Effective Invasiveness Recognition of Imbalanced Data by Semi-Automated Segmentations of Lung Nodules
Abstract
:1. Introduction
1.1. Background
- I.
- For technique, it further consists of two following sub-contributions, related to segmentation and classification, respectively.
- (1).
- In terms of semi-automated segmentation, a hybrid segmentation is proposed by fusing thresholding-based morphology and deep learning-based mask-RCNN. Basically, the thresholding-based morphology is the one with statistical thresholding and mathematical shaping, while the deep learning-based mask-RCNN is a region-based convolutional neural network with a fixed anchor. Finally, the better segmentation is derived by switching them.
- (2).
- In terms of invasiveness classification, a boosting ensemble classifier is constructed by equalized down-sampling (called BEED). Especially for imbalance data, the equalized down-sampling generates multiple balanced models, and then a group decision is performed to effectively recognize the invasiveness of early lung cancers.
- II.
- For novelty, most existing real systems mark the tumors as an initial segmentation by fully supervised learning. Then, it still needs to revise the segmentation. Otherwise, without initially automated segmentations, the manual cost is very high. These problems motivate us to conduct a semi-automated segmentation for a convenient usage. In addition to usage convenience, the semi-automated method is more effective than the fully automated ones because it employs click information to achieve a more accurate segmentation.
- III.
- For application, the proposed semi-automated segmentation satisfies the real need of generating a massive training dataset for deep learning. Additionally, the proposed invasiveness recognition can be materialized in real medical systems for effective treatments.
- IV.
- For extension, the proposed ideas of semi-automated segmentation and equalized down-sampling can be extended to other medical fields also with imbalance data such as liver tumor, brain tumor, and so on.
1.2. Related Work
1.2.1. Deep Learning on Object Segmentation
1.2.2. Biomedical Image Recognition and Segmentation
1.2.3. Invasiveness Recognition of Lung Nodules
2. Materials and Methods
2.1. Materials
2.2. Methods
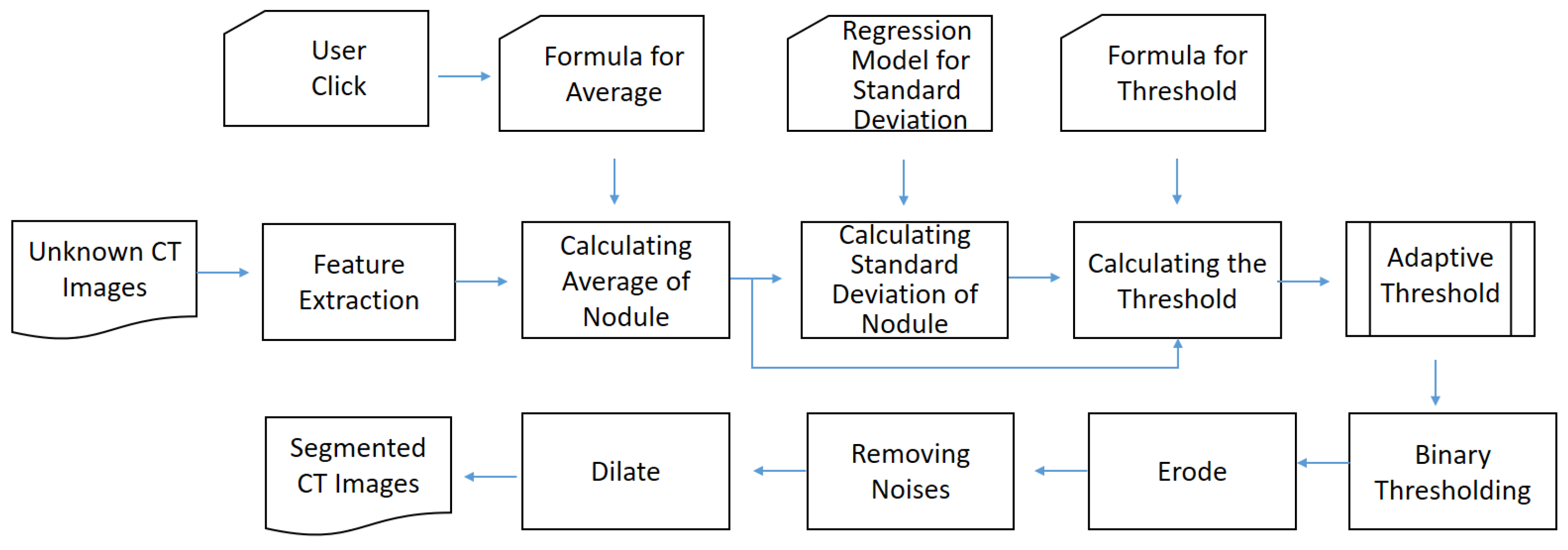
2.2.1. Overview of the Proposed Approach
- I.
