A Radiotherapy Dose Map-Guided Deep Learning Method for Predicting Pathological Complete Response in Esophageal Cancer Patients after Neoadjuvant Chemoradiotherapy Followed by Surgery
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
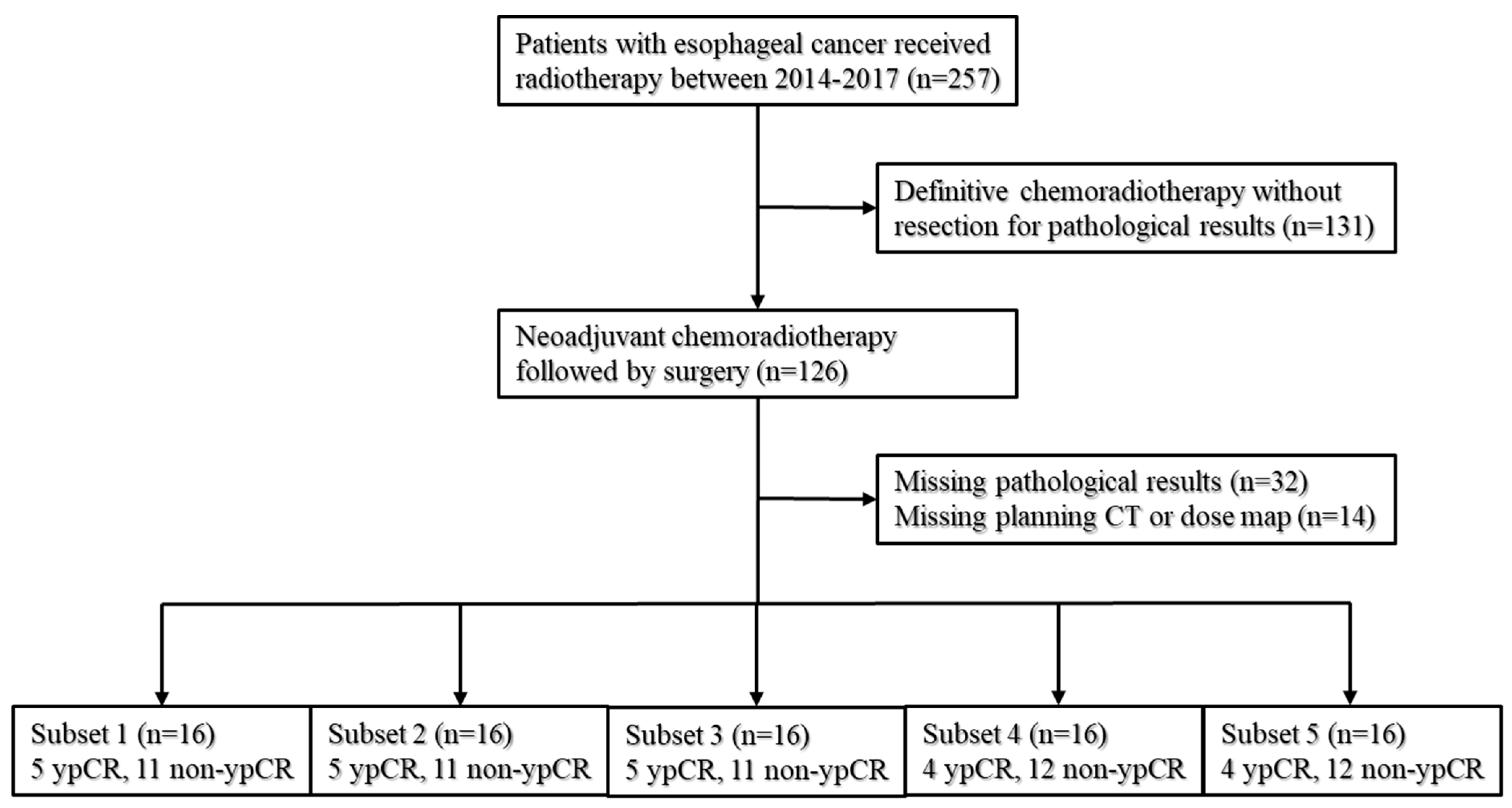
2.1. Patients
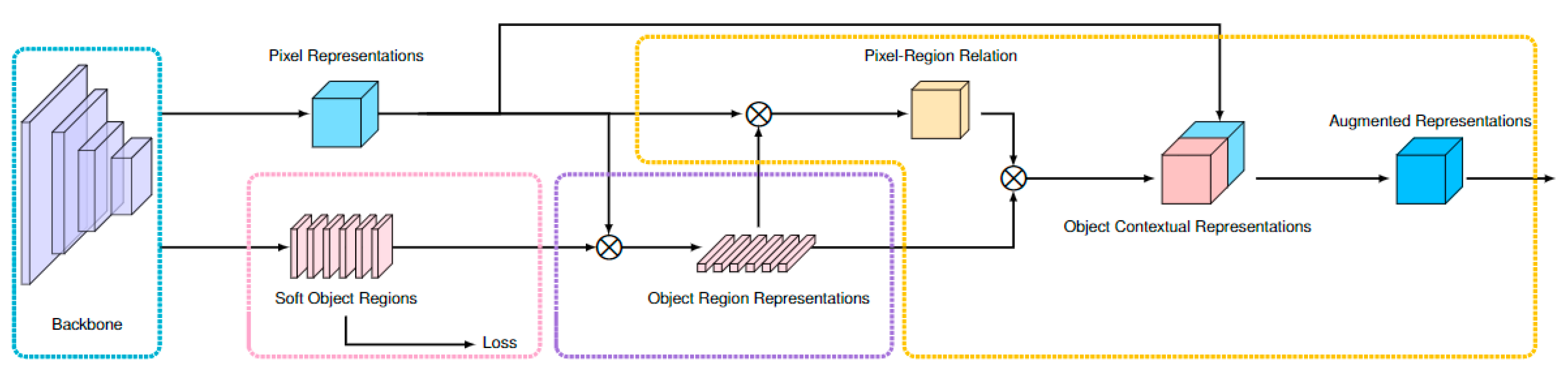
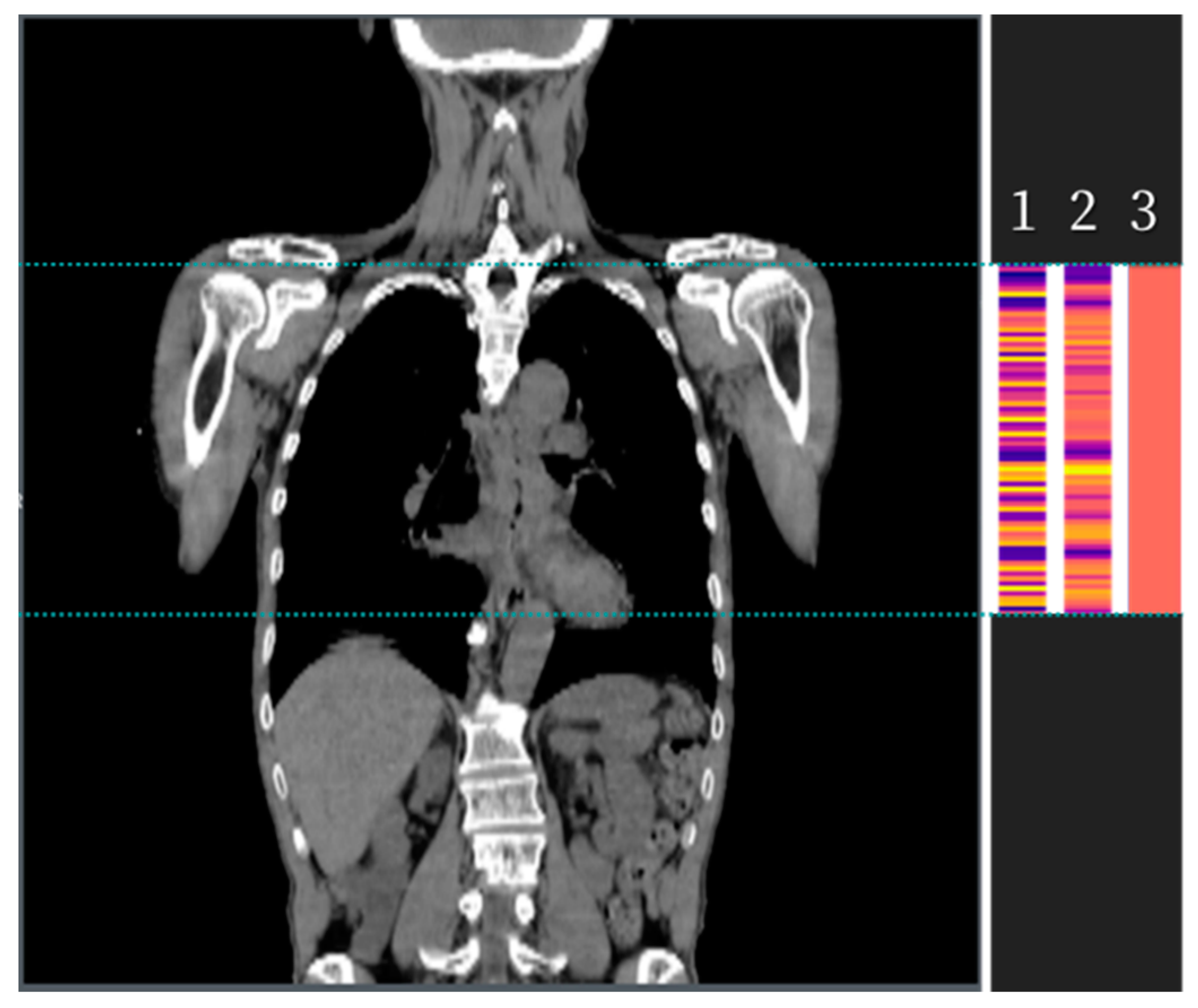
2.2. Development of the Algorithm
2.3. Statistical Analysis
2.4. Five-Fold Cross-Validation
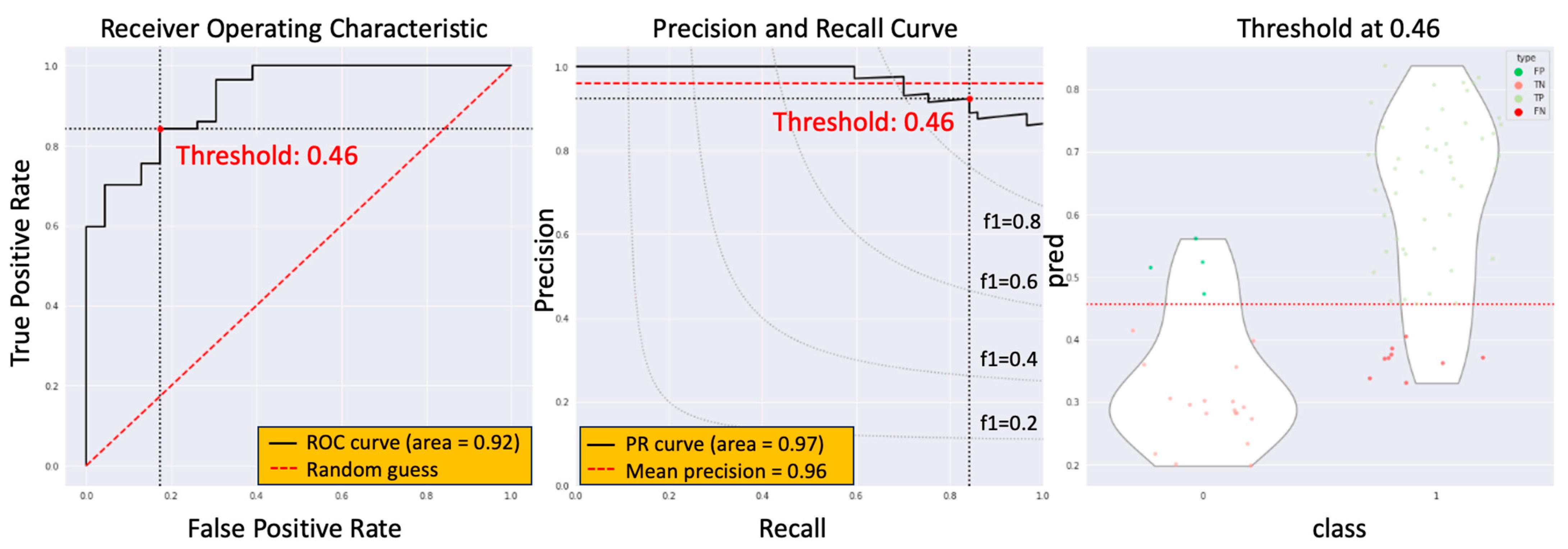
3. Results
3.1. Baseline Characteristics
3.2. The Comparison of Three Methods in the Patient Summary
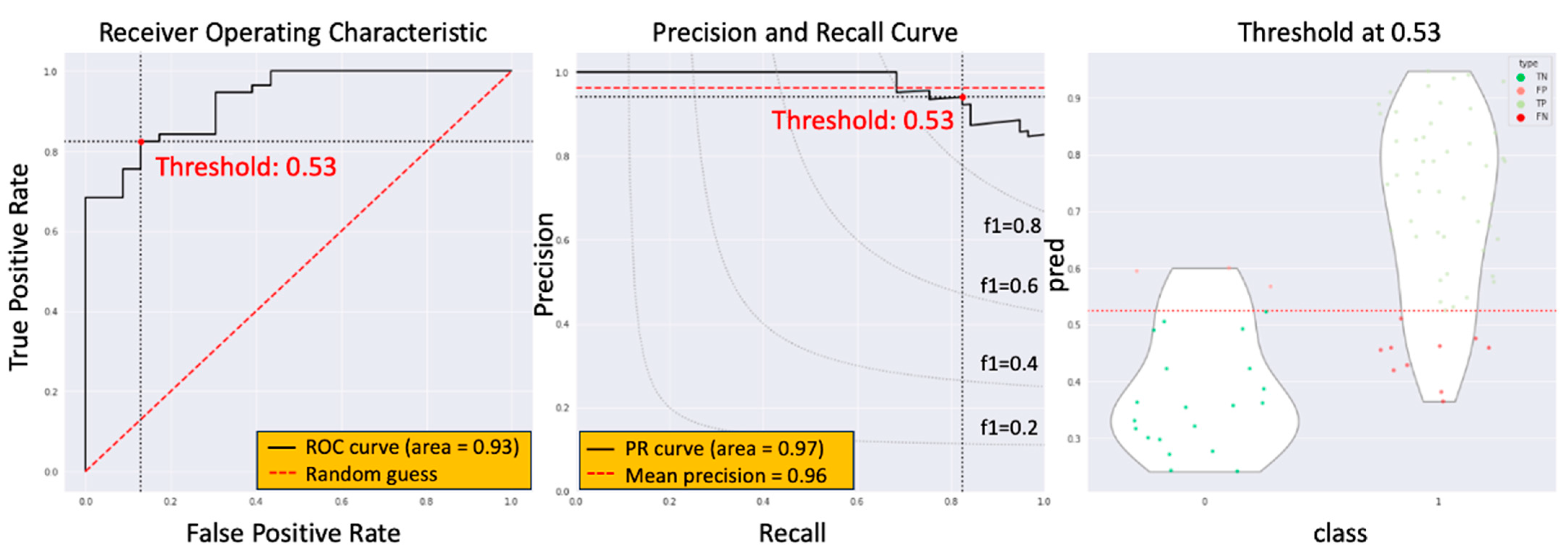
3.3. The Model Performance of DCRNet in Pathological Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, A.K.; El-Serag, H.B. Esophageal carcinoma. N. Engl. J. Med. 2014, 371, 2499–2509. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.; van Lanschot, J.J.B.; Hulshof, M.C.C.M.; van Hagen, P.