Lysine Acetylome of Breast Cancer-Derived Small Extracellular Vesicles Reveals Specific Acetylation Patterns for Metabolic Enzymes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culturing and sEV Isolation
2.2. Quantification of sEVs by Nanoparticle Tracking Analysis (NTA)
2.3. Characterization of EVs Protein Markers
2.4. Sample Preparation for Acetylomics
2.5. Protein Reduction, Alkylation and Enzymatic Digestion
2.6. Enrichment of Acetylated Peptides
2.7. Nano-LC-MS/MS
2.8. MS Spectra Processing
2.9. Data Filtering and Acetylation Site Localization
2.10. Disease and Functional Annotation Analysis
2.11. Data Availability
2.12. Aldolase Activity Assay
2.13. Glyceraldehyde 3 Phosphate Dehydrogenase Activity Assay
2.14. Phosphoglycerate Kinase Activity Assay
2.15. Enolase Activity Assay
2.16. Pyruvate Kinase Activity Assay
2.17. Statistical Analysis
3. Results
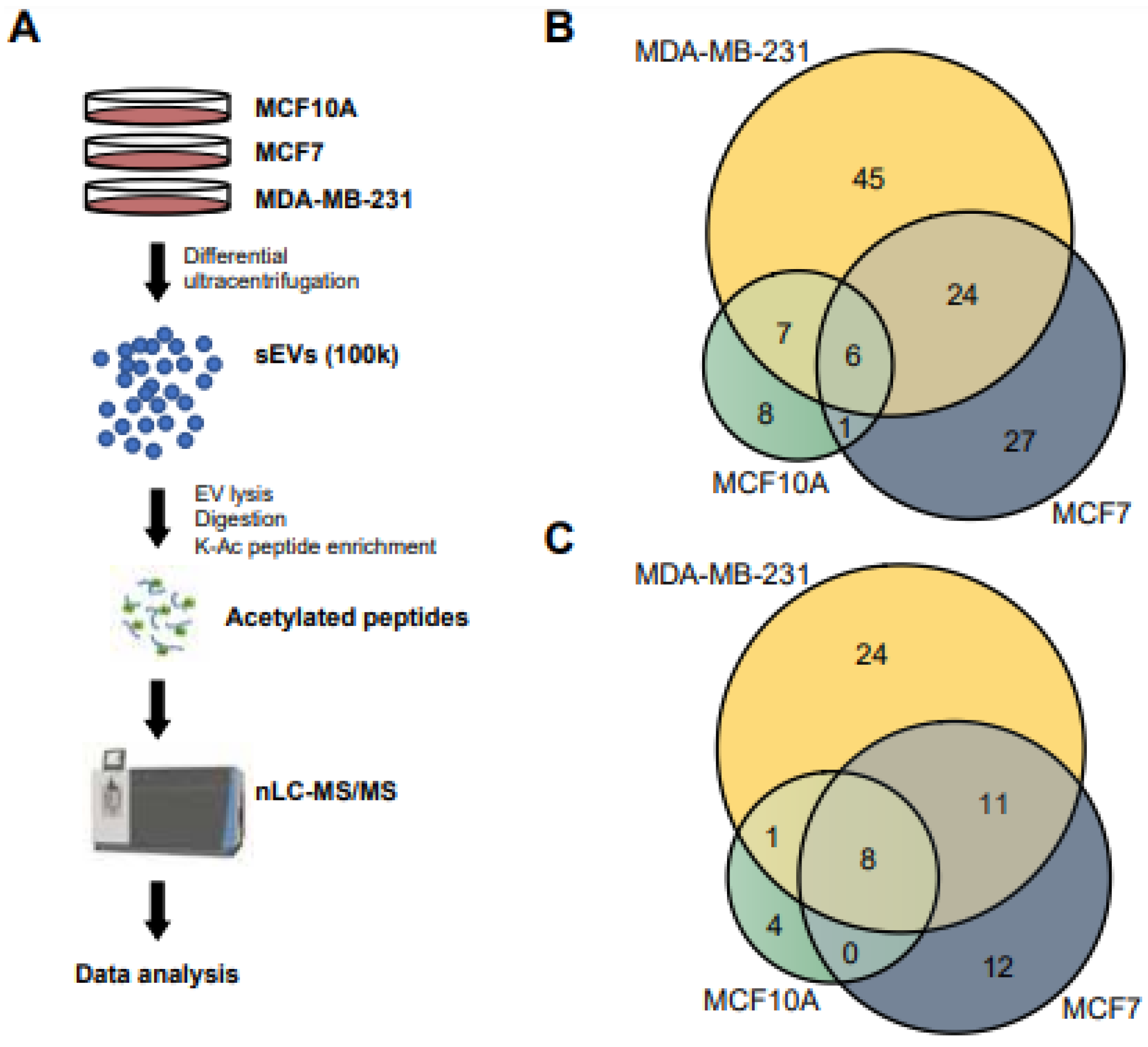
3.1. Isolation of sEV
3.2. Overall Acetylome Profiling
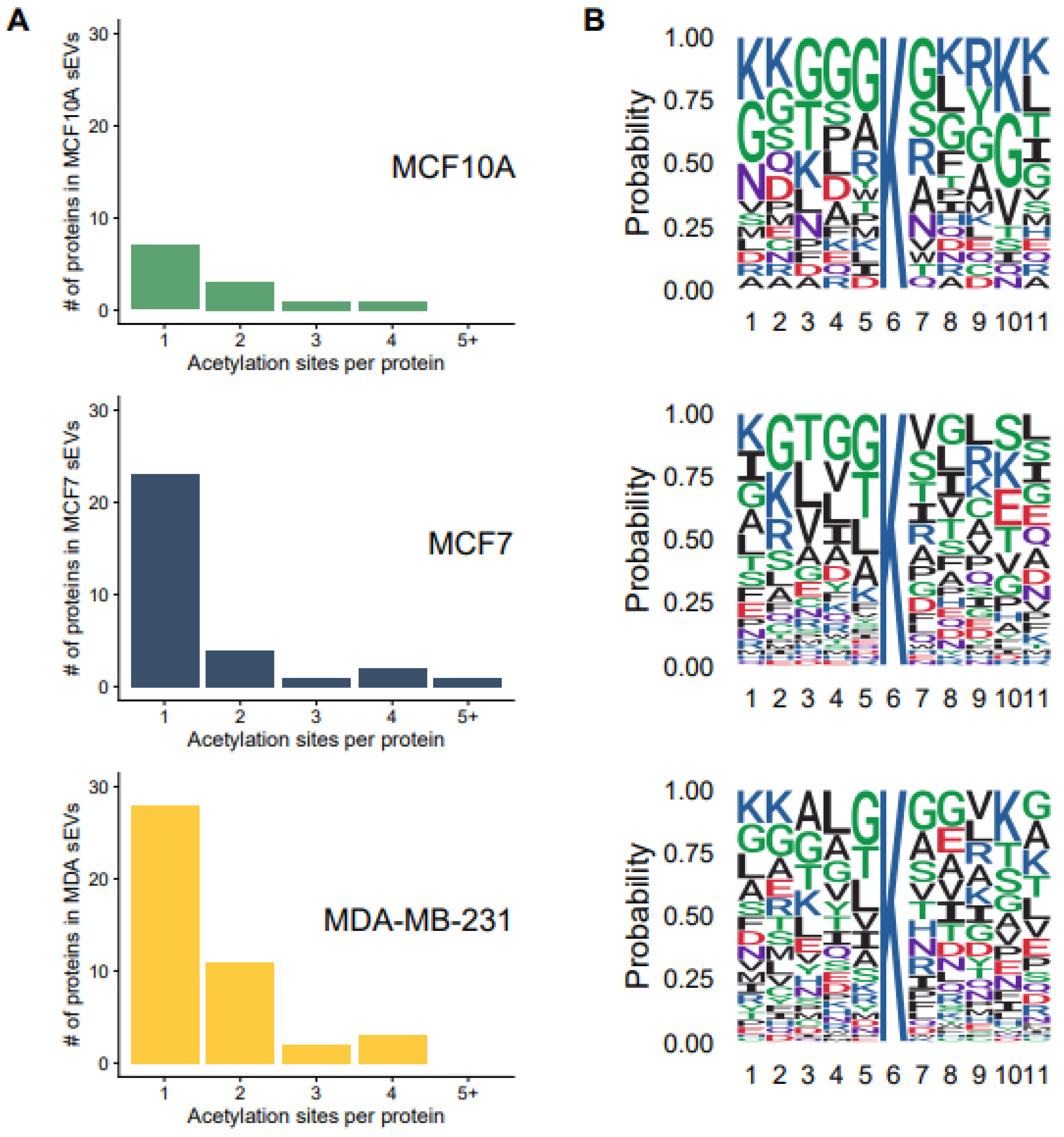
3.3. Acetylation Site Distributions and Motifs for the Identified Acetylation Sites
3.4. Functional and Pathway Analysis of Identified Acetylated Proteins
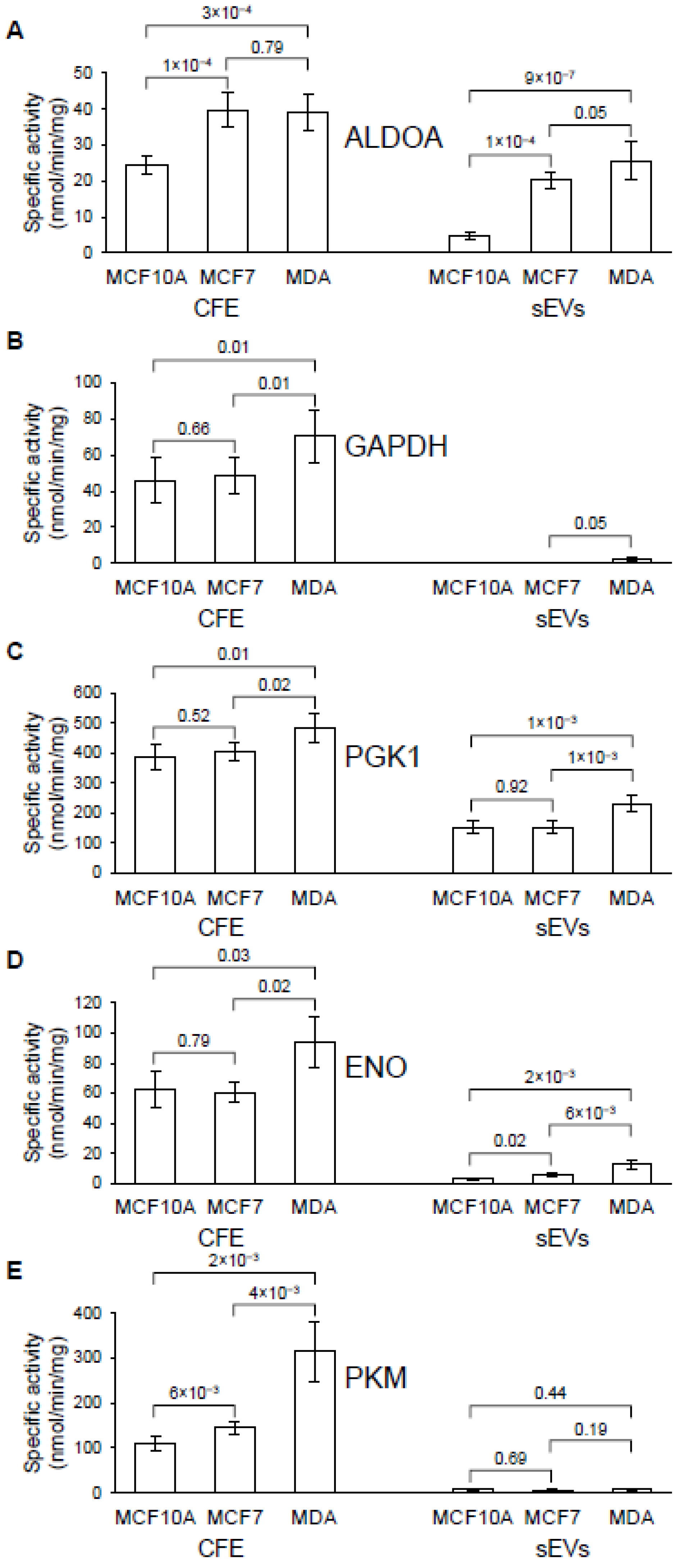
3.5. Analysis of ALDOA, GAPDH, PGK 1, ENO and PKM in Cells and Their sEVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kruger, S.; Abd Elmageed, Z.Y.; Hawke, D.H.; Wörner, P.M.; Jansen, D.A.; Abdel-Mageed, A.B.; Alt, E.U.; Izadpanah, R. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer 2014, 14, 44. [Google Scholar] [CrossRef] [Green Version]
- Palazzolo, G.; Albanese, N.N.; DI Cara, G.; Gygax, D.; Vittorelli, M.L.; Pucci-Minafra, I. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer Res. 2012, 32, 847–860. [Google Scholar] [PubMed]
- Hurwitz, S.N.; Rider, M.A.; Bundy, J.L.; Liu, X.; Singh, R.K.; Meckes, D.G., Jr. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 2016, 7, 86999–87015. [Google Scholar] [CrossRef] [PubMed]
- Demory Beckler, M.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell Proteomics 2013, 12, 343–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, B.; Peng, P.; Chen, S.; Li, L.; Zhang, M.; Cao, D.; Yang, J.; Li, H.; Gui, T.; Li, X.; et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteomics 2013, 80, 171–182. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers. Sci. Rep. 2020, 10, 13572. [Google Scholar] [CrossRef] [PubMed]