- Offline preprocessing: In this stage, lungs are partitioned from the known computed tomography (CT) images first. Next, the necessary components are generated for online recognition, including adaptive threshold, the anchor-fixed Mask-RCNN model, and invasiveness recognition model. For the adaptive threshold, it is determined by two statistical regressions. For Mask-RCNN, it is trained with a fixed anchor. For the invasiveness recognition model, the features are extracted and filtered first. Then, a set of balanced classification models is trained by equalized down-sampling.
- II.
- Online recognition: If the offline preprocessing is completed, the online recognition starts with a submission of unknown CT images. Next, the user will click the target nodules. Then, the system attempts to segment the nodules from unknown images by thresholding. If the result is null, the segmentation will be finished by Mask-RCNN. Finally, based on the segmented nodules, the invasiveness will be recognized by the boosting an ensemble classification model called BEED.
2.2.2. Lung Segmentation
2.2.3. Offline Preprocessing
Determining the Threshold Formula
Training the Anchor-Fixed Mask-RCNN
Training the Invasiveness Recognition Model
2.2.4. Online Recognition
Thresholding-Based Morphology for Semi-Automated Segmentation
Deep Learning-Based Mask-RCNN for Semi-Automated Segmentation
Invasiveness Recognition
3. Results
3.1. Experimental Settings
3.2. Experiments on Semi-Automated Segmentation
3.2.1. Results of Lung Segmentation
3.2.2. Ablation Study
3.2.3. Comparisons with Existing Semi-Automated Segmentation Methods
3.2.4. Illustrative Examples of Segmentation Results
3.3. Experiments on Invasiveness Recognition
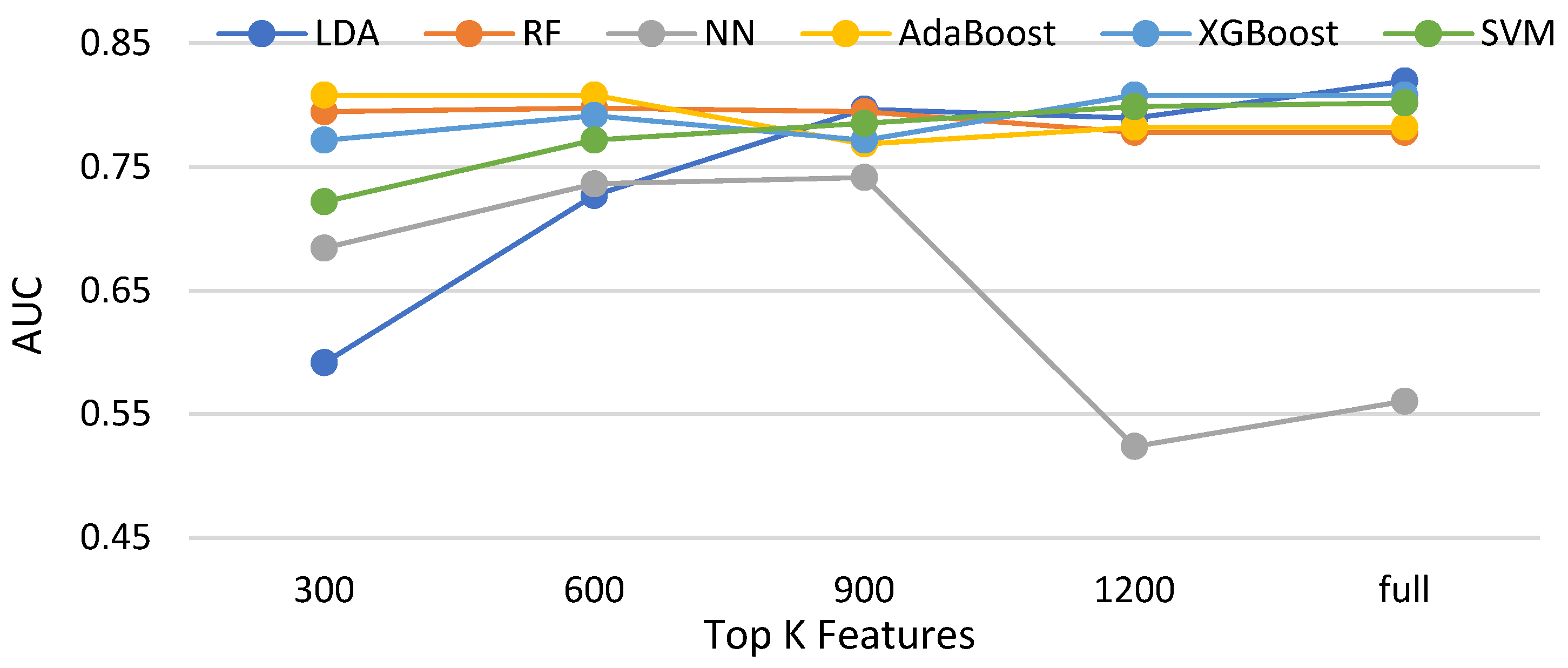
3.3.1. Effectiveness of Feature Selections for Compared Classifiers without Data Balancing
3.3.2. Comparisons of Balancing and Unbalancing Methods for Selected Classifiers
4. Discussion
- I.