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.L.; van Laarhoven, H.W.M.; Nieuwenhuijzen, G.A.P.; Hospers, G.A.P.; Bonenkamp, J.J.; et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): Long-term results of a randomised controlled trial. Lancet Oncol. 2015, 16, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, H.; Chen, Y.; Zhu, C.; Fang, W.; Yu, Z.; Mao, W.; Xiang, J.; Han, Y.; Chen, Z.; et al. Neoadjuvant chemoradiotherapy followed by surgery versus surgery alone for locally advanced squamous cell carcinoma of the esophagus (NEOCRTEC5010): A phase III multicenter, randomized, open-label clinical trial. J. Clin. Oncol. 2018, 36, 2796–2803. [Google Scholar] [CrossRef] [PubMed]
- de Gouw, D.J.; Klarenbeek, B.R.; Driessen, M.; Bouwense, S.A.; van Workum, F.; Fütterer, J.J.; Rovers, M.M.; Broek, R.P.T.; Rosman, C. Detecting Pathological Complete Response in Esophageal Cancer after Neoadjuvant Therapy Based on Imaging Techniques: A Diagnostic Systematic Review and Meta-Analysis. J. Thorac. Oncol. 2019, 14, 1156–1171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tan, S.; Chen, W.; Kligerman, S.; Kim, G.; D’Souza, W.D.; Suntharalingam, M.; Lu, W. Modeling pathologic response of esophageal cancer to chemoradiation therapy using spatial-temporal 18F-FDG PET features, clinical parameters, and demographics. Int. J. Radiat. Oncol. Biol. Phys. 2014, 88, 195–203. [Google Scholar] [CrossRef]
- Beukinga, R.J.; Hulshoff, J.B.; van Dijk, L.V.; Muijs, C.T.; Burgerhof, J.G.; Kats-Ugurlu, G.; Slart, R.H.; Slump, C.H.; Mul, V.E.; Plukker, J.T. Predicting Response to Neoadjuvant Chemoradiotherapy in Esophageal Cancer with Textural Features Derived from Pretreatment 18F-FDG PET/CT Imaging. J. Nucl. Med. 2017, 58, 723–729. [Google Scholar] [CrossRef]
- Yang, Z.; He, B.; Zhuang, X.; Gao, X.; Wang, D.; Li, M.; Lin, Z.; Luo, R. CT-based radiomic signatures for prediction of pathologic complete response in esophageal squamous cell carcinoma after neoadjuvant chemoradiotherapy. J. Radiat. Res. 2019, 60, 538–545. [Google Scholar] [CrossRef]
- Ypsilantis, P.-P.; Siddique, M.; Sohn, H.-M.; Davies, A.; Cook, G.; Goh, V.; Montana, G. Predicting Response to Neoadjuvant Chemotherapy with PET Imaging Using Convolutional Neural Networks. PLoS ONE 2015, 10, e0137036. [Google Scholar] [CrossRef]