- Risha, Y.; Susevski, V.; Hüttmann, N.; Poolsup, S.; Minic, Z.; Berezovski, M.V. Breast Cancer-Derived Microvesicles Are the Source of Functional Metabolic Enzymes as Potential Targets for Cancer Therapy. Biomedicines 2021, 9, 107. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, A.S.; Baeta, H.; Silva, B.C.; Moraes, M.C.S.; Bodo, C.; Beck, H.C.; Rodriguez, M.S.; Saraswat, M.; Pandey, A.; Matthiesen, R. Extra-cellular vesicles carry proteome of cancer hallmarks. Front. Biosci. (Landmark Ed.) 2020, 25, 398–436. [Google Scholar] [CrossRef]
- Brzozowski, J.S.; Jankowski, H.; Bond, D.R.; McCague, S.B.; Munro, B.R.; Predebon, M.J.; Scarlett, C.J.; Skelding, K.A.; Weidenhofer, J. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018, 17, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Greening, D.W.; Zhu, H.J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef] [Green Version]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Di Vizio, D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minic, Z.; Hüttmann, N.; Poolsup, S.; Li, Y.; Susevski, V.; Zaripov, E.; Berezovski, M.V. Phosphoproteomic Analysis of Breast Cancer-Derived Small Extracellular Vesicles Reveals Disease-Specific Phosphorylated Enzymes. Biomedicines 2022, 10, 408. [Google Scholar] [CrossRef] [PubMed]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int. J. Mol. Sci. 2019, 20, 3153. [Google Scholar] [CrossRef] [Green Version]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef] [Green Version]
- Yi, Y.W.; Kang, H.J.; Kim, H.J.; Kong, Y.; Brown, M.L.; Bae, I. Targeting mutant p53 by a SIRT1 activator YK-3-237 inhibits the proliferation of triple-negative breast cancer cells. Oncotarget 2013, 4, 984–994. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.S.; Hyun, C.L.; Park, I.A.; Kim, J.Y.; Chung, Y.R.; Im, S.A.; Lee, K.H.; Moon, H.G.; Ryu, H.S. SIRT1 induces tumor invasion by targeting epithelial mesenchymal transition-related pathway and is a prognostic marker in triple negative breast cancer. Tumour Biol. 2016, 37, 4743–4753. [Google Scholar] [CrossRef]