- For the mathematical morphology, a further concern needs to be clarified here. In the morphology, the object is reshaped by an erode and a dilate. The primary idea is to delete the noises and to restore the original shape. However, a potential question might thereby be caused: what if varying the numbers of erodes or dilates? Figure 18 shows the answer that the morphology fusing of one erode and one dilate is better than the others. This is because two dilates are too many for one erode. In contrast, for two erodes, two dilates recover the deleted but not complete. Additionally, the morphology with one erode and one dilate is cheaper than the others.
- II.
- In Equation (4), the parameter α determines the threshold highly related to the initial segmented area in binary thresholding. A small threshold might lead to a high recall and low precision. Otherwise, high precision and a low recall might be caused. Therefore, an extended issue for the impact of α is investigated here. Figure 19 shows the effectiveness of the proposed method under different settings of α in terms of precision, recall, and dice, which reaches the best dice as α = 2, with a balance between precisions and recalls. It is obvious that the recall increases as α increases. This is because the segmented area increases simultaneously. However, a larger α will cause a lower precision. This is why the α is set as 2 in this paper.
- III.
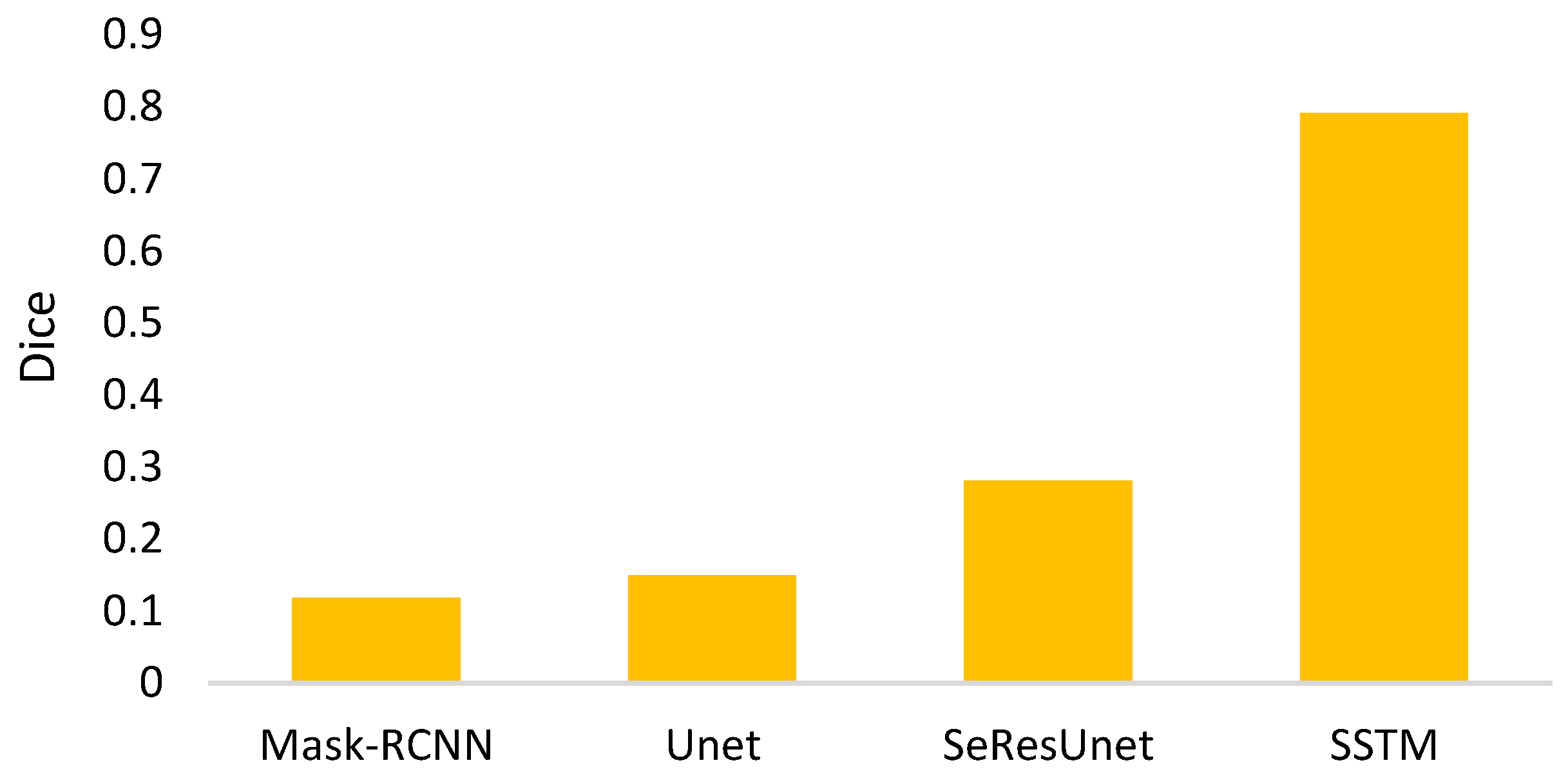
- The goal of semi-automated segmentation is to provide the doctors with an efficient and effective tool for marking the nodules. Actually, most existing marking systems perform the fully automated segmentation as an initial mark. Then, it is revised by the doctor. Hence, a potential question for effectiveness differences of the proposed semi-automated segmentation and fully automated ones needs to be replied. For this question, three recent fully automated segmentation methods, including Mask-RCNN [7], Unet [24], and SeResUnet [34] were compared with the proposed method SSTM. Figure 20 reveals that the proposed SSTM achieves much better dice than the fully automated methods, reaching a dice improvement of 392.3%. The first potential reason is that the training data for the compared methods are not enough. Second, additional click information is very helpful to segmentation. In summary, this result says that the proposed idea is robust if facing small data. Moreover, it is easy and cheap. Note that all methods were executed with the same experimental settings.