- Hou, Z.; Li, S.; Ren, W.; Liu, J.; Yan, J.; Wan, S. Radiomic analysis in T2W and SPAIR T2W MRI: Predict treatment response to chemoradiotherapy in esophageal squamous cell carcinoma. J. Thorac. Dis. 2018, 10, 2256–2267. [Google Scholar] [CrossRef]
- Hou, Z.; Ren, W.; Li, S.; Liu, J.; Sun, Y.; Yan, J.; Wan, S. Radiomic analysis in contrast-enhanced CT: Predict treatment response to chemoradiotherapy in esophageal carcinoma. Oncotarget 2017, 8, 104444–104454. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xie, C.; Yang, H.; Ho, J.W.K.; Wen, J.; Han, L.; Chiu, K.W.H.; Fu, J.; Vardhanabhuti, V. Assessment of Intratumoral and Peritumoral Computed Tomography Radiomics for Predicting Pathological Complete Response to Neoadjuvant Chemoradiation in Patients With Esophageal Squamous Cell Carcinoma. JAMA Netw. Open 2020, 3, e2015927. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.B.; Xiao, L.; Patel, V.R.; Maru, D.M.; Correa, A.M.; Amlashi, F.G.; Liao, Z.; Komaki, R.; Lin, S.H.; Skinner, H.D.; et al. Pathological complete response in patients with esophageal cancer after the trimodality approach: The association with baseline variables and survival-The University of Texas MD Anderson Cancer Center experience. Cancer 2017, 123, 4106–4113. [Google Scholar] [CrossRef]
- Shen, J.; AME Thoracic Surgery Collaborative Group; Kong, M.; Yang, H.; Jin, K.; Chen, Y.; Fang, W.; Yu, Z.; Mao, W.; Xiang, J.; et al. Pathological complete response after neoadjuvant treatment determines survival in esophageal squamous cell carcinoma patients (NEOCRTEC5010). Ann. Transl. Med. 2021, 9, 1516. [Google Scholar] [CrossRef]
- Yap, W.-K.; Chang, Y.-C.; Hsieh, C.-H.; Chao, Y.-K.; Chen, C.-C.; Shih, M.-C.; Hung, T.-M. Favorable versus unfavorable prognostic groups by post-chemoradiation FDG-PET imaging in node-positive esophageal squamous cell carcinoma patients treated with definitive chemoradiotherapy. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 689–698. [Google Scholar] [CrossRef]