- Wang, C.; Yang, W.; Dong, F.; Guo, Y.; Tan, J.; Ruan, S.; Huang, T. The prognostic role of Sirt1 expression in solid malignancies: A meta-analysis. Oncotarget 2017, 8, 66343–66351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.W.; Li, W.Q.; Wan, G.X.; Li, Y.X.; Du, X.M.; Li, Y.C.; Li, F. Correlation and prognostic value of SIRT1 and Notch1 signaling in breast cancer. J. Exp. Clin. Cancer Res. 2014, 33, 97. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.R.; Kim, H.; Park, S.Y.; Park, I.A.; Jang, J.J.; Choe, J.Y.; Jung, Y.Y.; Im, S.A.; Moon, H.G.; Lee, K.H.; et al. Distinctive role of SIRT1 expression on tumor invasion and metastasis in breast cancer by molecular subtype. Hum. Pathol. 2015, 46, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.I.; Nirzhor, S.S.R.; Akter, R. A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets. Biomolecules 2018, 8, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef] [Green Version]
- Sebastián, C.; Zwaans, B.M.; Silberman, D.M.; Gymrek, M.; Goren, A.; Zhong, L.; Ram, O.; Truelove, J.; Guimaraes, A.R.; Toiber, D.; et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 2012, 151, 1185–1199. [Google Scholar] [CrossRef] [Green Version]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, C.; Weinert, B.T.; Nishida, Y.; Verdin, E.; Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 536–550. [Google Scholar] [CrossRef]
- Verdin, E.; Ott, M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015, 16, 258–264. [Google Scholar] [CrossRef]
- Evjenth, R.; Hole, K.; Karlsen, O.A.; Ziegler, M.; Arnesen, T.; Lillehaug, J.R. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J. Biol. Chem. 2009, 284, 31122–31129. [Google Scholar] [CrossRef] [Green Version]
- Starheim, K.K.; Arnesen, T.; Gromyko, D.; Ryningen, A.; Varhaug, J.E.; Lillehaug, J.R. Identification of the human N(alpha)-acetyltransferase complex B (hNatB): A complex important for cell-cycle progression. Biochem. J. 2008, 415, 325–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, P.G.; Chaerkady, R.; Zhang, Z.; Davidson, N.E.; Pandey, A. Monoclonal antibody cocktail as an enrichment tool for acetylome analysis. Anal Chem. 2011, 83, 3623–3626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Bao, H.; Liu, L.; Zhu, W.; Zhang, L.; Yue, L. Systematic analysis of lysine acetylome and succinylome reveals the correlation between modification of H2A.X complexes and DNA damage response in breast cancer. Oncol. Rep. 2020, 43, 1819–1830. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, Q.; Kang, Y.; Xu, S.; Pang, D. Unconventional protein post-translational modifications: The helmsmen in breast cancer. Cell Biosci. 2022, 12, 22. [Google Scholar] [CrossRef]