- IV.
- The final issue to discuss in this paper is the scalability of the proposed methods, showing the capability of handling the data size variation. It can be interpreted by two categories, namely nodule segmentation and invasiveness recognition. Whether for nodule segmentation or invasiveness recognition, the training data sizes were set from 70% to 90% in this evaluation. Figure 21 and Figure 22 show the related results in terms of dice, AUCs, accuracies, sensitivities, and specificities, respectively. Although the larger training data sizes for all measures achieve the better results, the differences are not significant. It delivers an aspect that the proposed method is not very sensitive to the training data size.
5. Research Limitation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Reports. 2023. Available online: https://www.who.int/health-topics/cancer#tab=tab_1 (accessed on 8 October 2023).
- Uijlings, J.R.; Van De Sande, K.E.; Gevers, T.; Smeulders, A.W. Selective Search for Object Recognition. Int. J. Comput. Vis. 2013, 104, 154–171. [Google Scholar] [CrossRef]
- Sermanet, P.; Eigen, D.; Zhang, X.; Mathieu, M.; Fergus, R.; LeCun, Y. OverFeat: Integrated Recognition, Localization and Detection using Convolutional Networks. arXiv 2013, arXiv:1312.6229. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Region-Based Convolutional Networks for Accurate Object Detection and Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Honolulu, HI, USA, 21–26 July 2017; pp. 2961–2969. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7263–7271. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolov3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Jocher, G. YOLOv5 by Ultralytics. 2023. Available online: https://github.com/ultralytics/yolov5 (accessed on 5 August 2023).
- Li, C.; Li, L.; Jiang, H.; Weng, K.; Geng, Y.; Li, L.; Ke, Z.; Li, Q.; Cheng, M.; Nie, W.; et al. YOLOv6: A Single-Stage Object Detection Framework for Industrial Applications. arXiv 2022, arXiv:2209.02976. [Google Scholar]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar]
- Jocher, G.; Chaurasia, A.; Qiu, J. YOLO by Ultralytics. 2023. Available online: https://github.com/ultralytics/ultralytics (accessed on 5 August 2023).