- Yuan, Y.; Chen, X.; Chen, X.; Wang, J. Segmentation Transformer: Object-Contextual Representations for Semantic Segmentation. arXiv 2019, arXiv:1909.11065. [Google Scholar]
- Sun, K.; Zhao, Y.; Jiang, B.; Cheng, T.; Xiao, B.; Liu, D.; Mu, Y.; Wang, X.; Liu, W.; Wang, J. High-Resolution Representations for Labeling Pixels and Regions. arXiv 2019, arXiv:1904.04514. [Google Scholar]
- Shrivastava, A.; Gupta, A.; Girshick, R. Training Region-based Object Detectors with Online Hard Example Mining. arXiv 2016, arXiv:1604.03540. [Google Scholar]
- Delong, E.R.; Delong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.-S.; Xu, H.-Y.; Du, Z.-S.; Li, X.-Y.; Wu, S.-X.; Huang, H.-C.; Lin, L.-X. Impact of sex on the prognosis of patients with esophageal squamous cell cancer underwent definitive radiotherapy: A propensity score-matched analysis. Radiat. Oncol. 2019, 14, 74. [Google Scholar] [CrossRef]
- Ministry of Health and Welfare Taiwan. Cancer Registry Annual Report, 2020. 2022. Available online: https://www.hpa.gov.tw/Pages/Detail.aspx?nodeid=269&pid=16434 (accessed on 15 October 2023).