- Rey, M.; Irondelle, M.; Waharte, F.; Lizarraga, F.; Chavrier, P. HDAC6 is required for invadopodia activity and invasion by breast tumor cells. Eur. J. Cell Biol. 2011, 90, 128–135. [Google Scholar] [CrossRef]
- Riolo, M.T.; Cooper, Z.A.; Holloway, M.P.; Cheng, Y.; Bianchi, C.; Yakirevich, E.; Ma, L.; Chin, Y.E.; Altura, R.A. Histone deacetylase 6 (HDAC6) deacetylates survivin for its nuclear export in breast cancer. J. Biol. Chem. 2012, 287, 10885–10893. [Google Scholar] [CrossRef] [Green Version]
- Malonia, S.K.; Yadav, B.; Sinha, S.; Lazennec, G.; Chattopadhyay, S. Chromatin remodeling protein SMAR1 regulates NF-κB dependent Interleukin-8 transcription in breast cancer. Int. J. Biochem. Cell Biol. 2014, 55, 220–226. [Google Scholar] [CrossRef]
- Chang, Y.W.; Chen, H.A.; Tseng, C.F.; Hong, C.C.; Ma, J.T.; Hung, M.C.; Wu, C.H.; Huang, M.T.; Su, J.L. De-acetylation and degradation of HSPA5 is critical for E1A metastasis suppression in breast cancer cells. Oncotarget 2014, 5, 10558–10570. [Google Scholar] [CrossRef] [Green Version]
- You, D.; Zhao, H.; Wang, Y.; Jiao, Y.; Lu, M.; Yan, S. Acetylation Enhances the Promoting Role of AIB1 in Breast Cancer Cell Proliferation. Mol. Cells 2016, 39, 663–668. [Google Scholar] [CrossRef] [Green Version]
- Kawai, H.; Li, H.; Avraham, S.; Jiang, S.; Avraham, H.K. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int. J. Cancer 2003, 107, 353–358. [Google Scholar] [CrossRef]
- Liu, B.; Wang, T.; Wang, H.; Zhang, L.; Xu, F.; Fang, R.; Li, L.; Cai, X.; Wu, Y.; Zhang, W.; et al. Oncoprotein HBXIP enhances HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen resistance in breast cancer. J. Hematol. Oncol. 2018, 11, 26. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Mo, Y.; Li, M.T.; Zou, S.W.; Cheng, Z.L.; Sun, Y.P.; Xiong, Y.; Guan, K.L.; Lei, Q.Y. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J. Clin. Investig. 2014, 124, 5453–5465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Holloway, M.P.; Ma, L.; Cooper, Z.A.; Riolo, M.; Samkari, A.; Elenitoba-Johnson, K.S.; Chin, Y.E.; Altura, R.A. Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J. Biol. Chem. 2010, 285, 36129–36137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Sun, J.; Lungchukiet, P.; Quarni, W.; Yang, S.; Zhang, X.; Bai, W. Fe65 Suppresses Breast Cancer Cell Migration and Invasion through Tip60 Mediated Cortactin Acetylation. Sci. Rep. 2015, 5, 11529. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 28 March 2023).