- Aghamohammadi, A.; Ranjbarzadeh, R.; Naiemi, F.; Mogharrebi, M.; Dorosti, S.; Bendechache, M. TPCNN: Two-path convolutional neural network for tumor and liver segmentation in CT images using a novel encoding approach. Expert Syst. Appl. 2021, 183, 115406. [Google Scholar]
- Amin, J.; Sharif, M.; Gul, N.; Yasmin, M.; Shad, S.A. Brain tumor classification based on DWT fusion of MRI sequences using convolutional neural network. Pattern Recognit. Lett. 2020, 129, 115–122. [Google Scholar] [CrossRef]
- Qi, A.; Zhao, D.; Yu, F.; Heidari, A.A.; Wu, Z.; Cai, Z.; Alenezi, F.; Mansour, R.F.; Chen, H.; Chen, M. Directional mutation and crossover boosted ant colony optimization with application to COVID-19 X-ray image segmentation. Comput. Biol. Med. 2022, 148, 105810. [Google Scholar] [CrossRef]
- Wang, M.; Jin, R.; Lu, J.; Song, E.; Ma, G. Automatic CT liver Couinaud segmentation based on key bifurcation detection with attentive residual hourglass-based cascaded network. Comput. Biol. Med. 2022, 144, 105363. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.; Su, J.H.; Liao, Y.W.; Chang, C.D.; Cheng, Y.F.; Chang, W.C.; Chen, B.H. High-Performance Scaphoid Fracture Recognition via Effectiveness Assessment of Artificial Neural Networks. Appl. Sci. 2021, 11, 8485. [Google Scholar] [CrossRef]
- Vankdothu, R.; Hameed, M.A. Brain tumor MRI images identification and classification based on the recurrent convolutional neural network. Meas. Sens. 2022, 24, 100412. [Google Scholar] [CrossRef]
- Hart, S.N.; Flotte, W.; Andrew, F.; Shah, K.K.; Buchan, Z.R.; Mounajjed, T.; Flotte, T.J. Classification of Melanocytic Lesions in Selected and Whole-Slide Images via Convolutional Neural Networks. J. Pathol. Inform. 2019, 10, 5. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the 18th International Conference on Medical Image Computing and Computer, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, and 8th International Workshop, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; pp. 3–11. [Google Scholar]
- Roy, A.G.; Navab, N.; Wachinger, C. Concurrent Spatial and Channel Squeeze & Excitation in Fully Convolutional Networks. In Proceedings of the 21st International Conference Medical Image Computing and Computer Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 421–429. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Ni, Z.L.; Bian, G.B.; Zhou, X.H.; Hou, Z.G.; Xie, X.L.; Wang, C.; Zhou, Y.J.; Li, R.Q.; Li, Z. RAUNet: Residual Attention U-Net for Semantic Segmentation of Cataract Surgical Instruments. In Proceedings of the International Conference on Neural Information Processing, Vancouver, BC, Canada, 8–14 December 2019; pp. 139–149. [Google Scholar]
- Saumiya, S.; Franklin, S.W. Residual Deformable Split Channel and Spatial U-Net for Automated Liver and Liver Tumour Segmentation. J. Digit. Imaging 2023, 36, 2164–2178. [Google Scholar] [CrossRef]
- Ni, Y.; Yang, Y.; Zheng, D.; Xie, Z.; Huang, H.; Wang, W. The Invasiveness Classification of Ground-Glass Nodules Using 3D Attention Network and HRCT. J. Digit. Imaging 2020, 33, 1144–1154. [Google Scholar] [CrossRef]
- Qiu, L.; Zhang, X.; Mao, H.; Fang, X.; Ding, W.; Zhao, L.; Chen, H. Comparison of Comprehensive Morphological and Radiomics Features of Subsolid Pulmonary Nodules to Distinguish Minimally Invasive Adenocarcinomas and Invasive Adenocarcinomas in CT Scan. Front. Oncol. 2022, 11, 691112. [Google Scholar] [CrossRef]
- Kao, T.N.; Hsieh, M.S.; Chen, L.W.; Yang, C.F.J.; Chuang, C.C.; Chiang, X.H.; Chen, Y.-C.; Lee, Y.-H.; Hsu, H.-H.; Chen, C.-M.; et al. CT-Based Radiomic Analysis for Preoperative Prediction of Tumor Invasiveness in Lung Adenocarcinoma Presenting as Pure Ground-Glass Nodule. Cancers 2022, 14, 5888. [Google Scholar] [CrossRef]
- Sun, Y.; Li, C.; Jin, L.; Gao, P.; Zhao, W.; Ma, W.; Tan, M.; Wu, W.; Duan, S.; Shan, Y.; et al. Radiomics for lung adenocarcinoma manifesting as pure ground-glass nodules: Invasive prediction. Eur. Radiol. 2020, 30, 3650–3659. [Google Scholar] [CrossRef]
- Song, F.; Song, L.; Xing, T.; Feng, Y.; Song, X.; Zhang, P.; Zhang, T.; Zhu, Z.; Song, W.; Zhang, G. A Multi-Classification Model for Predicting the Invasiveness of Lung Adenocarcinoma Presenting as Pure Ground-Glass Nodules. Front. Oncol. 2022, 12, 800811. [Google Scholar] [CrossRef]
- Kaggle. Data Science Bowl 2017. Available online: https://www.kaggle.com/c/data-science-bowl-2017 (accessed on 7 June 2022).