- Wang, S.; Zheng, R.; Arnold, M.; Abnet, C.; Zeng, H.; Zhang, S.; Chen, R.; Sun, K.; Li, L.; An, L.; et al. Global and national trends in the age-specific sex ratio of esophageal cancer and gastric cancer by subtype. Int. J. Cancer 2022, 151, 1447–1461. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | Category | All Patients, n (%) | ypCR, n (%) | Non-ypCR, n (%) | p |
|---|---|---|---|---|---|
| Sex | Male | 76 (95) | 20 (87.0) | 56 (98.2) | 0.04 |
| Female | 4 (5) | 3 (13.0) | 1 (1.8) | ||
| Age (year) | Mean (SD) | 55.68 ± 9.5 | 55.48 ± 7.3 | 55.76 ± 10.3 | 0.28 |
| Location | Proximal | 5 (8.9) | 3 (13.1) | 2 (6.1) | 0.16 |
| Middle | 23 (41.1) | 12 (52.2) | 11 (33.3) | ||
| Distal | 28 (50) | 8 (34.8) | 20 (60.6) | ||
| Clinical stage | II | 5 (6.3) | 2 (8.7) | 3 (5.3) | 0.70 |
| III | 57 (71.2) | 17 (73.9) | 40 (70.1) | ||
| IVA | 18 (22.5) | 4 (17.4) | 14 (24.6) | ||
| Total radiation dose (Gy) | 44.0 ± 2.1 | 44.5 ± 2.4 | 43.8 ± 2.8 | 0.81 | |
| OS (month) | 24.1 ± 13.7 | 27.7 ± 16.2 | 22.6 ± 14.8 | 0.08 |
| AUC (95% CI) | ypCR | ypT0 | ypN0 |
|---|---|---|---|
| HRN + DCR | 0.928 (0.884–0.972) | 0.939 (0.928–0.950) | 0.891 (0.881–0.901) |
| HRN | 0.865 (0.856–0.873) | 0.877 (0.865–0.889) | 0.859 (0.846–0.872) |
| RN + DCR | 0.829 (0.809–0.849) | 0.845 (0.828–0.862) | 0.813 (0.799–0.827) |
| RN | 0.763 (0.751–0.775) | 0.775 (0.759–0.791) | 0.751 (0.733–0.769) |
| EN + DCR | 0.836 (0.818–0.854) | 0.847 (0.825–0.869) | 0.825 (0.806–0.844) |
| EN | 0.766 (0.749–0.783) | 0.780 (0.756–0.804) | 0.752 (0.736–0.768) |
| DN + DCR | 0.832 (0.812–0.852) | 0.844 (0.826–0.862) | 0.820 (0.803–0.837) |
| DN | 0.761 (0.742–0.780) | 0.776 (0.756–0.796) | 0.746 (0.731–0.761) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yap, W.-K.; Hsiao, I.-T.; Yap, W.-L.; Tsai, T.-Y.; Lu, Y.-A.; Yang, C.-K.; Peng, M.-T.; Su, E.-L.; Cheng, S.-C. A Radiotherapy Dose Map-Guided Deep Learning Method for Predicting Pathological Complete Response in Esophageal Cancer Patients after Neoadjuvant Chemoradiotherapy Followed by Surgery. Biomedicines 2023, 11, 3072. https://doi.org/10.3390/biomedicines11113072
Yap W-K, Hsiao I-T, Yap W-L, Tsai T-Y, Lu Y-A, Yang C-K, Peng M-T, Su E-L, Cheng S-C. A Radiotherapy Dose Map-Guided Deep Learning Method for Predicting Pathological Complete Response in Esophageal Cancer Patients after Neoadjuvant Chemoradiotherapy Followed by Surgery. Biomedicines. 2023; 11(11):3072. https://doi.org/10.3390/biomedicines11113072
Chicago/Turabian StyleYap, Wing-Keen, Ing-Tsung Hsiao, Wing-Lake Yap, Tsung-You Tsai, Yi-An Lu, Chan-Keng Yang, Meng-Ting Peng, En-Lin Su, and Shih-Chun Cheng. 2023. "A Radiotherapy Dose Map-Guided Deep Learning Method for Predicting Pathological Complete Response in Esophageal Cancer Patients after Neoadjuvant Chemoradiotherapy Followed by Surgery" Biomedicines 11, no. 11: 3072. https://doi.org/10.3390/biomedicines11113072