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- The gene ontology consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Carlson, M. Org.Hs.Eg.Db: Genome Wide Annotation for Human; R Package Version 3.8.2; 2019. [Google Scholar] [CrossRef]
- Li, T.; Lu, D.; Yao, C.; Li, T.; Dong, H.; Li, Z.; Xu, G.; Chen, J.; Zhang, H.; Yi, X.; et al. Kansl1 haploinsufficiency impairs autophagosome-lysosome fusion and links autophagic dysfunction with Koolen-de Vries syndrome in mice. Nat. Commun. 2022, 13, 931. [Google Scholar] [CrossRef]
- Kim, S.M.; Ha, E.; Kim, J.; Cho, C.; Shin, S.J.; Seo, J.H. NAA10 as a New Prognostic Marker for Cancer Progression. Int. J. Mol. Sci. 2020, 21, 8010. [Google Scholar] [CrossRef] [PubMed]
- Di Martile, M.; Del Bufalo, D.; Trisciuoglio, D. The multifaceted role of lysine acetylation in cancer: Prognostic biomarker and therapeutic target. Oncotarget 2016, 7, 55789–55810. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, L.; Fan, Z.; Xue, W.; Shen, Z.; Yuan, Y.; Sun, X.; Wang, D.; Lian, J.; Wang, L.; et al. Hypoxia-induced GBE1 expression promotes tumor progression through metabolic reprogramming in lung adenocarcinoma. Signal Transduct. Target. Ther. 2020, 5, 54. [Google Scholar] [CrossRef]
- Phan, L.M.; Yeung, S.C.; Lee, M.H. Cancer metabolic reprogramming: Importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014, 11, 1–19. [Google Scholar] [CrossRef]
- Gwangwa, M.V.; Joubert, A.M.; Visagie, M.H. Crosstalk between the Warburg effect, redox regulation and autophagy induction in tumourigenesis. Cell Mol. Biol. Lett. 2018, 23, 20. [Google Scholar] [CrossRef] [Green Version]
- Vinaik, R.; Barayan, D.; Auger, C.; Abdullahi, A.; Jeschke, M.G. Regulation of glycolysis and the Warburg effect in wound healing. JCI Insight 2020, 5, e138949. [Google Scholar] [CrossRef]
- Hennipman, A.; Smits, J.; van Oirschot, B.; van Houwelingen, J.C.; Rijksen, G.; Neyt, J.P.; Van Unnik, J.A.; Staal, G.E. Glycolytic enzymes in breast cancer, benign breast disease and normal breast tissue. Tumour Biol. 1987, 8, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bai, Y.; Gao, Y.; Li, D.; Chen, L.; Zhou, C.; Feng, M.; Chen, X.; Jin, W.; Cao, Y. Systematic Analysis Uncovers Associations of PGK1 with Prognosis and Immunological Characteristics in Breast Cancer. Dis. Markers 2021, 2021, 7711151. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z. Proteomic Studies of the Effects of Different Stress Conditions on Central Carbon Metabolism in Microorganisms. J. Proteom. Bioinform. 2015, 8, 80–90. [Google Scholar]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miwa, N.; Uebi, T.; Kawamura, S. S100-annexin complexes—Biology of conditional association. FEBS J. 2008, 275, 4945–4955. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Naka, M.; Muneyuki, M.; Tanaka, T. Ca2+-dependent inhibition of actin-activated myosin ATPase activity by S100C (S100A11), a novel member of the S100 protein family. Biochem. Biophys. Res. Commun. 2000, 267, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.J.; Rescher, U.; Gerke, V.; Moss, S.E. Annexin-actin interactions. Traffic 2004, 5, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Fernández, L.; Menéndez, S.T.; Otero-Rosales, M.; Montoro-Jiménez, I.; Hermida-Prado, F.; García-Pedrero, J.M.; Álvarez-Teijeiro, S. Pathobiological functions and clinical implications of annexin dysregulation in human cancers. Front. Cell Dev. Biol. 2022, 10, 1009908. [Google Scholar] [CrossRef] [PubMed]
- Mallawaaratchy, D.M.; Buckland, M.E.; McDonald, K.L.; Li, C.C.; Ly, L.; Sykes, E.K.; Christopherson, R.I.; Kaufman, K.L. Membrane proteome analysis of glioblastoma cell invasion. J. Neuropathol. Exp. Neurol. 2015, 74, 425–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallawaaratchy, D.M.; Hallal, S.; Russell, B.; Ly, L.; Ebrahimkhani, S.; Wei, H.; Christopherson, R.I.; Buckland, M.E.; Kaufman, K.L. Comprehensive proteome profiling of glioblastoma-derived extracellular vesicles identifies markers for more aggressive disease. J. Neurooncol. 2017, 131, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, C.N.; Tu, Y.; Langenbach, S.; Baloyan, D.; Pattison, A.D.; Lock, P.; Britt, K.L.; Lehmann, B.D.; Beilharz, T.H.; Ernst, M.; et al. Annexin A1 Is Required for Efficient Tumor Initiation and Cancer Stem Cell Maintenance in a Model of Human Breast Cancer. Cancers 2021, 13, 1154. [Google Scholar] [CrossRef]
- Silva-Oliveira, R.; Pereira, F.F.; Petronilho, S.; Martins, A.T.; Lameirinhas, A.; Constâncio, V.; Caldas-Ribeiro, I.; Salta, S.; Lopes, P.; Antunes, L.; et al. Clinical Significance of ARID1A and ANXA1 in HER-2 Positive Breast Cancer. J. Clin. Med. 2020, 9, 3911. [Google Scholar] [CrossRef]