- Jha, D. ResUNetPlusPlus. Available online: https://github.com/DebeshJha/ResUNetPlusPlus/blob/master/m_resunet.py (accessed on 6 January 2023).
- Shur, J.D.; Doran, S.J.; Kumar, S.; ap Dafydd, D.; Downey, K.; O’Connor, J.P.B.; Papanikolaou, N.; Messiou, C.; Koh, D.-M.; Orton, M.R. Radiomics in Oncology: A Practical Guide. Radiographics 2021, 41, 1717–1732. [Google Scholar] [CrossRef]
- Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillon-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Osher, S.; Fedkiw, R.; Piechor, K. Level set methods and dynamic implicit surfaces. Appl. Mech. Rev. 2004, 57, B15. [Google Scholar] [CrossRef]
- Huang, Z.K.; Chau, K.W. A new image thresholding method based on Gaussian mixture model. Appl. Math. Comput. 2008, 205, 899–907. [Google Scholar] [CrossRef]
- Chaki, N.; Shaikh, S.H.; Saeed, K.; Chaki, N.; Shaikh, S.H.; Saeed, K. A Comprehensive Survey on Image Binarization Techniques; Springer: Delhi, India, 2004; pp. 5–15. [Google Scholar]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Cuevas, A.; Febrero, M.; Fraiman, R. An anova test for functional data. Comput. Stat. Data Anal. 2004, 47, 111–122. [Google Scholar] [CrossRef]
