPK3, and MLKL | In vitro | [163] |
| AgNPs | Trigger mixed cell death, disrupt the antioxidant, and induce oxidative and nitro-oxidative mechanisms, increasing RIP-1, RIP-3, and MLKL | In vitro/in vitro | [165,166,167] |
| IMB5036 | Trigger mixed cell death, increasing expression of RIPK1, RIPK3, and MLKL | In vivo/in vitro | [168] |
| MLN8237 (alisertib) | Inhibition AURKA | In vivo/in vitro | [173] |
| CCT137690 | Inhibition AURKA | In vivo/in vitro | [171] |
| AdipoRon | Producing superoxide, activating RIPK1 | In vivo/in vitro | [174,175] |
| Vanadium compound | Inhibition of the cell cycle, increasing ROS upregulating RIPK1 and RIPK3 | In vitro | [176] |
| GEM + MLN8237 | Increase GEM sensitivity | Clinical (phase I) | [177] |
| GEM + SK | Regulating the expression of RIP1/RIP3 | In vivo/in vitro | [178] |
| GEM + GAC0003A4 | Impair cholesterol and phospholipid metabolism | In vitro | [63] |
| BV6 + 2′3′-cGAMP | MLKL phosphorylation, stimulation of NF-κB, type I interferons (IFNs), TNFα, and IFN-regulatory factor 1 (IRF1) signaling pathways | In vitro | [179] |
| Agent | Mechanisms of Effect | Stage of Treatment | Ref. |
|---|---|---|---|
| VX-765 | Inhibition caspase-1 | In vivo/in vitro | [68] |
| NAC | ROS scavenger | In vivo/in vitro | [68] |
| Dasatinib | Inhibition Src, increasing ceramide levels | In vivo/in vitro | [201] |
| ceramidase inhibitor B13 | Increasing ASAH2 and ceramide levels | In vivo/in vitro | [201] |
| SEP | Damaging DNA, mitochondrial superoxide anion radical formation induces PANoptosis | In vivo/in vitro | [202,203] |
| Steroidal saponins PPI/CCRIS/PSV | Caspase-3-mediated cleavage of GSDME | In vivo/in vitro | [204] |
| LY364947 + ultrasound | TGF-β receptor inhibitor, produced ROS, caused GSDME to be cleaved by caspase-3, broke the dense TME | In vivo | [205] |
| ALA-lipid/PLGA MBs-mediated SDT | Producing ROS | In vivo/in vitro | [206] |
| PDT + TBD-3C | Convert cold TME to hot TME | In vivo/in vitro | [207] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barar, E.; Shi, J. Genome, Metabolism, or Immunity: Which Is the Primary Decider of Pancreatic Cancer Fate through Non-Apoptotic Cell Death? Biomedicines 2023, 11, 2792. https://doi.org/10.3390/biomedicines11102792
Barar E, Shi J. Genome, Metabolism, or Immunity: Which Is the Primary Decider of Pancreatic Cancer Fate through Non-Apoptotic Cell Death? Biomedicines. 2023; 11(10):2792. https://doi.org/10.3390/biomedicines11102792
Chicago/Turabian StyleBarar, Erfaneh, and Jiaqi Shi. 2023. "Genome, Metabolism, or Immunity: Which Is the Primary Decider of Pancreatic Cancer Fate through Non-Apoptotic Cell Death?" Biomedicines 11, no. 10: 2792. https://doi.org/10.3390/biomedicines11102792