- Christensen, M.V.; Høgdall, C.K.; Jochumsen, K.M.; Høgdall, E.V.S. Annexin A2 and cancer: A systematic review. Int. J. Oncol. 2018, 52, 5–18. [Google Scholar] [CrossRef] [Green Version]
- Rodrigo Tapia, J.P.; Pena Alonso, E.; García-Pedrero, J.M.; Florentino Fresno, M.; Suárez Nieto, C.; Owen Morgan, R.; Fernández, M.P. Annexin A2 expression in head and neck squamous cell carcinoma. Acta Otorrinolaringol. Esp. 2007, 58, 257–262. [Google Scholar] [CrossRef]
- Yee, D.S.; Narula, N.; Ramzy, I.; Boker, J.; Ahlering, T.E.; Skarecky, D.W.; Ornstein, D.K. Reduced annexin II protein expression in high-grade prostatic intraepithelial neoplasia and prostate cancer. Arch. Pathol. Lab. Med. 2007, 131, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Yang, L.; Wang, Y.; Yuan, J.; Chen, T.; Cai, X. Down-regulation of annexin II in prostate cancer is associated with Gleason score, recurrence, metastasis and poor prognosis. Mol. Med. Rep. 2010, 3, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Smitherman, A.B.; Mohler, J.L.; Maygarden, S.J.; Ornstein, D.K. Expression of annexin I, II and VII proteins in androgen stimulated and recurrent prostate cancer. J. Urol. 2004, 171, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, L.D.; Mansheim, K.; Maji, S.; Nandy, R.; Lewis, C.M.; Vishwanatha, J.K.; Chaudhary, P. Clinical Significance of Annexin A2 Expression in Breast Cancer Patients. Cancers 2020, 13, 2. [Google Scholar] [CrossRef]
- Long, Y.; Chong, T.; Lyu, X.; Chen, L.; Luo, X.; Faleti, O.D.; Deng, S.; Wang, F.; He, M.; Qian, Z.; et al. FOXD1-dependent RalA-ANXA2-Src complex promotes CTC formation in breast cancer. J. Exp. Clin. Cancer Res. 2022, 41, 301. [Google Scholar] [CrossRef]
- Peng, B.; Guo, C.; Guan, H.; Liu, S.; Sun, M.-Z. Annexin A5 as a potential marker in tumors. Clin. Chim. Acta 2014, 427, 42–48. [Google Scholar] [CrossRef]
- Sato, H.; Ogata, H.; De Luca, L.M. Annexin V inhibits the 12-O-tetradecanoylphorbol-13-acetate-induced activation of Ras/extracellular signal-regulated kinase (ERK) signaling pathway upstream of Shc in MCF-7 cells. Oncogene 2000, 19, 2904–2912. [Google Scholar] [CrossRef] [Green Version]
- Korolkova, O.Y.; Widatalla, S.E.; Whalen, D.S.; Nangami, G.N.; Abimbola, A.; Williams, S.D.; Beasley, H.K.; Reisenbichler, E.; Washington, M.K.; Ochieng, J.; et al. Reciprocal expression of Annexin A6 and RasGRF2 discriminates rapidly growing from invasive triple negative breast cancer subsets. PLoS ONE 2020, 15, e0231711. [Google Scholar] [CrossRef] [Green Version]