- Han, H.; Wang, W.-Y.; Mao, B.-H. Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning. In Proceedings of the International Conference on Intelligent Computing, Hefei, China, 23–26 August 2005; pp. 878–887. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]



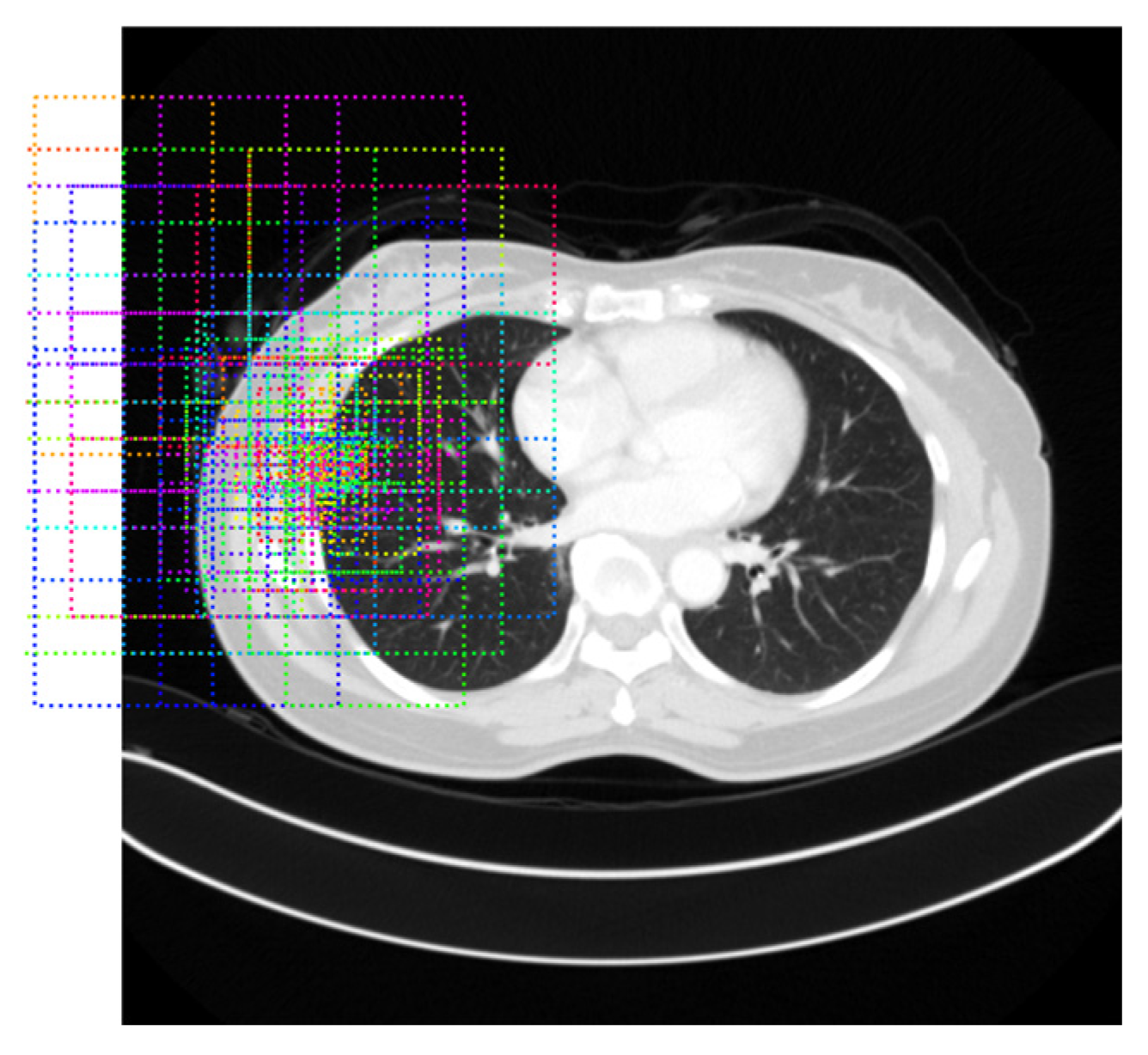






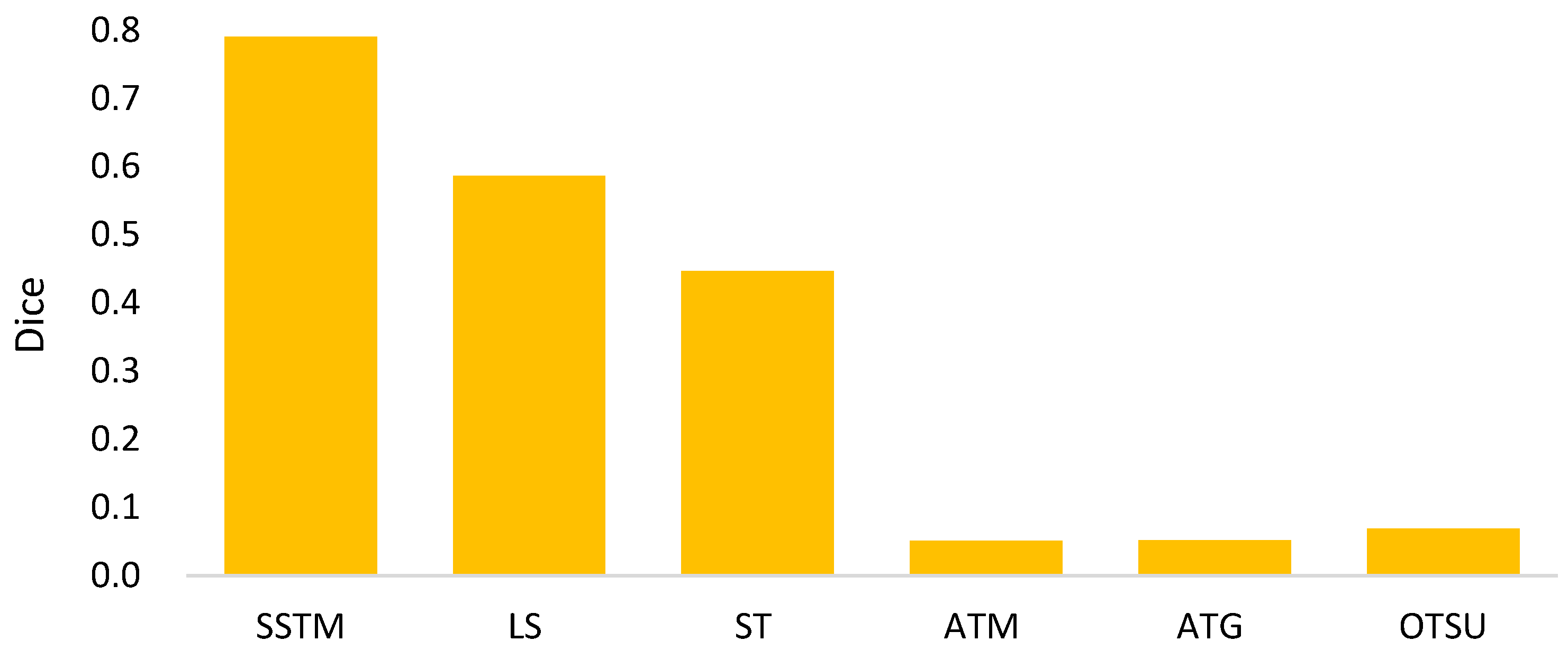








| Fold # | Dice | Standard Deviation |
|---|---|---|
| Fold 1 | 0.991 | 0.104842 |
| Fold 2 | 0.982 | 0.241201 |
| Fold 3 | 0.984 | 0.247245 |
| Fold 4 | 0.990 | 0.046181 |
| Fold 5 | 0.983 | 0.198752 |
| Average | 0.986 | 0.167644 |
| Method | Terminology |
|---|---|
| Proposed Fusion of TM and MR | SSTM |
| Level-Set [39] | LS |
| Static Threshold | ST |
| Adaptive Threshold by Mean [40] | ATM |
| Adaptive Threshold by Gaussian [41] | ATG |
| Adaptive Threshold by OTSU [42] | OTSU |
| Classifier | Terminology |
|---|---|
| Linear Discriminant Analysis | LDA |
| Random Forest | RF |
| Neural Network | NN |
| AdaBoost | AdaBoost |
| XGBoost | XGBoost |
| Support Vector Machine | SVM |
| Accuracy | AUC | Sensitivity | Specificity | ||
|---|---|---|---|---|---|
| RF (Information Gain 300) | BEED (proposed) | 0.9 | 0.859 * | 0.919 | 0.8 * |
| SMOTE | 0.895 | 0.816 | 0.931 | 0.7 | |
| Imbalanced | 0.9 | 0.778 | 0.956 * | 0.6 | |
| LDA (Full Features) | BEED (proposed) | 0.853 | 0.764 | 0.894 | 0.633 |
| SMOTE | 0.663 | 0.624 | 0.681 | 0.567 | |
| Imbalanced | 0.879 | 0.82 | 0.906 | 0.733 | |
| XGBoost (ANOVA 1200) | BEED (proposed) | 0.884 | 0.782 | 0.931 | 0.633 |
| SMOTE | 0.874 | 0.79 | 0.913 | 0.667 | |
| Imbalanced | 0.911 * | 0.811 | 0.956 * | 0.667 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tung, Y.-C.; Su, J.-H.; Liao, Y.-W.; Lee, Y.-C.; Chen, B.-A.; Huang, H.-M.; Jhang, J.-J.; Hsieh, H.-Y.; Tong, Y.-S.; Cheng, Y.-F.; et al. Effective Invasiveness Recognition of Imbalanced Data by Semi-Automated Segmentations of Lung Nodules. Biomedicines 2023, 11, 2938. https://doi.org/10.3390/biomedicines11112938
Tung Y-C, Su J-H, Liao Y-W, Lee Y-C, Chen B-A, Huang H-M, Jhang J-J, Hsieh H-Y, Tong Y-S, Cheng Y-F, et al. Effective Invasiveness Recognition of Imbalanced Data by Semi-Automated Segmentations of Lung Nodules. Biomedicines. 2023; 11(11):2938. https://doi.org/10.3390/biomedicines11112938
Chicago/Turabian StyleTung, Yu-Cheng, Ja-Hwung Su, Yi-Wen Liao, Yeong-Chyi Lee, Bo-An Chen, Hong-Ming Huang, Jia-Jhan Jhang, Hsin-Yi Hsieh, Yu-Shun Tong, Yu-Fan Cheng, and et al. 2023. "Effective Invasiveness Recognition of Imbalanced Data by Semi-Automated Segmentations of Lung Nodules" Biomedicines 11, no. 11: 2938. https://doi.org/10.3390/biomedicines11112938