- Cluntun, A.A.; Huang, H.; Dai, L.; Liu, X.; Zhao, Y.; Locasale, J.W. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab. 2015, 3, 10. [Google Scholar] [CrossRef] [Green Version]
- Elsheikh, S.E.; Green, A.R.; Rakha, E.A.; Powe, D.G.; Ahmed, R.A.; Collins, H.M.; Soria, D.; Garibaldi, J.M.; Paish, C.E.; Ammar, A.A.; et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009, 69, 3802–3809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abell, A.N.; Jordan, N.V.; Huang, W.; Prat, A.; Midland, A.A.; Johnson, N.L.; Granger, D.A.; Mieczkowski, P.A.; Perou, C.M.; Gomez, S.M.; et al. MAP3K4/CBP-regulated H2B acetylation controls epithelial-mesenchymal transition in trophoblast stem cells. Cell Stem Cell 2011, 8, 525–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]


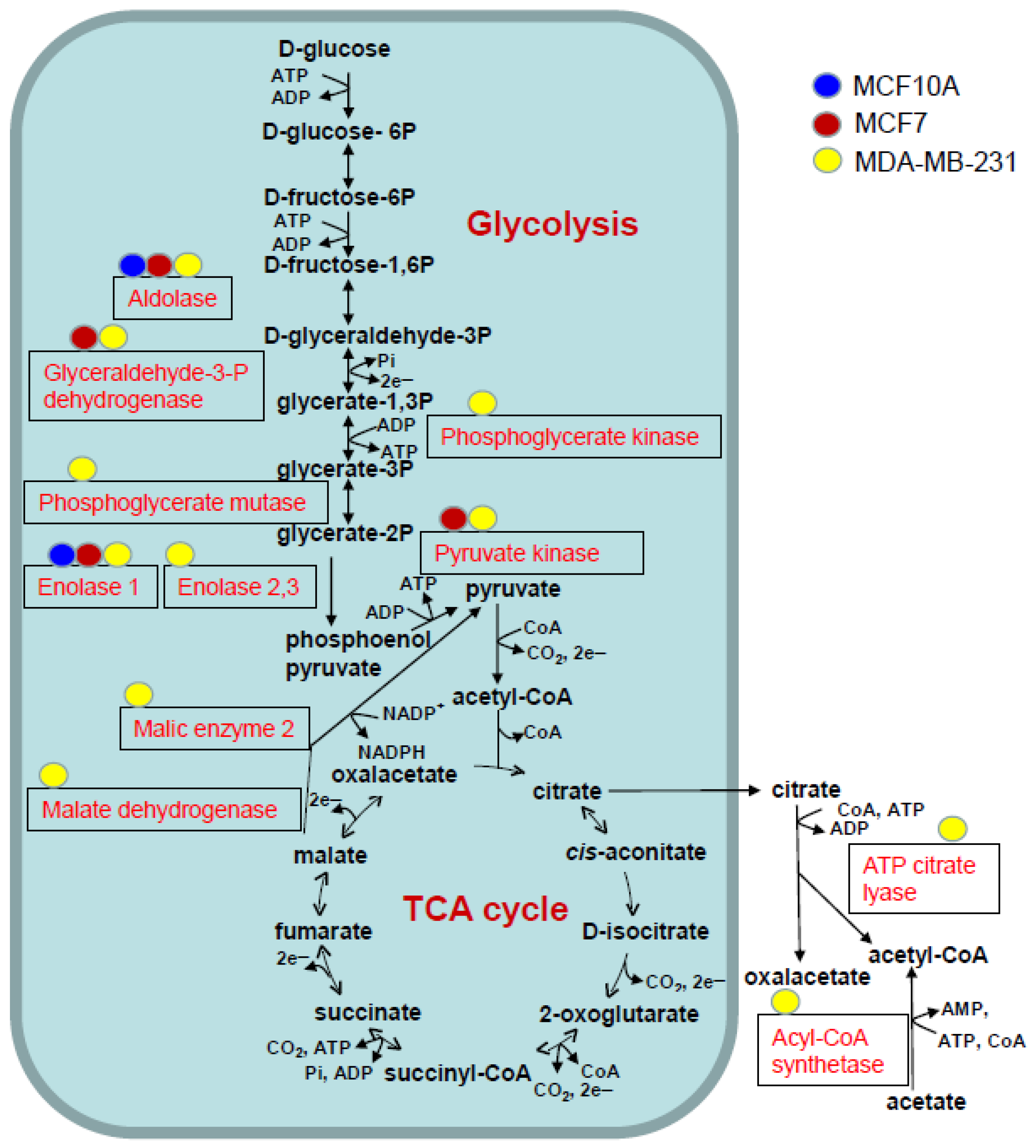
| Enzyme | Gene | Acetylation Sites | MCF10A sEV | MCF7 sEV | MDA-MB-231 sEV |
|---|---|---|---|---|---|
| Position of Acetylation | |||||
| Aldolase | ALDOA | 3 | K-147 | K-42, K-147, K-230 | K-147, K-230 |
| Glyceraldehyde-3-P-dehydrogenase | GAPDH | 3 | K-219 | K-61, K-194, K-219 | |
| Phosphoglycerate kinase 1 | PGK1 | 1 | K-131 | ||
| phosphoglycerate mutase 1 | PGAM1 | 1 | K-100 | ||
| Enolase 1 | ENO1 | 4 | K-343 | K-343 | K-60, K-193, K-203, K-343 |
| Enolase 2 | ENO2 | 1 | K-343 | ||
| Enolase 3 | ENO3 | 2 | K-60, K394 | ||
| pyruvate kinase M1/2 | PKM | 1 | K-433 | K-433 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minic, Z.; Li, Y.; Hüttmann, N.; Uppal, G.K.; D’Mello, R.; Berezovski, M.V. Lysine Acetylome of Breast Cancer-Derived Small Extracellular Vesicles Reveals Specific Acetylation Patterns for Metabolic Enzymes. Biomedicines 2023, 11, 1076. https://doi.org/10.3390/biomedicines11041076
Minic Z, Li Y, Hüttmann N, Uppal GK, D’Mello R, Berezovski MV. Lysine Acetylome of Breast Cancer-Derived Small Extracellular Vesicles Reveals Specific Acetylation Patterns for Metabolic Enzymes. Biomedicines. 2023; 11(4):1076. https://doi.org/10.3390/biomedicines11041076
Chicago/Turabian StyleMinic, Zoran, Yingxi Li, Nico Hüttmann, Gurcharan K. Uppal, Rochelle D’Mello, and Maxim V. Berezovski. 2023. "Lysine Acetylome of Breast Cancer-Derived Small Extracellular Vesicles Reveals Specific Acetylation Patterns for Metabolic Enzymes" Biomedicines 11, no. 4: 1076. https://doi.org/10.3390/biomedicines11041076






